Abstract
Introduction
Stem cells have been evaluated as a potential therapeutic approach for several neurological disorders of the central and peripheral nervous system as well as for traumatic brain and spinal cord injury. Currently, the lack of a reliable and safe method to accurately and non-invasively locate the site of implantation and track the migration of stem cells in vivo hampers the development of stem cell therapy and its clinical application.
In this report, we present data that demonstrate the feasibility of using the human sodium iodide symporter (hNIS) as a reporter gene for tracking neural stem cells (NSCs) after transplantation in the brain by using single-photon emission tomography/computed tomography (SPECT/CT) imaging.
Methods
NSCs were isolated from the hippocampus of adult rats (Hipp-NSCs) and transduced with a lentiviral vector containing the hNIS gene. Hipp-NSCs expressing the hNIS (NIS-Hipp-NSCs) were characterized in vitro and in vivo after transplantation in the rat brain and imaged by using technetium-99m (99mTc) and a small rodent SPECT/CT apparatus. Comparisons were made between Hipp-NSCs and NIS-Hipp-NSCs, and statistical analysis was performed by using two-tailed Student’s t test.
Results
Our results show that the expression of the hNIS allows the repeated visualization of NSCs in vivo in the brain by using SPECT/CT imaging and does not affect the ability of Hipp-NSCs to generate neuronal and glial cells in vitro and in vivo.
Conclusions
These data support the use of the hNIS as a reporter gene for non-invasive imaging of NSCs in the brain. The repeated, non-invasive tracking of implanted cells will accelerate the development of effective stem cell therapies for traumatic brain injury and other types of central nervous system injury.
Introduction
During the past decade, several reports have explored the therapeutic potential of neural stem cells (NSCs) for the treatment of neurological disorders of the central nervous system (CNS) (i.e., macular degeneration, Parkinson’s disease, Alzheimer’s disease, and multiple sclerosis) and for traumatic brain and spinal cord injury [1–8]. However, the successful development of stem cell therapy and its translation to the clinical setting is currently hampered by the lack of a reliable and safe method to accurately monitor the location, migration, and phenotypical differentiation of transplanted cells. So far, the majority of preclinical studies on stem cell fate have relied on ex vivo histological analysis of green fluorescent protein (GFP) or β-galactosidase expression in the grafted cells. These analyses require the euthanasia of the animals at each time point studied and therefore are laborious and time-consuming. Recently, the development of in vivo non-invasive imaging technologies has provided the means to monitor the delivery, grafting, and survival of stem cells. Current imaging modalities to monitor cells in the brain include magnetic resonance imaging (MRI), single-photon emission tomography (SPECT), positron emission tomography (PET), bioluminescence, and fluorescence imaging [9].
The use of fluorescence and luciferase for bioluminescence imaging is an excellent tool to monitor grafted cells in small animals but is not translatable to the human patient. MRI and SPECT/PET are non-invasive imaging modalities that are suited for human use. Although MRI has a higher spatial resolution than SPECT/PET, the sensitivity of detection is higher for SPECT/PET than MRI (SPECT/PET: 10−10–10−12 M levels of probe; MRI: 10−3–10−5 M levels of probe). Additionally, PET has the ability to detect reporter genes [10, 11].
The use of reporter genes to track stem cell fate in vivo is particularly appealing, as it is the only method that allows studying stem cell survival (only viable cells will be able to express the reporter protein), proliferation (the reporter gene will be passed on to daughter cells, and the corresponding imaging signals will increase in intensity), and death (cells that are apoptotic or dead will not be able to express the reporter protein). Moreover, a reporter gene can be placed under cell-specific promoters (e.g., neuron-specific or glia-specific), thus allowing monitoring of the fate of the transplanted cells within the host tissue [12, 13].
When clinicians decide on which reporter gene to use for imaging stem cells, important factors to be considered are (1) biological distribution of the gene, (2) availability of the probes, and (3) effect of the expression of the reporter gene on the physiology of the cells.
In this respect, the sodium iodide symporter (NIS) reporter gene represents a good choice for imaging stem cells in the brain because (1) it is not expressed in the brain, (2) the radio probes for NIS are readily available in most nuclear medicine clinics and no radio synthesis is required, (3) the metabolism and clearance in the body of both radiodiodide and technetium-99m (99mTc) are well known, and (4) the imaging potentials of NIS have been shown in vitro and in vivo [14, 15].
The use of the NIS to monitor in vivo the delivery, grafting, and phenotypical differentiation of cells after transplantation has recently been investigated in particular in cardiovascular research [16–18]. The NIS has also been used to monitor trafficking of immune cells in vivo. Seo et al. reported how transfecting immortalized macrophage cell lines with the hNIS allowed monitoring of their migration toward areas of inflammation in vivo in nude mice by using PET imaging [19].
Up until now, however, no studies have been reported on the use of the NIS for imaging NSCs in the brain in vivo. Here, we report on the development of the use of the NIS for imaging NSCs in the brain.
Methods
Animals
Male Sprague–Dawley rats (200–350 g) were used in all of the experiments described. Experimental protocols were approved by the Institutional Animal Care and Use Committee at the University of Texas Medical Branch, Galveston, in accordance with the guidelines provided by the National Institutes of Health.
Isolation and in vitro expansion of rat hippocampal neural stem cells
Cell culture reagents were obtained from Invitrogen (Carlsbad, CA, USA), except where noted. Adult male Sprague–Dawley rats (200–250 g) were anesthetized with isofluorane (4 % by inhalation) and euthanized by decapitation. The brain was rapidly removed, and the hippocampi were identified and dissected out. The hippocampi from three or four rats were collected into a 50-ml Falcon tube containing sterile Dulbecco’s modified Eagle’s medium/F12 (DMEM/F12) medium with antibiotics (penicillin and streptomycin) and kept on ice. The tissue was minced into small pieces in cold Hanks’ Balanced Salt Solution (HBSS) medium containing 1 mM EGTA in a sterile Petri dish. The tissue pieces were transferred into a sterile 50-ml tube containing HBSS (without Ca2+ and Mg2+) with 0.1 % collagenase/dispase, 0.01 % DNase I; 1 ml for 100 mg of tissue) and incubated for 30 min at 37 °C. The tissue was triturated every 10 min by using a sterile disposable 5-ml pipette. At the end of the incubation, the cell suspension was centrifuged at 200×g for 5 min at room temperature (RT). The pellet was resuspended in HBSS containing 0.025 % trypsin/ethylenediaminetetraacetic acid and incubated for 10 min at 37 °C. An equal volume of DMEM/F12, 2 mM L-glutamine, 10 % fetal bovine serum (FBS), and antibiotics was added to stop the digestion and the cell suspension was centrifuged at 200×g for 5 min. The pellet was resuspended in sterile DMEM/F12 with L-glutamine, 10 % FBS, and antibiotics; triturated by using a 1-ml tip; and centrifuged. This step was repeated two to four times until a single cell suspension was obtained. The cell suspension was filtered through a 70-μm Falcon filter, centrifuged at 200×g for 5 min, and resuspended in complete growth medium: neurobasal A medium containing the serum-free supplement B27 (without retinoic acid), 2 mM L-glutamine, 20 ng/ml of epidermal growth factor (EGF), 20 ng/ml of basic fibroblast growth factor-2 (FGF-2), and penicillin/streptomycin. The cells were plated in an uncoated T25 flask at approximately 1×106 cells per 10 ml of complete growth medium. The medium was changed after 24 h and every other day after that.
Coating of plates with poly-ornithine and laminin
Stock solutions were prepared as follows: poly-L-ornithine (Sigma-Aldrich, St. Louis, MO, USA, catalog number P3655) was reconstituted with sterile distilled water at a concentration of 10 mg/ml, and aliquots were stored at −20 °C; laminin (1 mg/ml; Invitrogen) was stored at −20 °C.
The poly-L-ornithine stock solution was diluted 1:1000 in sterile distilled water to a final working concentration of 10 μg/ml and added to the plates as follow: 1.5 ml/well of six-well plates; 0.2 ml/well of eight-well chambered slides (Lab-Tek II Chambered Slide System; Nalgene Nunc International, Naperville, IL, USA). Plates and slides were incubated overnight at RT and then washed twice with distilled water, incubated with laminin overnight at RT (10 μg/ml in Dulbecco’s phosphate-buffered saline (D-PBS) without Ca2+ and Mg2+; 1 ml/well of six-well plates and 0.2 ml/well of eight-well chambered slides), and stored at −20 °C until ready to use. The plates were washed with D-PBS once before use.
Induction of differentiation
Hipp-NSCs were dissociated into single cells by using StemPro Accutase (Invitrogen) in accordance with the instructions of the manufacturer and counted on a hemocytometer. The cells were plated onto poly-ornithine/laminin-coated plates as follows: 125,000 cells per well into a six-well plate and 10,000 cells per well into eight-well chambered slides. To induce differentiation, the cells were cultured for 8–10 days in neurobasal A medium containing B27 with retinoic acid, 0.5 mM L-glutamine, 1 % FBS, and penicillin/streptomycin. The medium was changed every other day.
Lentivirus packaging
The full-length human sodium iodide symporter cDNA (hNIS, a kind gift from Sissy M. Jhiang) was cloned into the pCDH-CMV-MCS-EF1-copGFP cDNA cloning and expression lentivector (System Biosciences, Mountain View, CA, USA). The lentivector product was packaged into VSV-G pseudotyped viral particles by System Biosciences (titer 3.12×109 infectious units/ml).
Transduction of Hipp-NSCs
Adult rat Hipp-NSCs were dissociated into single cells by using StemPro Accutase (Invitrogen) in accordance with the instructions of the manufacturer and counted on a hemocytometer. The cells were divided into sterile 1.5-ml Eppendorf tubes (100,000 cells per tube in 250 μl of complete growth medium). Pseudovirus particles packaged with the lentivector containing the hNIS were added to each tube at 5, 10, 20, and 40 multiplicity of infection, and the tubes were incubated at 37 °C in a 5 % CO2 incubator for 30 min. The cells were then transferred to a 24-well plate (250 μl of cells was added to 750 μl of complete growth medium per well) and placed in a 5 % CO2 incubator at 37 °C. After 48 h, the cells were collected into a sterile 15-ml conical tube, centrifuged at 200×g for 5 min, resuspended in complete growth medium, and plated out into a sterile T75 flask. The medium was changed every other day. Expression of GFP (indicative of transduction efficiency) was monitored by using an inverted fluorescent microscope equipped with a 488-nm excitation filter (Nikon Eclipse TS100; Nikon, Tokyo, Japan).
Fluorescence-activated cell sorting
Transduced Hipp-NSCs were dissociated into single cells by using StemPro Accutase, resuspended in sterile D-PBS containing 1 % bovine serum albumin at a concentration of 10–20×106 cells/ml, and passed through a 70-μm Falcon filter. The cells were kept on ice and brought to the University of Texas Medical Branch (UTMB) Flow Cytometry Core Facility, where they were run through a FACSAria cell sorter (BD Biosciences, San Jose, CA, USA). The cells were sorted on the basis of the expression of GFP and collected into a 15-ml tube containing 1 ml of complete growth medium with 10 mM HEPES. The collected cells were centrifuged and replated in fresh complete growth medium.
In vitro uptake of technetium-99m
99mTc was obtained from the Department of Nuclear Medicine at UTMB. Cells (Hipp-NSCs expressing the hNIS and naïve Hipp-NSCs not transduced) were plated out into a six-well plate at a density of 500,000 cells/ml (1.5×106 cells per well) in complete growth medium. The cells were incubated with 99mTc (3–5 μCi/well) for 30 min at 37 °C in a 5 % CO2 incubator and then washed with ice-cold PBS two times by collection and centrifugation at 200×g for 5 min. At the end of the last centrifugation, the cells were lysed in 500 μl of NaOH (0.33 M) containing 1 % SDS. Counts per minute (CPMs) were read on a gamma counter. Baseline readings were obtained from tubes containing NaOH-1 % SDS and no cells. Data were corrected by subtracting the baseline reading averaged from three separate measurements from the sample CPMs.
Cell proliferation assay
Cell proliferation was measured by using the CellTiter 96 Aqueous non-radioactive cell proliferation assay (Promega Corporation, Madison, WI, USA). This assay is based on the principle that a tetrazolium compound ([3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] (MTS)) is converted by viable cells into a soluble formazan product that can be measured by reading the absorbance at 490 nm. Briefly, Hipp-NSCs and Hipp-NSCs transduced with the hNIS were dissociated into single cells by using StemPro Accutase, plated out into a 96-well plate (10,000 and 20,000 cells per well) in complete growth medium (100 μl/well), and cultured for 24–48 h. At the desired time point, 20 μl of MTS/PMS solution (prepared in accordance with the instructions of the manufacturer) was added to each well and the plate was incubated for 3–4 h at 37 °C in a humidified incubator, 5 % CO2 atmosphere. The plate was read in an enzyme-linked immunosorbent assay plate reader (Molecular Devices, Sunnyvale, CA, USA) at 490 nm. Background absorbance was measured from wells that were loaded with complete growth medium only and to which MTS/PMS was added.
Total protein extraction and Western Blot Analysis
Cells growing as neurospheres in complete growth medium were collected in a 15-ml conical tube and centrifuged at 200×g for 5 min at RT. The cells were resuspended in D-PBS and centrifuged again. RIPA lysis buffer containing protease inhibitor cocktail, phosphatase inhibitors cocktail, and 100 μM phenylmethylsulfonyl fluoride (PMSF) (all from Sigma-Aldrich) was added to the cell pellet (150–250 μl of lysis buffer for 2–5×106 cells). The lysed cells were transferred to 1.5-ml Eppendorf tubes, vortexed, and incubated on ice for 5 min and then quickly frozen in liquid nitrogen and stored at −80 °C. Cells growing on poly-ornithine/laminin-coated plates were detached by using StemPro Accutase, collected into 15-ml tubes, and centrifuged at 200×g for 5 min. The cell pellet was washed in D-PBS and lysed in RIPA lysis buffer as described above. Total protein content was measured by using a Pierce BCA protein assay kit (Thermo Scientific, Rockford, IL, USA).
The protein samples were processed for SDS-polyacrylamide gel electrophoresis (PAGE) with XCell SureLock® Mini-Cell (Invitrogen). After electrophoresis, proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories, Hercules, CA, USA) overnight at 4 °C. Blots were incubated in blocking buffer (5 % non-fat dry milk in Tris-buffered saline containing 0.1 % Tween 20; TBS-T) for 1 h at RT and probed with primary antibodies diluted in blocking buffer overnight at 4 °C. After three washes in TBS-T, the blots were incubated with horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology, Inc., Danvers, MA, USA; 1:200 dilution in blocking buffer) for 1 h at RT.
Detection was performed using ECL plus kit (Amersham, GE Healthcare, Little Chalfont, UK) followed by exposure of x-ray films (Bioexpress, Kaysville, UT, USA) that were developed with M35A-X-OMAT Processor (Kodak, Rochester, NY, USA). Band intensities were quantified by using ImageJ software, and data were normalized to the expression of the housekeeping protein GAPDH (glyceraldehyde 3-phosphate dehydrogenase).
Immunofluorescence analysis
Cells growing on poly-ornithine/laminin-coated chambered slides were washed in PBS (pH 7.4) and fixed in 4 % paraformaldehyde (pH 7.4 in PBS) for 15 min at RT. After two washes in PBS, the cells were blocked and permeabilized in PBS containing 5 % normal goat serum and 0.3 % triton X-100 for 30 min at RT. The cells were incubated with primary antibodies (Table 1) diluted in 1.5 % normal goat serum in PBS overnight at 4 °C in a humid chamber. After three washes in PBS, the cells were incubated with secondary antibodies (1:400; Alexa-conjugated anti-mouse and anti-rabbit; Invitrogen) in 1.5 % normal goat serum in PBS for 1 h at RT in a humid chamber. The cells were washed two times in PBS and once in tap water before being coverslipped with mounting medium containing DAPI (4′,6-diamidino-2-phenylindole), a fluorescent stain that binds strongly to A-T-rich regions in DNA and is used to label cell nuclei (Vector laboratories, Burlingame, CA, USA). The slides were viewed with an Olympus BX51 fluorescence microscope equipped with a cooled charge-coupled device camera (Microfire; Optronics, Goleta, CA, USA) and image acquisition software (PictureFrame; Optronics).
Table 1.
List of antibodies used for the phenotypical characterization of Hipp-NSCs and hNIS-Hipp-NSCs
| Primary antibody | Specificity | Dilution | Source |
|---|---|---|---|
| Mouse anti-nestin | Neural progenitor cells | 1:5000 WB | Millipore |
| 1:200 IF | |||
| Rabbit anti-SOX2 | Neural progenitor cells | 1:2000 WB | Abcam |
| 1:100 IF | |||
| Mouse anti-βIII-tubulin | Immature and mature neurons | 1:5000 WB | Promega |
| 1:1000 IF | |||
| Mouse anti-NeuN | Mature neurons | 1:2000 WB | Millipore |
| 1:1000 IF | |||
| Rabbit anti-GFAP | Astrocytes | 1:5000 WB | Dako |
| 1:1000 IF | |||
| Mouse anti-oligodendrocyte specific protein | Oligodendrocytes | 1:200,000 IF | Millipore |
| Mouse anti-hNIS | Human NIS | 1:100 IF | Abcam |
Hipp-NSC hippocampus-derived neural stem cell, hNIS-Hipp-NSC hippocampus-derived neural stem cell expressing the human sodium iodide symporter, WB Western blot, IF immunofluorescence, anti-GFAP anti-glial fibrillary acidic protein
Injection of Hipp-NSCs in the hippocampus
Hipp-NSCs were labeled with the cell tracker CMFDA (Invitrogen), suspended at a concentration of 50,000 cells/μl in D-PBS, and kept on ice until ready to inject. Experimental recipient adult male rats were deeply anesthetized with isoflurane (2–4 %) and intubated with an endotracheal tube (6.5-French tubing attached to 16-gauge needle adapter) attached to a ventilator (tidal volume set at 12–15 cc/kg and breath rate set at approximately 28–30 breaths per minute). The anesthetized rats were placed at the flat-skull position on a small animal stereotaxic apparatus, and a craniotomy was performed. The cells were injected by using a 33-gauge cannulae supported by the outside 26-gauge guide cannulae connected with a Hamilton syringe. The cells were injected into the hippocampal CA1—anterior-posterior (AP): −3.6, medial-lateral (ML): +2.0, dorsal-ventral (DV): −2.6—and CA3 (AP: −3.6, ML: +3.6, DV: −3.6) regions (1 μl/site at a speed of 0.2 μl/min). The number of cells injected was chosen on the basis of the work by Gao et al. [20].
Injection of Hipp-NSCs in the lateral ventricles
Hipp-NSCs were suspended in D-PBS at a concentration of 2×105 cells/μl. Experimental recipient adult male rats were deeply anesthetized with isoflurane (2–4 %) and intubated with an endotracheal tube (6.5-French tubing attached to 16-gauge needle adapter) attached to a ventilator (tidal volume set at 12–15 cc/kg and breath rate set at approximately 28–30 breaths per minute). The anesthetized rats were placed at the flat-skull position on a small animal stereotaxic apparatus, and a craniotomy was performed. The injection site was drilled by using a Dremel drill 1.5 mm posterior to bregma and 1.2 mm to lateral right of sagittal suture for ventricular pressure verification, and injection of cells was performed (5 μl; 0.2 μl/min).
SPECT/CT imaging
Imaging was performed by using an Inveon™ PET-SPECT-CT system (Siemens Medical Solutions, Knoxville, TN, USA) equipped with two gamma cameras (for SPECT modality) and x-ray source and camera for CT imaging mounted on a rotating stage. The PET detector is mounted separately and was not used in this study. This scanner combines PET, SPECT, and CT modalities in one system under the control of a single workstation that allows integrated data acquisition and processing. CT imaging provides mostly anatomical reference information (where the gamma signal comes from), and SPECT provides functional information showing the intensity and the distribution of the isotope in the body. Injected in the animal, a gamma-emitted isotope serves as a source of signal, and the signal is detectable by the SPECT cameras. Since the isotope (in our study, 99mTc) is specifically attaching to the cells with NIS (naturally, to the thyroid cells; in our study, to the modified stem cells) and is not attaching to the other cells, it provides the imaging contrast between the signal (coming from the cells of interest) and background. Cameras are rotated around the animal and the signal collected for 3D reconstruction. In general, we followed the same CT/SPECT imaging as described by Terrovitis et al. [16].
In vivo SPECT imaging
The rats were anesthetized with isofluorane and positioned on the bed of the SPECT/CT module with their nose inside a mask connected to the isofluorane dispenser. High-resolution CT scans were performed before each SPECT session. The 99mTc signal was co-registered to the CT scan image by using data processing software Inveon Research Workspace (Siemens Medical Solutions). CT parameters were 2048×3072 pixels, 70 kV, 500 μA, and 520 steps, giving a 3D isotropic resolution of approximately 0.1 mm. SPECT parameters were 40 projections and 9-degree step, for a total of about 16 min for one scan.
Intracerebral cannulation for delivery of 99mTc in the brain
The rats were anesthetized with isoflurane (2–4 %) and intubated with an endotracheal tube (6.5-French tubing attached to 16-gauge needle adapter) attached to a ventilator (tidal volume set at 12–15 cc/kg and breath rate set at approximately 28–30 breaths per minute). The anesthetized rats were placed at the flat-skull position on a small animal stereotaxic apparatus, and a craniotomy was performed. A microdialysis CMA guide cannula/probe (BASi, West Lafayette, IN, USA) was inserted into the hippocampus (Bregma: −4.3 mm; ML: −3.5 mm; Dura: −3.2 mm).
Tissue processing and immunofluorescence analysis
At the desired time point after cell transplantation, the rats were anesthetized with pentobarbital and cardially perfused with heparinized saline followed by ice-cold 4 % paraformaldehyde (pH 7.4). The brains were removed and embedded in 20 % sucrose in PBS (pH 7.4). Sections (20 μm) were cut on a cryostat, collected on glass microscope slides (Superfrost Plus; Thermo Fisher Scientific Inc., Marietta, OH, USA), and stored at −20 °C. For immunofluorescence analysis of grafted cells, the sections were hydrated in PBS and incubated in PBS containing 10 % normal goat serum and 0.3 % Triton X-100 for 30 min at RT. The sections were incubated with primary antibodies diluted in PBS containing 1.5 % normal goat serum, overnight at 4 °C and with secondary antibodies (594 Alexa-conjugated, Invitrogen; 1:400 dilution in PBS with 1.5 % normal goat serum) for 1 h at RT. After washing in PBS, the sections were rinsed in tap water and coverslipped with mounting medium with DAPI (Vector Laboratories).
Statistical analysis
Data were expressed as the mean ± standard error. Statistical analysis between two independent experimental groups was performed by using two-tailed Student’s t test. Results were considered significant for P values of less than 0.05.
Results
Expansion and characterization of Hipp-NSCs
NSCs were isolated from the hippocampus of adult rats and maintained in culture in serum-free neurobasal A medium supplemented with B27 and the growth factors EGF and FGF-2 (complete growth medium). Under these conditions, the cells actively proliferate and form a characteristic spheroid group of cells that express the NSC marker nestin (Fig. 1a, b). NSCs are defined not only by their ability to proliferate and generate daughter cells that are unspecialized and express specific stem cell markers but also by their ability to generate neurons and glia. When Hipp-NSCs were plated onto poly-ornithine/laminin-coated plates in serum-free medium containing B27, retinoic acid, and no growth factors (EGF and FGF-2), they attached to the substrate and underwent rapid morphological changes (Fig. 1c). Immuncytochemical analysis of cells grown for 8 days under differentiating conditions confirmed that both neurons and glia were generated from Hipp-NSCs (Fig. 1d).
Fig. 1.
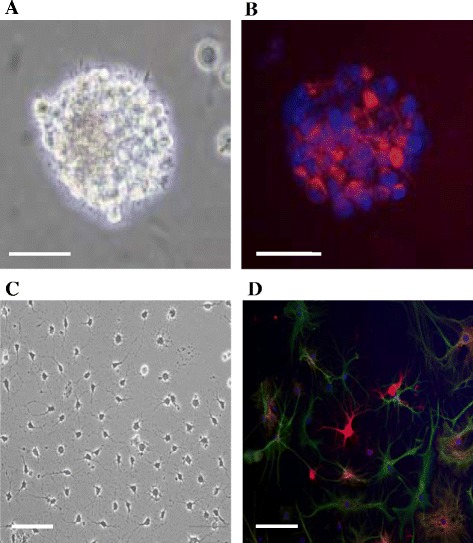
Hipp-NSCs form neurospheres and generate neurons and glia in vitro. a A typical neurosphere formed by Hipp-NSCs growing in serum-free medium with EGF and FGF-2. b Immunofluorescence analysis of nestin expression (red) in Hipp-NSC neurospheres. Nuclei are stained blue with DAPI. c Phase-contrast photograph of Hipp-NSCs growing on poly-ornithine-coated plates in differentiation medium for 8 days. d Immunofluorescence analysis of Hipp-NSCs after 8 days of differentiation showing the expression of the neuronal marker βIII-tubulin (red) and of the glial marker GFAP (green). Nuclei are stained blue with DAPI. Scale bars = 50 μm. DAPI 4′,6-diamidino-2-phenylindole, EGF epidermal growth factor, FGF-2 fibroblast growth factor-2, GFAP glial fibrillary acidic protein, Hipp-NSC hippocampus-derived neural stem cell
Generation of Hipp-NSCs expressing the hNIS
Hipp-NSCs were transduced by using pseudoviral particles packaged with a lentivector containing the hNIS and the reporter gene GFP (Fig. 2a, b). The expression of GFP and fluorescence-activated cell sorting was used to select out Hipp-NSCs expressing the hNIS (Fig. 2c, d). The expression of the hNIS in the sorted cells was confirmed by immunocytochemical analysis (Fig. 2e). We also determined whether the expression of the hNIS would be retained after differentiation. Hipp-NSCs transduced with the hNIS were cultured onto poly-ornithine/laminin-coated plates in differentiation medium for 7 days. The hNIS was maintained in the differentiated cells as shown by the expression of GFP and by immunofluorescence analysis by using a specific antibody against hNIS (Fig. 2f, g).
Fig. 2.
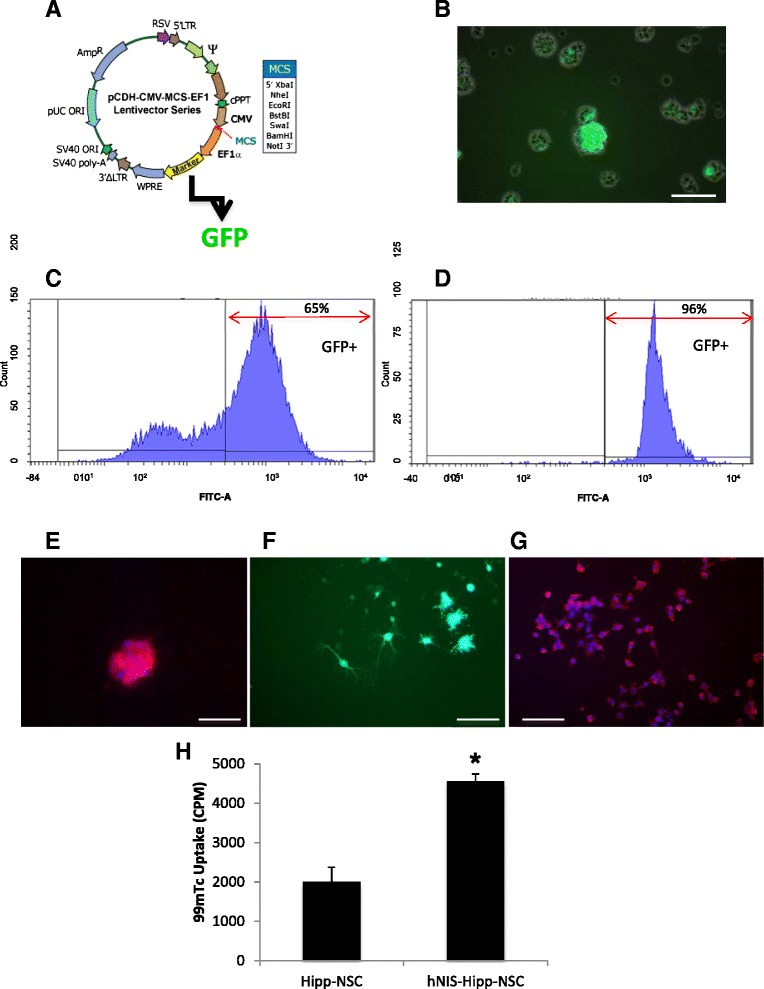
Generation of Hipp-NSCs expressing the hNIS. a Map of the HIV-based lentivector CD11B-1 (System Biosciences). The hNIS cDNA was cloned in the multiple cloning site (MSC) located downstream of the CMV promoter. Downstream of the expression cassette for the hNIS, an EF1 promoter drives the expression of GFP. b Expression of GFP in Hipp-NSCs transduced with pseudoviral particles for 48 h. The fluorescent image is superimposed on the phase-contrast image. c Flow cytometric analysis of Hipp-NSCs transduced with the hNIS showing the expression of GFP (indicative of the expression of hNIS) in 62 % of the cells before sorting and in 96 % of the cells after sorting (d). e Immunofluorescence analysis of the expression of the hNIS (in red) in Hipp-NSCs transduced with the lentivector and selected by sorting for GFP. Nuclei are stained blue with DAPI. f Expression of GFP in differentiated Hipp-NSCs transduced with the hNIS. g Immunoreactivity for the hNIS in differentiated NIS-Hipp-NSCs. Nuclei are counterstained blue with DAPI. h Uptake of 99mTc in NIS-Hipp-NSCs was measured as counts per minute (CPM) in a gamma counter and corrected by subtracting background CPM. N = 3; *P < 0.01 by Student’s t test. Scale bars = 100 μm. CMV cytomegalovirus, DAPI 4′,6-diamidino-2-phenylindole, GFP green fluorescent protein, Hipp-NSC hippocampus-derived neural stem cell, hNIS human sodium iodide symporter, NIS-Hipp-NSC hippocampus-derived neural stem cell expressing the human sodium iodide symporter, 99m Tc technetium-99m
To determine whether the hNIS expressed by Hipp-NSCs is functional, we tested the ability of the transduced cells to transport 99mTc, a gamma-emitting radioisotope that is used by the NIS instead of endogenous iodide. A significant increase in CPM was measured in Hipp-NSCs expressing the hNIS as compared with control Hipp-NSCs not expressing the hNIS and exposed to 99mTc (Fig. 2h).
Characterization of Hipp-NSCs expressing the hNIS in vitro
To assess the effect of the hNIS on cell viability, Hipp-NSCs were plated in a 96-well plate in neurobasal A medium containing EGF and FGF (complete growth medium) at two different densities (10,000 and 20,000 cells per well) and cultured for 24 h. To measure proliferation, the cells were plated out at a density of 10,000 cells per well and cultured for 24 and 48 h. Cell viability was assessed by using the MTS assay (CellTiter 96 Aqueous non-radioactive cell proliferation assay; Promega Corporation). No significant differences were detected between Hipp-NSCs expressing the hNIS (NIS-Hipp-NSCs) and Hipp-NSCs in their naïve form (not expressing the hNIS) (Fig. 3a, b). To test the effect of 99mTc incorporation on cell viability, Hipp-NSCs and NIS-Hipp-NSCs were exposed to 99mTc (100 μCi added to the cell culture medium) for 30 min. The cells were washed, resuspended in proliferation medium, plated out in a 96-well plate (10,000 cells per well), and cultured for 48 h. Cell viability was assessed 48 h later by using the MTS assay (CellTiter 96 Aqueous non-radioactive cell proliferation assay). No significant differences were detected between Hipp-NSCs and NIS-Hipp-NSCs, indicating that the incorporation of 99mTc in NIS-Hipp-NSCs does not reduce cell viability. In addition to studying the effect of the hNIS on Hipp-NSC viability and proliferation, we studied whether NIS-Hipp-NSCs retain their characteristic stem cell signature. This was done by analyzing the expression of NSC markers by Western blot analysis. There was no difference in the expression of nestin and SOX2 between Hipp-NSCs transduced with the hNIS (NIS-Hipp-NSCs) and naïve Hipp-NSCs (Fig. 3c, d).
Fig. 3.
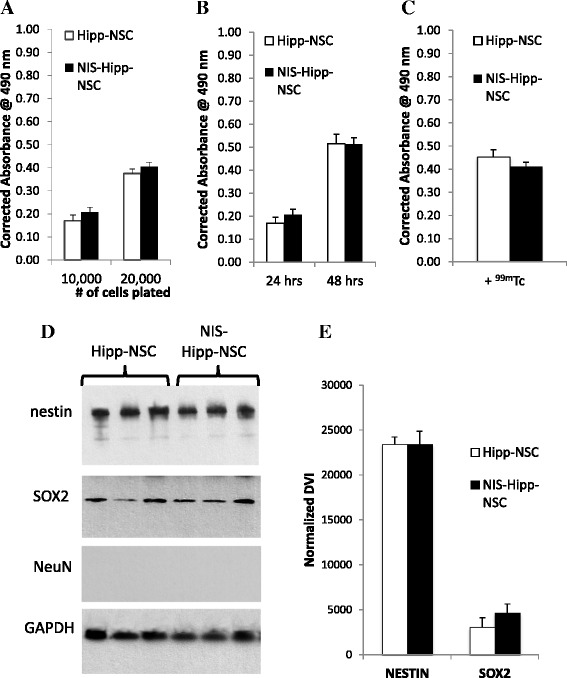
NIS-Hipp-NSCs are viable, proliferate, and express NSC markers. MTS-based analysis of cell viability (a) and proliferation (b) of Hipp-NSCs transduced with the hNIS (NIS-Hipp-NSCs) compared with naïve Hipp-NSCs. N = 3; P > 0.05 by Student’s t test. c Hipp-NSCs and NIS-Hipp-NSCs were exposed to 99mTc for 30 min. MTS-based analysis of cell viability was performed 48 h later. N = 3; P > 0.05 by Student’s t test. d Western blot analysis of total protein extracts from NIS-Hipp-NSCs and naïve Hipp-NSCs grown under proliferating conditions. e Densitometric analysis. The expression of GAPDH was used to normalize the densitometry value index (DVI). N = 3; P > 0.05 by Student’s t test. GAPDH glyceraldehyde 3-phosphate dehydrogenase, Hipp-NSC hippocampus-derived neural stem cell, hNIS human sodium iodide symporter, MTS 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt, NIS-Hipp-NSC hippocampus-derived neural stem cell expressing the human sodium iodide symporter, NSC neural stem cell, 99m Tc technetium-99m
Differentiation potential of Hipp-NSCs expressing the hNIS
To test the effect of hNIS on the ability of Hipp-NSCs to generate neurons and glia, the cells were plated out onto poly-ornithine/laminin-coated plates and cultured for 8 days in neurobasal medium containing B27 with retinoic acid and 1 % FBS (differentiation medium). Neuronal and glial differentiation was assessed by immunofluorescence and Western blot analysis. The results show that the expression of hNIS does not affect the ability of Hipp-NSCs to generate neurons and glia in vitro (Fig. 4).
Fig. 4.
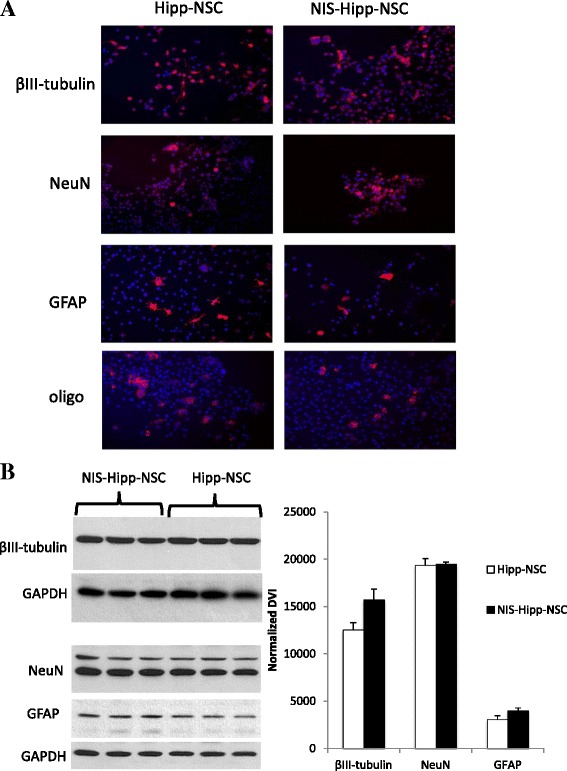
Differentiation potential of NIS-Hipp-NSCs. a Immunofluorescence analysis of Hipp-NSCs transduced with the hNIS (NIS-Hipp-NSCs) and of naïve Hipp-NSCs cultured in differentiation medium for 7 days. Nuclei are counterstained blue with DAPI. b Western blot analysis of total protein extracts from differentiated NIS-Hipp-NSCs and naïve Hipp-NSCs. The expression of GAPDH was used to normalize the densitometry value index (DVI). N = 3; *P < 0.05 by Student’s t test. DAPI 4′,6-diamidino-2-phenylindole, GAPDH glyceraldehyde 3-phosphate dehydrogenase, Hipp-NSC hippocampus-derived neural stem cell, hNIS human sodium iodide symporter, NIS-Hipp-NSC hippocampus-derived neural stem cell expressing the human sodium iodide symporter
SPECT/CT imaging of Hipp-NSCs expressing the hNIS
In vitro imaging
Hippocampus-derived NSCs can be visualized by using SPECT imaging by providing 99mTc to the cells and measuring the gamma-ray emission from the cells that have taken up the 99mTc via the NIS. Hipp-NSCs stably expressing the NIS were incubated in vitro with 99mTc and imaged by placing the cells in an Eppendorf tube inside the SPECT/CT imaging module. Control cells (Hipp-NSCs not expressing the NIS) were also incubated with 99mTc and imaged (Fig. 5a).
Fig. 5.
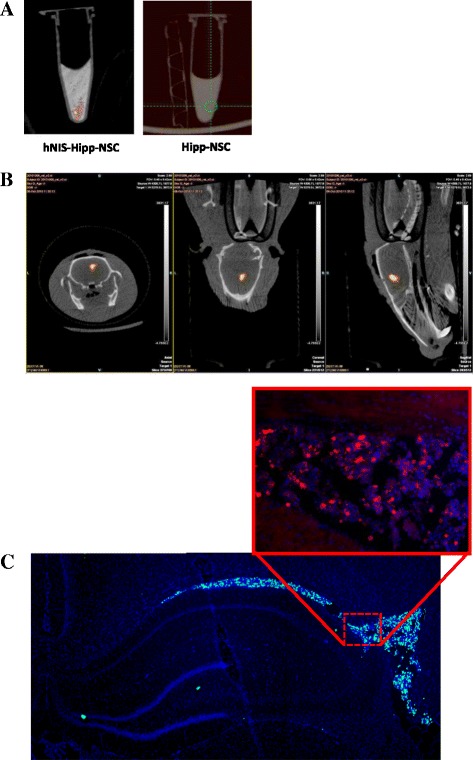
SPECT imaging of NIS-Hipp-NSCs. a SPECT signal from hNIS-Hipp-NSCs and Hipp-NSCs that were incubated with 99mTc for 30 min and washed in phosphate-buffered saline before imaging. The SPECT signal is shown superimposed on the CT scan image. b SPECT signal recorded from NIS-Hipp-NSCs that were incubated with 99mTc in vitro before transplantation in the right lateral ventricle. Transverse, coronal, and sagittal orientations of the brain are shown. c Analysis of the brain shows grafted cells (identified by the expression of GFP) in the ventricle. The expression of the NIS in the grafted cells was confirmed by immunofluorescence by using an antibody against hNIS (inset). CT computed tomography, GFP green fluorescent protein, Hipp-NSC hippocampus-derived neural stem cell, hNIS human sodium iodide symporter, NIS sodium iodide symporter, NIS-Hipp-NSC hippocampus-derived neural stem cell expressing the human sodium iodide symporter, SPECT single-photon emission tomography, 99m Tc technetium-99m
In vivo imaging
To assess whether grafted cells can be visualized by using SPECT/CT imaging in the live animal, in a first set of experiments we incubated NIS-Hipp-NSCs in vitro with 99mTc before transplantation in the rat brain. The cells were injected in the left lateral ventricle (1×106 cells) (Fig. 5b) or in the hippocampus (two injection sites per hippocampus; amount of cells per site ranging from 440,000 to 55,000 cells) (Fig. 6a), and the rats were subjected to SPECT imaging. The SPECT signal was superimposed to the CT scan to localize the cells within the brain. Quantification of the signal in the hippocampus was performed by measuring the average number of pixels in the region of interest by using image analysis software (Image J) and correlated to the number of cells injected (Fig. 6b). Post-imaging analysis of the rat brains confirmed the presence of the cells by immunofluorescence analysis (Fig. 6c).
Fig. 6.
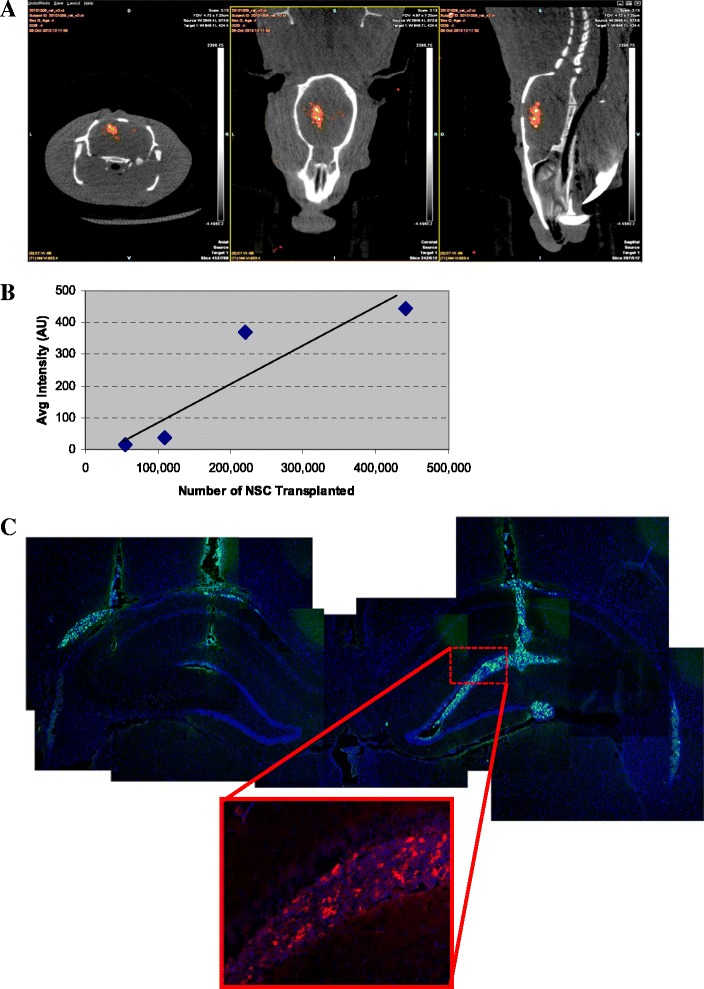
In vivo SPECT imaging of NIS-Hipp-NSCs injected in the brain hippocampus. a SPECT signal recorded from NIS-Hipp-NSCs that were incubated with 99mTc in vitro before transplantation in the hippocampus (two sites/hippocampus). Transverse, coronal, and sagittal orientations of the brain are shown. b Correlation between the number of cells injected in the hippocampus and the signal detected by SPECT imaging. Correlation coefficient r = 0.9566; two-tailed P value = 0.0108. c Analysis of the brain shows grafted cells (identified by the expression of GFP) in the hippocampus. The expression of the NIS in the grafted cells was confirmed by immunofluorescence by using an antibody against hNIS (inset). GFP green fluorescent protein, Hipp-NSC hippocampus-derived neural stem cell, hNIS human sodium iodide symporter, NIS sodium iodide symporter, NIS-Hipp-NSC hippocampus-derived neural stem cell expressing the human sodium iodide symporter, SPECT single-photon emission tomography, 99m Tc technetium-99m
In a second set of experiments, NIS-Hipp-NSCs were injected through an intracranial cannula implanted in the rat brain. For these experiments, NIS-Hipp-NSCs were resuspended in PBS and injected in the brain (1–2×105 cells in 5 μl at 0.2 μl/min). At various time points after cell injection (2 and 24 h), 99mTc was delivered to the brain via the cannula (0.5 mCi in 5 μl at a speed of 0.5 μl/min), and the rats were subjected to SPECT/CT imaging 4 h later (Fig. 7).
Fig. 7.
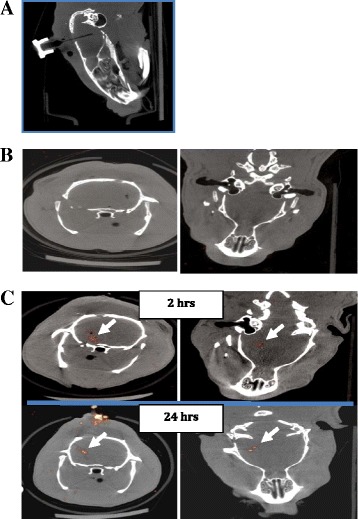
Repetitive in vivo SPECT imaging of NIS-Hipp-NSCs. a CT scan of the rat head showing the placement of the cannula used to deliver 99mTc into the brain. b SPECT imaging of the brain of a rat that had received 99mTc and no cells. c SPECT signal recorded from the rat brain 2 and 24 h after NIS-Hipp-NSC intracranial injection following delivering of 99mTc. Transverse and coronal slice orientations are shown. CT computed tomography, NIS-Hipp-NSC hippocampus-derived neural stem cell expressing the human sodium iodide symporter, SPECT single-photon emission tomography, 99m Tc technetium-99m
These data demonstrate that Hipp-NSCs expressing the hNIS can be visualized in vivo in the rat brain. Specifically, these data show adequate spatial resolution and signal detection sensitivity of the SPECT imaging system and the feasibility to repetitively image grafted NSCs in the rat brain.
Characterization of Hipp-NSCs expressing the hNIS after transplantation in the rat brain
To determine whether the expression of the hNIS would alter the ability of Hipp-NSCs to graft and survive after transplantation in the intact rat brain, Hipp-NSCs expressing the hNIS were injected in the hippocampus of adult rats and the animals were euthanized 2 weeks later. Control groups consisted of rats that were injected with Hipp-NSCs without the hNIS. Grafted cells in the brain were localized on the basis of the presence of CMFDA (a cell tracker dye that was loaded in the cells before transplantation). Both Hipp-NSCs and Hipp-NSCs expressing the hNIS were found grafted in the hippocampus 2 weeks after transplantation. Grafted cells were localized in the pyramidal layer (CA1) and some cells were also visible between the CA1 and granular layer. Phenotypical characterization of the grafted cells performed by immunofluorescence analysis revealed that some of the grafted cells had differentiated into mature neuronal cells expressing NeuN (Fig. 8).
Fig. 8.
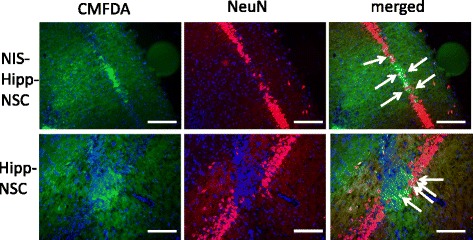
In vivo SPECT imaging of NIS-Hipp-NSCs injected in the brain hippocampus. Representative cross-sections of rat brain 2 weeks after transplantation of NIS-Hipp-NSCs or Hipp-NSCs in the hippocampus (CA1 region). Grafted cells are located on the basis of the expression of the cell tracker dye CMFDA. Mature neurons are identified on the basis of the expression of NeuN. Merged images show that some of the grafted cells express the neuronal marker NeuN (arrows). Cell nuclei are stained blue with DAPI. Scale bar = 50 μm. DAPI 4′,6-diamidino-2-phenylindole, Hipp-NSC hippocampus-derived neural stem cell, NIS-Hipp-NSC hippocampus-derived neural stem cell expressing the human sodium iodide symporter, SPECT single-photon emission tomography
Discussion
NIS expression in NSCs
The first objective of this work was to determine the effect of NIS expression on the biology of NSCs. The analysis of the effect of the expression of NIS on Hipp-NSCs was directed to both the basic biological properties as well as to their ability to respond to the host brain tissue in vivo. Our data show that the expression of NIS in Hipp-NSCs does not change their proliferation rate in vitro. The ability to proliferate is a key functional signature of NSCs and is of particular importance in clinical applications because in vitro NSC expansion is necessary to generate a clinically useful number of cells for transplantation. Biochemical expression of proteins that are characteristically found in NSCs (SOX2 and nestin) was also not affected by the NIS. Although the expression of SOX2 and nestin is commonly used to identify NSCs, a more complete analysis of their biochemical profile and of their karyotype should be performed in future studies. Nonetheless, the results shown in this work demonstrate that the exogenous expression of NIS in Hipp-NSCs does not alter the basic biological properties of the cells. This is in agreement with other studies that have characterized the use of the NIS in other types of stem cells.
Possibly the most important feature of NSCs, and one that makes these cells so appealing for use in replacement therapy of the nervous system, is their ability to generate neurons and glia. This important feature is maintained in Hipp-NSCs that express the NIS, as demonstrated by the expression of proteins that are known markers of differentiated phenotypes—βIII-tubulin and NeuN for neurons and glial fibrillary acidic protein (GFAP) and oligodendrocyte-specific protein for glia—analyzed by using both immunofluorescence and Western blot techniques. Additionally, no differences were found between Hipp-NSCs expressing the NIS and naïve Hipp-NSCs when comparing the expression of neuronal and glial markers by Western blot analysis. This is true both in vitro as well as in vivo in the rat brain, where Hipp-NSCs expressing the NIS where found grafted in the hippocampus 2 weeks after transplantation with at least some cells showing expression of neuronal markers. Although the studies presented in this report are limited to a 2-week follow-up, a large body of literature testifies to the safety of 99mTc accumulation in cells in vivo [16, 21–23]. Specifically, NIS-based imaging has been validated and used in humans [24, 25], thus supporting the translational potential of this reporter system.
Molecular imaging of NSCs using the NIS
The main goal of this work was to develop an imaging system that will allow tracking NSCs after transplantation in the brain. We have provided several pieces of evidence, in vitro and in vivo, that the expression of the reporter gene hNIS allows the visualization of live and viable Hipp-NSCs by using SPECT imaging. We have first demonstrated that the expression of the hNIS in Hipp-NSCs is functional in that it allows for the selective uptake of 99mTc inside the cells. 99mTc is a gamma-emitting radioisotope that is taken up by the NIS in place of iodide (the endogenous, biological ligand of the NIS). Moreover, 99mTc is widely available and currently approved by the US Food and Drug Administration in combination with SPECT imaging for diagnostic purposes.
In the in vivo experiments, 99mTc was delivered directly in the brain parenchyma where the cells were grafted. The results demonstrate that the Hipp-NSCs grafted in the brain are viable, can take up 99mTc via the NIS, and can be visualized by SPECT imaging.
A successful imaging system needs to have sufficient signal sensitivity to detect grafted cells and sufficient spatial resolution to determine their location within the tissue/organ. The data presented in this article demonstrate that NIS combined with SPECT/CT imaging possesses enough spatial resolution and sensitivity to detect viable grafted cells in the brain. Specifically, when Hipp-NSCs were injected at different sites within the hippocampus and at different densities, discrete regions of positive signals were detected and a correlation between the signal intensity and the number of cells injected could be derived. Another important result of this work is the demonstration that Hipp-NSCs can be visualized repetitively over time on the same rat. This is a critical aspect of a reporter gene imaging system because, by allowing the visualization of grafted cells over time and for as long as they are viable, it sets this system apart from other imaging methodologies.
Limitations
One limitation of using the hNIS reporter system for imaging cells in the brain parenchyma is the relatively low permeability of 99mTc through the blood–brain barrier (BBB). In this report, therefore, we opted to use an intracranial cannula to deliver 99mTc directly to the brain in order to bypass any potential issue with 99mTc penetration through the BBB. This allowed us to provide proof-of-principle data demonstrating the feasibility of tracking grafted NSCs in the brain by using the NIS reporter system and SPECT detection. More clinically relevant and non-invasive delivery routes for 99mTc have been reported, including intravenous injections to image intracranial gliomas in experimental animals [26, 27], injection into the olfactory route [28, 29], encapsulation of 99mTc into liposomes, or nanoparticles targeted to the CNS via specific BBB transporters or receptor systems [30–32]. In future studies, we will test these routes of delivery; however, even as it stands, our data show that delivery of 99mTc via implanted intracranial cannulas is a valuable method for tracking NSCs in the brain in preclinical animal models of disease that will allow researchers to follow cell grafts over time in the same animals. This not only will provide valuable information on the location and viability of the cells and their biological activity but also will significantly shorten the time and reduce the number of experimental animals needed to perform the studies, thus expediting the translation of stem cell therapy to the clinical setting.
Although here we used NSCs isolated from the hippocampus of adult rats (Hipp-NSCs), it has to be noted that more easily accessible sources of NSCs (i.e., induced pluripotent stem cells, embryonic stem cell-derived NSCs, and NSCs derived from mesenchymal stem cells isolated from bone marrow, fat, or umbilical cord) have been described and are clinically attractive sources of cells as they allow autologous transplantation, an appealing approach that eliminates the need for immunosuppressive therapies. Because NSCs share common basic biological properties, the results of this work are significant, as they lay the groundwork for the use of the NIS to image NSCs and can very likely be extrapolated to other sources of NSCs.
Additionally, it is important to notice that in this study the cells were transplanted in naïve/uninjured rats. The response of Hipp-NSCs to an injured (i.e., after traumatic brain injury) or diseased brain will likely differ. However, a detailed analysis of grafting and differentiation potentials of Hipp-NSCs in the injured and uninjured brain goes beyond the scope of this work and will be the focus of future studies.
Conclusions
In this report, we have characterized the NIS as a reporter gene for imaging NSCs in the brain. Developing non-invasive imaging of grafted cells has emerged as a fundamental tool to advance the field of stem cell therapy. The ability to track cells in vivo after transplantation on the same animal over time will allow a reduction in the number of experimental animals needed to determine the optimal source of cells as well as the best route and site of delivery that will produce a functional recovery in animal models of disease. This will significantly shorten the time between experimental animal work and translation to the clinical setting.
Acknowledgments
This study was supported by grant 1UL1RR029876-01 from the National Center for Research Resources, National Institutes of Health and by the Moody Project for Translational Traumatic Brain Injury Research. We thank Sissy M. Jhiang for providing the full-length hNIS cDNA, Lori Follis for preparing 99mTc, Elizabeth Bishop for helping with the cell culture of Hipp-NSCs, Donald J. Deyo for providing valuable expertise and assistance with the intraventricular injection procedure, and Christine Courteau-Butler for editorial support.
Abbreviations
- AP
Anterior-posterior
- BBB
Blood–brain barrier
- CNS
Central nervous system
- CPM
Counts per minute
- CT
Computed tomography
- DAPI
4′,6-diamidino-2-phenylindole
- DMEM
Dulbecco’s modified Eagle’s medium
- D-PBS
Dulbecco’s phosphate-buffered saline
- DV
Dorsal-ventral
- EGF
Epidermal growth factor
- FBS
Fetal bovine serum
- FGF-2
Fibroblast growth factor-2
- GFP
Green fluorescent protein
- HBSS
Hanks’ Balanced Salt Solution
- Hipp-NSC
Hippocampus-derived neural stem cell
- hNIS
Human sodium iodide symporter
- ML
Medial-lateral
- MRI
Magnetic resonance imaging
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- NIS
Sodium iodide symporter
- NIS-Hipp-NSC
Hippocampus-derived neural stem cell expressing the human sodium iodide symporter
- NSC
Neural stem cell
- PBS
Phosphate-buffered saline
- PET
Positron emission tomography
- PMS
Phenazine methosulfate
- SPECT
Single-photon emission tomography
- TBS-T
Tris-buffered saline containing 0.1 % Tween 20
- Tc
Technetium-99m
- UTMB
University of Texas Medical Branch
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
M-AM designed and planned the experiments, isolated and cultured Hipp-NSCs, performed immunofluorescence and Western blotting analysis, analyzed the data, and wrote the manuscript. DRB performed the cloning of the hNIS into the lentivector. MAP performed the cell transplantation procedures and cannulated the rats. JW operated the SPECT/CT imaging system. IP operated the SPECT/CT imaging system and calculated required 99mTc dose. MM participated in the design of the imaging studies. HLH participated in the design of the study and in the cloning of the hNIS into the lentivector. All authors read and approved the final manuscript.
Contributor Information
Maria-Adelaide Micci, Email: mmicci@utmb.edu.
Debbie R. Boone, Email: drkenned@utmb.edu
Margaret A. Parsley, Email: maparsle@utmb.edu
Jingna Wei, Email: jiwei@utmb.edu.
Igor Patrikeev, Email: igpatrik@utmb.edu.
Massoud Motamedi, Email: mmotamed@utmb.edu.
Helen L. Hellmich, Email: hhellmic@utmb.edu
References
- 1.Cummings BJ, Uchida N, Tamaki SJ, Salazar DL, Hooshmand M, Summers R, et al. Human neural stem cells differentiate and promote locomotor recovery in spinal cord-injured mice. Proc Natl Acad Sci U S A. 2005;102:14069–74. doi: 10.1073/pnas.0507063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain KK. Cell therapy for CNS trauma. Mol Biotechnol. 2009;42:367–76. doi: 10.1007/s12033-009-9166-8. [DOI] [PubMed] [Google Scholar]
- 3.Pluchino S, Zanotti L, Deleidi M, Martino G. Neural stem cells and their use as therapeutic tool in neurological disorders. Brain research. Brain Res Rev. 2005;48:211–9. doi: 10.1016/j.brainresrev.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Martino G, Pluchino S. The therapeutic potential of neural stem cells. Nat Rev Neurosci. 2006;7:395–406. doi: 10.1038/nrn1908. [DOI] [PubMed] [Google Scholar]
- 5.Ourednik J, Ourednik V. Graft-induced plasticity in the mammalian host CNS. Cell Transplant. 2004;13:307–18. doi: 10.3727/000000004783983963. [DOI] [PubMed] [Google Scholar]
- 6.Ourednik V, Ourednik J. Graft/host relationships in the developing and regenerating CNS of mammals. Ann N Y Acad Sci. 2005;1049:172–84. doi: 10.1196/annals.1334.016. [DOI] [PubMed] [Google Scholar]
- 7.Conti L, Reitano E, Cattaneo E. Neural stem cell systems: diversities and properties after transplantation in animal models of diseases. Brain Pathol. 2006;16:143–54. doi: 10.1111/j.1750-3639.2006.00009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baghbaderani BA, Mukhida K, Hong M, Mendez I, Behie LA. A review of bioreactor protocols for human neural precursor cell expansion in preparation for clinical trials. Curr Stem Cell Res Ther. 2011;6:229–54. doi: 10.2174/157488811796575378. [DOI] [PubMed] [Google Scholar]
- 9.Zhao C, Tian M, Zhang H. In vivo stem cell imaging. Open Nucl Med J. 2010;2:171–7. doi: 10.2174/1876388X01002010171. [DOI] [Google Scholar]
- 10.Schaller B. Usefulness of positron emission tomography in diagnosis and treatment follow-up of brain tumors. Neurobiol Dis. 2004;15:437–48. doi: 10.1016/j.nbd.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 11.Spiriev T, Sandu N, Schaller B. Molecular imaging and tracking stem cells in neurosciences. Methods Mol Biol. 2013;1052:195–201. doi: 10.1007/7651_2013_27. [DOI] [PubMed] [Google Scholar]
- 12.Brader P, Serganova I, Blasberg RG. Noninvasive molecular imaging using reporter genes. J Nucl Med. 2013;54:167–72. doi: 10.2967/jnumed.111.099788. [DOI] [PubMed] [Google Scholar]
- 13.Hwang-do W, Kang JH, Jeong JM, Chung JK, Lee MC, Kim S, et al. Noninvasive in vivo monitoring of neuronal differentiation using reporter driven by a neuronal promoter. Eur J Nucl Med Mol Imaging. 2008;35:135–45. doi: 10.1007/s00259-007-0561-8. [DOI] [PubMed] [Google Scholar]
- 14.Penheiter AR, Russell SJ, Carlson SK. The sodium iodide symporter (NIS) as an imaging reporter for gene, viral, and cell-based therapies. Curr Gene Ther. 2012;12:33–47. doi: 10.2174/156652312799789235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Portulano C, Paroder-Belenitsky M, Carrasco N. The Na+/I- symporter (NIS): mechanism and medical impact. Endocr Rev. 2014;35:106–49. doi: 10.1210/er.2012-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terrovitis J, Kwok KF, Lautamaki R, Engles JM, Barth AS, Kizana E, et al. Ectopic expression of the sodium iodide symporter enables imaging of transplanted cardiac stem cells in vivo by single-photon emission computed tomography or positron emission tomography. J Am Coll Cardiol. 2008;52:1652–60. doi: 10.1016/j.jacc.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higuchi T, Anton M, Saraste A, Dumler K, Pelisek J, Nekolla SG, et al. Reporter gene PET for monitoring survival of transplanted endothelial progenitor cells in the rat heart after pretreatment with VEGF and atorvastatin. J Nucl Med. 2009;50:1881–6. doi: 10.2967/jnumed.109.067801. [DOI] [PubMed] [Google Scholar]
- 18.Templin C, Zweigerdt R, Schwanke K, Olmer R, Ghadri JR, Emmert MY, et al. Transplantation and tracking of human-induced pluripotent stem cells in a pig model of myocardial infarction: assessment of cell survival, engraftment, and distribution by hybrid single photon emission computed tomography/computed tomography of sodium iodide symporter transgene expression. Circulation. 2012;126:430–9. doi: 10.1161/CIRCULATIONAHA.111.087684. [DOI] [PubMed] [Google Scholar]
- 19.Seo JH, Jeon YH, Lee YJ, Yoon GS, Won DI, Ha JH, et al. Trafficking macrophage migration using reporter gene imaging with human sodium iodide symporter in animal models of inflammation. J Nucl Med. 2010;51:1637–43. doi: 10.2967/jnumed.110.077891. [DOI] [PubMed] [Google Scholar]
- 20.Gao J, Prough DS, McAdoo DJ, Grady JJ, Parsley MO, Ma L, et al. Transplantation of primed human fetal neural stem cells improves cognitive function in rats after traumatic brain injury. Exp Neurol. 2006;201:281–92. doi: 10.1016/j.expneurol.2006.04.039. [DOI] [PubMed] [Google Scholar]
- 21.Beasley TM, Palmer HE, Nelp WB. Distribution and excretion of technetium in humans. Health Phys. 1966;12:1425–35. doi: 10.1097/00004032-196610000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Miyagawa M, Beyer M, Wagner B, Anton M, Spitzweg C, Gansbacher B, et al. Cardiac reporter gene imaging using the human sodium/iodide symporter gene. Cardiovasc Res. 2005;65:195–202. doi: 10.1016/j.cardiores.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Shin JH, Chung JK, Kang JH, Lee YJ, Kim KI, Kim CW, et al. Feasibility of sodium/iodide symporter gene as a new imaging reporter gene: comparison with HSV1-tk. Eur J Nucl Med Mol Imaging. 2004;31:425–32. doi: 10.1007/s00259-003-1394-8. [DOI] [PubMed] [Google Scholar]
- 24.Barton KN, Stricker H, Brown SL, Elshaikh M, Aref I, Lu M, et al. Phase I study of noninvasive imaging of adenovirus-mediated gene expression in the human prostate. Mol Ther. 2008;16:1761–9. doi: 10.1038/mt.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barton KN, Stricker H, Elshaikh MA, Pegg J, Cheng J, Zhang Y, et al. Feasibility of adenovirus-mediated hNIS gene transfer and 131I radioiodine therapy as a definitive treatment for localized prostate cancer. Mol Ther. 2011;19:1353–9. doi: 10.1038/mt.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho JY, Shen DH, Yang W, Williams B, Buckwalter TL, La Perle KM, et al. In vivo imaging and radioiodine therapy following sodium iodide symporter gene transfer in animal model of intracerebral gliomas. Gene Ther. 2002;9:1139–45. doi: 10.1038/sj.gt.3301787. [DOI] [PubMed] [Google Scholar]
- 27.Varma NR, Barton KN, Janic B, Shankar A, Iskander A, Ali MM, et al. Monitoring adenoviral based gene delivery in rat glioma by molecular imaging. World J Clin Oncol. 2013;4:91–101. doi: 10.5306/wjco.v4.i4.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Illum L. Nasal drug delivery--possibilities, problems and solutions. J Control Release. 2003;87:187–98. doi: 10.1016/S0168-3659(02)00363-2. [DOI] [PubMed] [Google Scholar]
- 29.Okuyama S. The first attempt at radioisotopic evaluation of the integrity of the nose-brain barrier. Life Sci. 1997;60:1881–4. doi: 10.1016/S0024-3205(97)00149-5. [DOI] [PubMed] [Google Scholar]
- 30.Singh R, Lillard JW., Jr Nanoparticle-based targeted drug delivery. Exp Mol Pathol. 2009;86:215–23. doi: 10.1016/j.yexmp.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olivier JC, Huertas R, Lee HJ, Calon F, Pardridge WM. Synthesis of pegylated immunonanoparticles. Pharm Res. 2002;19:1137–43. doi: 10.1023/A:1019842024814. [DOI] [PubMed] [Google Scholar]
- 32.Begley DJ. Delivery of therapeutic agents to the central nervous system: the problems and the possibilities. Pharmacol Ther. 2004;104:29–45. doi: 10.1016/j.pharmthera.2004.08.001. [DOI] [PubMed] [Google Scholar]


