Abstract
Background
In engineered strains of Escherichia coli, bioconversion efficiency is determined by not only metabolic flux but also the turnover efficiency of relevant pathways. Methyl-d-erythritol 4-phosphate (MEP)-dependent carotenoid biosynthesis in E. coli requires efficient turnover of precursors and balanced flux among precursors, cofactors, and cellular energy. However, the imbalanced supply of glyceraldehyde 3-phosphate (G3P) and pyruvate precursors remains the major metabolic bottleneck. To address this problem, we manipulated various genetic targets related to the Entner–Doudoroff (ED)/pentose phosphate (PP) pathways. Systematic target modification was conducted to improve G3P and pyruvate use and rebalance the precursor and redox fluxes.
Results
Carotenoid production was improved to different degrees by modifying various targets in the Embden–Meyerhof–Parnas (EMP) and ED pathways, which directed metabolic flux from the EMP pathway towards the ED pathway. The improvements in yield were much greater when the MEP pathway was enhanced. The coordinated modification of ED and MEP pathway targets using gene expression enhancement and protein coupling strategies in the pgi deletion background further improved carotenoid synthesis. The fine-tuning of flux at the branch point between the ED and PP pathways was important for carotenoid biosynthesis. Deletion of pfkAB instead of pgi reduced the carotenoid yield. This suggested that anaplerotic flux of G3P and pyruvate might be necessary for carotenoid biosynthesis. Improved carotenoid yields were accompanied by increased biomass and decreased acetate overflow. Therefore, efficient use of G3P and pyruvate precursors resulted in a balance among carotenoid biosynthesis, cell growth, and by-product metabolism.
Conclusions
An efficient and balanced MEP-dependent carotenoid bioconversion strategy involving both the ED and PP pathways was implemented by the coordinated modification of diverse central metabolic pathway targets. In this strategy, enhancement of the ED pathway for efficient G3P and pyruvate turnover was crucial for carotenoid production. The anaplerotic role of the PP pathway was important to supply precursors for the ED pathway. A balanced metabolic flux distribution among precursor supply, NADPH generation, and by-product pathways was established.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-015-0301-x) contains supplementary material, which is available to authorized users.
Keywords: Carotenoid biosynthesis, Methyl-d-erythritol 4-phosphate pathway, Genetic targets, Central metabolic pathways, Escherichia coli, Balance of precursors
Background
Bioconversion by engineered microbes in cell factories is a promising alternative strategy to produce valuable chemicals, many of which are expensive to produce in their native hosts or by chemical processes [1, 2]. To construct an engineered strain with optimal phenotypes, the first steps are to enhance native pathways or introduce superior heterogeneous pathways for biosynthesis of target chemicals. Another strategy that can be more important and laborious is to seek and integrate separate genetic modification targets that improve productivity, even though the mechanisms of these targets are sometimes poorly understood.
The heterologous production of carotenoids in Escherichia coli is a well-studied example of an engineered microbial cell factory in which there has been coordinated modification of multiple targets [3–5]. Carotenoids are isoprenoid pigments naturally produced by plants, algae, and photosynthetic bacteria. They have attracted much attention for industrial use because of their diverse physiological effects and their potential applications as antioxidants and nutraceuticals [6–8]. Carotenoids are structurally derived from two universal isoprene units; isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). There are two independent pathways for the biosynthesis of IPP and DMAPP: the mevalonate pathway from the precursor acetyl-CoA in eukaryotic and archaea cells, and the methyl-d-erythritol 4-phosphate pathway (MEP) pathway from the precursors pyruvate and glyceraldehyde-3-phosphate (G3P) in prokaryotic cells and plastids in plants. Bioconversion of carotenoids by engineered strain has been developed based on either of these two pathways [9, 10].
In E. coli, MEP pathway-dependent carotenoid biosynthesis was initially engineered by introducing the carotenoid synthesis gene cluster crtEBI and by enhancing the expressions of genes in the MEP pathway, such as dxs and idi [9]. The modification of chromosomal targets has been shown to further improve carotenoid production [3, 11, 12]. PykFA was the first gene target identified to improve carotenoid production. Deletion of pykFA reduced the flux from G3P to pyruvate, suggesting that the balance between G3P and pyruvate was important for carotenoid synthesis [13]. Additional rate-limiting targets for carotenoid biosynthesis were further explored using either of two major methods: systematic approaches using genome-scale modeling; and combinatorial approaches based on phenotypic diversification and screening. The targets included the global regulators Hnr and ClpXP; the central metabolism enzymes YtjC and AceE, which may directly affect precursor levels by catalyzing G3P- and pyruvate-related metabolic reactions; and GdhA and FdhF, which might indirectly contribute to cofactors or flux balance [5, 11, 12, 14]. Thus, as suggested, improving and balancing the precursor supply remains an obstacle for MEP pathway-based carotenoid production.
The redistribution of metabolic flux at the pathway level is an alternative strategy to improve chemical biosynthesis and precursor supply [15]. This strategy is always more complex, because desirable flux distribution is not usually achieved by modifying single genes. The MEP pathway begins with the condensation of G3P and pyruvate in equal amounts. These precursors are primarily supplied by the Embden–Meyerhof–Parnas pathway (EMP pathway). The Entner–Doudoroff pathway (ED pathway), as a variant glycolysis pathway, produces equal amounts of G3P and pyruvate. This superior stoichiometric feature makes the ED pathway a preferable route for precursor supply. The strategy to redistribute central metabolic pathways has been used to improve MEP-dependent carotenoid production by engineered microbes [16–18]. However, the exact flux profile is not well understood. Systematic modification and analysis of gene targets in the EMP, ED, and pentose phosphate (PP) pathways may provide new insights on the flux profile.
In addition to the precursor supply, cofactor generation might be another key factor for the MEP pathway and carotenoid biosynthesis. Cofactors such as NAPDH and ATP are necessary for MEP reactions. Furthermore, the bioconversion efficiency could be affected by the accumulation of by-products such as acetate. Thus, it is necessary to elucidate flux distribution among precursor supply, cofactor generation, and by-product pathways. To this end, we implemented an engineering strategy that combined systematic genetic target modification and central metabolic flux redistribution. Multiple gene targets were either knocked out or overexpressed to redistribute the fluxes of the EMP, ED, and PP pathways. The effects of these modifications on the supply of G3P and pyruvate precursors, redox generation, and the metabolism of by-products were analyzed. These results will provide guidance for further MEP-pathway engineering studies.
Results
Analysis of metabolic engineering targets within EMP, PP, and ED pathways
Pyruvate and G3P are the two substrates of 1-deoxy-d-xylulose 5-phosphate synthase (encoded by dxs), the first enzyme in the MEP pathway. The favorable stoichiometric ratio of G3P and pyruvate generated via the ED pathway inspired the attempt to develop a non-EMP pathway route for MEP-dependent carotenoid bioconversion. In the EMP pathway, there are three steps in the conversion of the starting compound glucose-6-phosphate (G6P) into G3P, the precursor of the MEP pathway. Phosphoglucose isomerase (encoded by pgi) catalyzes the first glycolytic reaction at the branch point of the EMP and ED/PP pathways. The following two reaction steps are catalyzed by 6-phosphofructokinase I/II (encoded by pfkA and pfkB) and fructose bisphosphate aldolase II/I (encoded by fbaA and fbaB), respectively. Theoretically, blocking either of these genes would direct carbon flux from the EMP pathway to the ED/PP pathway, resulting in enhanced flux through the ED pathway. However, deletion of pgi disrupted the recycling of F6P or G3P to the oxidative PP pathway, resulting in decreased NADPH generation [19]. In the lower part of the EMP pathway, the conversion from G3P to pyruvate comprises five steps. Among the genes involved in these steps, ytjC and pykFA have been identified as targets for modification to improve MEP-dependent biosynthesis. The ED pathway is a shortcut for G3P and pyruvate generation. It produces the two MEP precursors in equal amounts via a four-step reaction from G6P. The ED pathway shares the first two steps with the PP pathway. The key gene targets within the ED and PP pathways are shown in Fig. 1. Compared with the ED pathway, the PP pathway generates one additional molecule of NADPH per molecule of glucose, which might be necessary for a MEP pathway reaction.
Fig. 1.
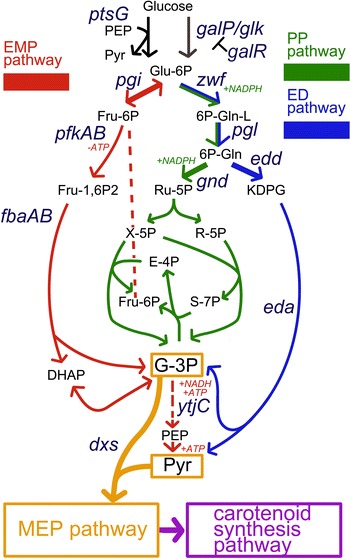
Genetic targets in central metabolic pathways for MEP-dependent carotenoid biosynthesis.
Analysis of targets responsible for directing flux from EMP pathway to ED/PP pathways
Initially, we constructed two reporter vectors for high carotenoid production, pLY036 and pLY10RK; their products were neurosporene and lycopene, respectively. The crtEBI cassette alone was insufficient for high carotenoid production (data not shown). Therefore, an additional E. coliidi gene was inserted downstream of crtEBI. The yields of neurosporene and lycopene from pLY036 and pLY10RK in wild E. coli BW25113 were 2.05 and 1.64 mg/g dry cell weight (DCW), respectively.
Various target genes within the EMP pathway were deleted to eliminate the flux of G3P and pyruvate through the EMP pathway, and the effects on neurosporene production were investigated. First, neurosporene production over time was compared between the Δpgi strain (P036) and the wild-type strain (W036) (Fig. 2a). The neurosporene yield of P036 remained stable during cell culture and was consistently greater than that of W036. The highest yield of P036 (after cell culture for 30 h) was approximately 25% greater than that of W036. The W036 strain produced the highest yield at the beginning of bioconversion, and then the yield gradually decreased. Considering cell growth and turnover, the decrease in neurosporene production per cell mass in W036 might be because neurosporene did not further accumulate during bioconversion. In contrast, the deletion of pgi might have resulted in the continued supply of optimal precursors or cofactors for the MEP pathway. The effects of deletions of other EMP genes were then investigated (Fig. 2b, biomass and carotenoids production of all strains in this study are summarized in Additional file 1: Table S1). Compared with the wild-type strain, the ΔpfkA and ΔpfkB strains (KA036 and KB036) and the ΔpfkAΔpfkB strain (KAB036) produced approximately 25% more neurosporene, similar to P036. As for the targets of fructose bisphosphate aldolase, only fbaB was a non-lethal deletion. However, deletion of fbaB (strain F036) had no effect on neurosporene production.
Fig. 2.
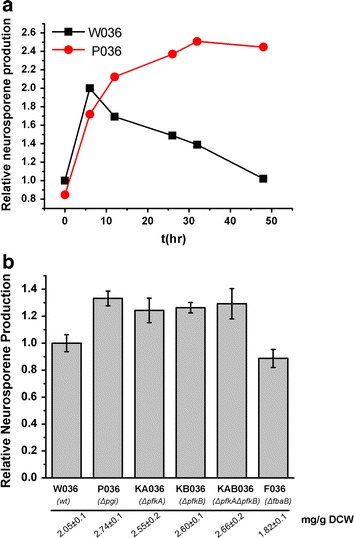
Effects of deleting different EMP pathway genes on neurosporene biosynthesis. a Relative neurosporene production of W036 (wild type) and P036 (Δpgi) over time; b relative neurosporene production of strains with deletions of different EMP pathway genes after bioconversion for 12 h.
To further investigate the effects of central flux redistribution on carotenoid biosynthesis, three branching point enzymes within the PP and ED pathways, zwf, edd, and eda, were each overexpressed in E. coli, and neurosporene production was compared among the overexpressing strains. As shown in Fig. 3, overexpression of Zwf (strain Z036) and Eda (strain E036) increased neurosporene yields by approximately 20 and 12%, respectively, while overexpression of edd in strain D036 decreased the yield by approximately 20%. The SDS-PAGE results (see Additional file 2: Figure S1) implied that edd overexpression might interfere with the normal expression of other genes such as crtI, resulting in decreased neurosporene production. These findings indicate that neurosporene biosynthesis could be improved by engineering target genes that could redistribute flux towards the ED/PP pathways, especially the ED pathway.
Fig. 3.
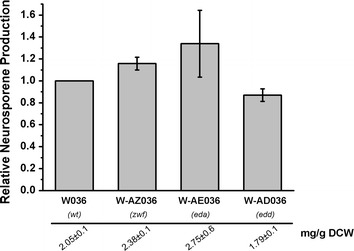
Effects of overexpression of different ED/PP pathway genes on neurosporene biosynthesis. Relative neurosporene production of strains overexpressing different ED/PP pathway genes after bioconversion for 12 h.
Efficient substrate use of G3P and pyruvate by overexpression of dxs in ED/PP pathways
To further study the effects of redistributed metabolic routes on carotenoid production, the pathways were modified in a MEP pathway-enhanced background. The first enzyme of the MEP pathway, Dxs, was overexpressed to increase the flux into the MEP pathway. This was achieved using three strategies: replacing the native promoter of the chromosomal dxs gene with the strong constitutive T5 promoter, which produced 3.20 mg/g DCW neurosporene (strain TX036); overexpressing the E. colidxs gene in a low copy-number plasmid (pSC101) under the control of the araBAD promoter (pSB1s-dxs), resulting in 5.03 mg/g DCW neurosporene production in the wild-type strain (SX036); and combining these two strategies, resulting in a neurosporene yield of 5.30 mg/g DCW (TX-SX036). These three strategies were also applied to the Δpgi strain (PTX036 for chromosomal T5 promoter dxs strain, P-SX036 for the plasmid dxs strain, PTX-SX036 for the combined strategy strain) and neurosporene production was compared with that of the wild-type strain. After bioconversion, the neurosporene yields of PTX036, P-SX036, and PTX-SX036 were 6.94, 9.40, and 10.71 mg/g DCW respectively, about 1.51–1.88 times more than that of the wild-type strain (Fig. 4a). This result implied that the substrate use of Dxs was significantly improved, possibly because of a balanced G3P and pyruvate supply from the ED pathway.
Fig. 4.
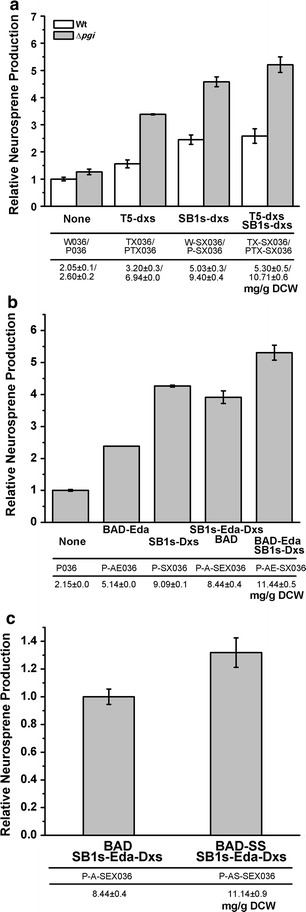
Enhanced precursor use by manipulating Dxs and Eda. a Dxs was expressed at different strengths, and neurosporene production was compared between Δpgi strain and wild-type strain after bioconversion for 12 h; b neurosporene production in strain simultaneously expressing Dxs and Eda after bioconversion for 12 h; c neurosporene production in strain in which Dxs and Eda were spatially coupled by protein scaffold after bioconversion for 12 h.
To further verify the role of the ED pathway, we attempted to improve the precursor supply by coordinated improvements to Eda and Dxs. First, we overexpressed Eda and Dxs simultaneously in the Δpgi strain. Eda and Dxs were expressed in the pBAD and pSB1s plasmids (P-AE036 and P-SX036, and co-transformation of the two plasmids for P-AE-SX036), respectively. As shown in Fig. 4b, the production of neurosporene was increased 25% by Eda enhancement, as compared with those of P-SX036 and P-AE-SX036. Second, a protein scaffold was used to spatially recruit Eda and Dxs through the interaction between tagged ligands. We designed a protein scaffold platform based on two proteins from Clostridium sp.; cellulosome cohesin and dockerin. Eda and Dxs were each fused with different dockerins and were co-expressed with scaffold protein (SS) containing a cohesin domain in the Δpgi strain. The function of SS was confirmed in a His-tag pull down assay (Additional file 3: Figure S2). Eda and Dxs co-localized with each other via the interaction between dockerins and SS. To balance the expression level with the scaffold, Eda and Dxs were co-transcribed under the control of the araBAD promoter using the low-copy pSB1s plasmid in the Δpgi strain (P-A-SEX036), compatible with SS expression using the pBAD plasmid. As shown in Fig. 4c, neurosporene production was increased by approximately 30% in the scaffold platform strain (P-AS-SEX036). These results confirmed the significant role of the ED pathway in carotenoid production.
Fine-tuning gnd expression to balance PP and ED pathway flux
The above results indicated that modification of ED targets resulted in more efficient precursor use by the MEP pathway. However, the ED pathway does not generate sufficient NADPH for MEP reactions. To explore the balance between the ED and PP pathways in this metabolic engineering route, either edd or gnd was knocked out in the Δpgi strain to construct PP-only and ED-only strains. The MEP pathway was enhanced by introducing pSB1s-dxs. In the bioconversion experiments, neurosporene production was significantly decreased in both PP-only (PD-SX036) and ED-only (PG-SX036) strains (Fig. 5a), indicating that both the PP pathway and the ED pathway were important for MEP-dependent biosynthesis. Thus, fine-tuning the flux between the ED pathway and the PP pathway was necessary to optimize the bioconversion process.
Fig. 5.
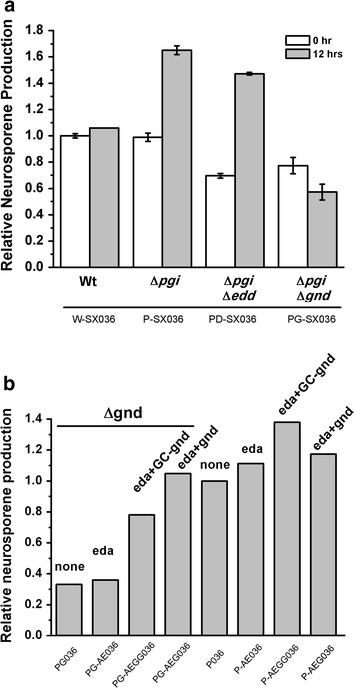
Roles of PP pathway and ED pathway in ED/PP pathway route and balance between PP and ED pathways. a Neurosporene production in strains ΔpgiΔedd and ΔpgiΔgnd after bioconversion for 0 or 12 h; b neurosporene production in strains with different gnd and eda engineering genotypes after bioconversion for 12 h.
Different flux distributions between the PP and ED pathways were achieved by coordinated engineering of gnd and eda expression. In this experiment, gnd was overexpressed at two different levels in either the wild-type or the Δgnd background: high-level expression was obtained using the araBAD promoter; low-level expression was obtained by introducing rare codons (Gly-Pro) at the N-terminal of Gnd peptides under the control of the araBAD promoter (Additional file 4: Figure S3). gnd was co-expressed with eda. The results indicated that moderate gnd expression combined with eda was most favorable for neurosporene synthesis in the wild-type background (Fig. 5b).
Anaplerotic role of PP pathway is important for carotenoid biosynthesis
Next, different targets were modified to determine the effects of PP pathway on neurosporene production. Compared with Δpgi, ΔpfkAB generated more NADPH through partial cyclization of the PP pathway [19]. ΔpfkA or ΔpfkAB have been reported as useful targets to improve NAPDH generation for chemical production [20, 21]. If the PP pathway contributes to neurosporene synthesis mainly by generating NAPDH, then further improvement of the NADPH supply will benefit neurosporene synthesis. To test this assumption, Δpgi was replaced by ΔpfkA or ΔpfkAB in an MEP-enhanced background. A novel plasmid pSB1s-XID was used to further improve the MEP flux, in which E. coliidi and ispDF were located downstream of dxs in pSB1s-dxs. The neurosporene yields of both KA-SXID036 (ΔpfkA) and KAB-SXID036 (ΔpfkAB) were lower than that of P-SXID036 (Δpgi) (Fig. 6). We suspected that the PP pathway might play additional roles other than simply generating NADPH for the ED pathway. We further deleted the lower EMP pathway targets, pfkA, pfkAB, fbaB, and ytjC in the Δpgi background, aiming to block the anaplerotic flux from the PP pathway towards G3P and pyruvate. Neurosporene production was decreased in all these strains (Fig. 6). Together, these results suggested that the PP pathway might play an anaplerotic role in supplying precursors for the ED pathway.
Fig. 6.
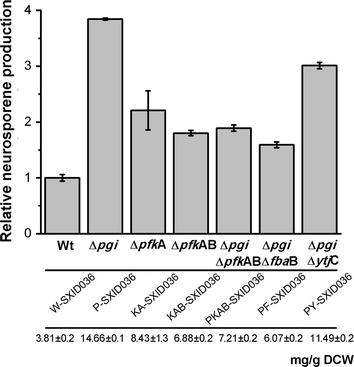
Flux balance within EMP pathway and its effects on carotenoid biosynthesis. Neurosporene production in strains with deletions of different EMP pathway targets after bioconversion for 12 h.
Enhanced carotenoid biosynthesis is accompanied with improved biomass and decreased acetate overflow
Redistribution of central metabolic flux always has complex effects. To investigate the balance between carotenoid biosynthesis and other cell metabolic pathways, batch fermentation for neurosporene production was conducted in a microbioreactor. As the MEP pathway flux still played a significant role in carotenoid production, the MEP pathway was further improved by replacing the chromosomal promoter with the T5 promoter to drive idi, resulting in PTXI036 and PTXI-SXID036 (containing pSB1s-dxs-idi-ispDF). Both biomass and neurosporene production were monitored in these strains. Surprisingly, the synthesis of neurosporene was beneficial for cell growth in the ED/PP pathway strains but not in the wild-type strain. As shown in Fig. 7 and Table 1, the strains with greater neurosporene synthesis showed greater cell growth and biomass, except for SXID036, which did not have modified central metabolic pathways. Thus, the inclusion of ED/PP pathways led to significant improvements in the total carotenoid titer.
Fig. 7.
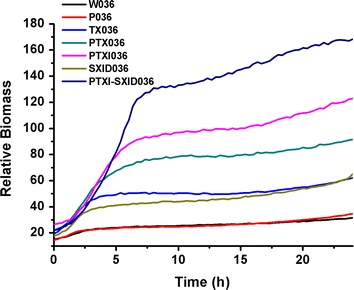
Batch fermentation to produce neurosporene in a microbioreactor. Change in relative biomass over time for strains W036 (BW25113 with pLY036), P036 (Δpgi with pLY036), TX036 (PT5-dxs with pLY036), PTX036 (Δpgi PT5-dxs with pLY036), PTXI036 (Δpgi PT5-dxs PT5-idi with pLY036), SXID036 (BW25113 with pSB1s-dxs-idi-ispDF and pLY036), PTXI-SXID036 (Δpgi PT5-dxs PT5-idi with pSB1s-dxs-idi-ispDF and pLY036).
Table 1.
Batch fermentation for neurosporene production in a microbioreactor
| Strain | Biomass (OD600 value) | Neurosporene production (mg neurosporene/g DCW) | Titer (mg neurosporene/L) |
|---|---|---|---|
| W036 | 8.91 | 1.76 | 4.7 |
| P036 | 10.35 | 1.83 | 5.68 |
| TX036 | 16.2 | 2.40 | 11.66 |
| PTX036 | 23.16 | 4.64 | 32.24 |
| PTXI036 | 30.35 | 5.35 | 48.71 |
| W-SXID036 | 17.3 | 4.53 | 23.51 |
| PTXI-SXID036 | 40.47 | 16.69 | 202.64 |
Bioconversion was performed in a microbioreactor for 24 h and biomass and neurosporene production were monitored.
Next, we analyzed metabolic intermediates to elucidate the coupling mechanism between biomass accumulation and carotenoid biosynthesis. The strains were cultured in flasks, and acetate accumulation and glucose consumption were monitored. The biomass results were similar to those observed in the microbioreactor experiments. Improvement of the MEP pathway in the Δpgi background resulted in significantly reduced acetate accumulation and enhanced glucose consumption (Table 2). Thus, we proposed that efficient use of glucose in the MEP pathway can establish a flux distribution that is favorable not only for carotenoid production but also for cell growth. This is probably because of the blockage of some by-product pathways, such as acetate overflow.
Table 2.
Acetate accumulation and glucose consumption of strains
| Strain | Biomass (OD600) | Acetate production (mg acetate/mL) | Residual glucose (mg glucose/mL) |
|---|---|---|---|
| W036 | 10 | 7.86 | 14.6 |
| P036 | 10.8 | 5.80 | 7.8 |
| W-SXID036 | 13.1 | 2.28 | 0.03 |
| PSX036 | 18.3 | 0.49 | 0 |
| PTXI-SXID036 | 22.6 | 0.21 | 0 |
Strains were cultured in flasks for bioconversion, and acetate accumulation and glucose consumption were monitored.
Combined modification of targets in ED/PP pathways for efficient lycopene production
Finally, we developed an optimally engineered strain based on the ED/PP pathway route (based on Δpgi, T5-dxs-idi, pBAD-eda-GCgnd, and pSB1s-dxs-idi-ispDF) for lycopene production using the reporter vector pLY10RK. To improve the total flux for lycopene production, glucose supply was optimized by engineering a glucose-uptake pathway. Namely, ptsG (encoding glucose PTS permease) was knocked out to reduce consumption of pyruvate and ATP. The non-phosphotransferase system was improved by inserting the P119-glk cassette (glucokinase gene glk under the control of the strong constitutive promoter P119) at the chromosomal galR locus [releasing the repression of the galactose repressor GalR to increase expression of galactose permease (GalP)]. Combining these targets, the resulting strain (PTXIGK-AEGG-SXID10RK) produced 17.8 ± 0.2 mg lycopene/g DCW in flask culture (Fig. 8). The lycopene production was about 3.6 times higher than that of the non ED/PP pathway route strain TXI-SXID10RK.
Fig. 8.
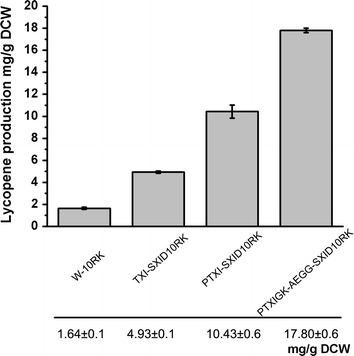
Lycopene production in strains with ED/PP pathway route. Lycopene production in wild-type strain (W-10RK, BW25113 with pLY10RK), non-ED/PP route strain (TXI-SXID10RK, PT5-dxs PT5-idi with pSB1s-dx -idi-ispDF and pLY10RK), ED/PP route strain (PTXI-SXID10RK, Δpgi PT5-dxs PT5-idi with pSB1s-dx -idi-ispDF and pLY10RK), and final optimal engineered strain (PTXIGK-AEGG-SXID10RK, ΔpgiPT5-dxsPT5-idiΔptsGgalR::P119-glk with pBAD-eda-GCgnd, pSB1s-dxs-idi-ispDF and pLY10RK) after bioconversion for 24 h.
Discussion and conclusions
An ideal microbial cell factory for bioconversion should maximize the target metabolic flux with an exact stoichiometric ratio of precursors and cofactors. However, this is hard to achieve because of the complexity of metabolic flux and its regulation. For MEP-dependent carotenoid biosynthesis, imbalanced precursor supply is one of the major metabolic bottlenecks, especially considering the limit of precursor (G3P) availability. Disruption of the EMP pathway target pykFA has been shown enhance lycopene production by retarding the catabolism of G3P to pyruvate [13]. In another study, the unexpected accumulation of indole may indicate that the pool of the G3P precursor was insufficient, because indole may be a by-product of an anaplerotic supply of G3P through the tryptophan biosynthesis pathway during MEP-dependent taxol production [22]. Research on non-EMP pathway precursor supply routes has mainly focused on the deletion of pgi [16, 18]. However, a parallel study has shown that lycopene yield was improved in a reverse redistribution pathway route with knocked-out zwf [17]. This indicated that detailed studies of both flux and targets were necessary. Herein, we conducted a systematic analysis and modification of genetic targets within central metabolic pathways for the redistribution of metabolic flux for MEP-dependent carotenoid biosynthesis.
As expected, the ED/PP pathway route created by deleting EMP targets and/or improving ED/PP targets enhanced carotenoid biosynthesis. Eda and Dxs seemed to be key targets for improving carotenoid biosynthesis. This was most likely because of their roles in the balance and efficient use of G3P and pyruvate precursors supplied by the ED pathway. Further metabolic engineering work will focus on coordinated modifications of Eda and Dxs. The flux balance between the ED and PP pathways was optimized by fine tuning gnd expression. Weak gnd expression was better for carotenoid synthesis, indicating the importance of the proper flux ratio between the ED and PP pathways. Although the ED pathway generated precursors with an exactly balanced ratio for MEP-dependent biosynthesis, the precursor balance still might be perturbed as both G3P and pyruvate could be metabolized in other cellular pathways. The results of this study implied that the anaplerotic role of the PP pathway might be important for precursor supply. The first clue was that the lower part of the MEP pathway was still necessary for ED/PP pathway route-dependent carotenoid biosynthesis. In addition, the carotenoid yields of Δpgi strains were greater than those of ΔpfkAB strains. The PP pathway routes achieved by Δpgi and ΔpfkAB might differ in their theoretical flux distribution [21]. Deletion of pgi would block the recycling of F6P and G3P into the oxidative PP pathway, and increase flux into the lower part of the EMP pathway (PP1, Fig. 9). However, pfkAB deletion might create a partial cyclization for the recovery of F6P from the PP pathway (PP2, Fig. 9). More NADPH was generated by the PP2 route than by the PP1 route, while the PP1 route supplied more G3P (Fig. 9). A pathway route based on Δpgi (ED + PP1) was more favorable for MEP-dependent biosynthesis. This suggested that the major role of the PP pathway was to sustain the G3P pool, although NADPH generated by the PP pathway was also important because the ED pathway was redox imbalanced for MEP-pathway reactions. However, detailed flux and metabolite analyses are required to determine the exact metabolic profile.
Fig. 9.
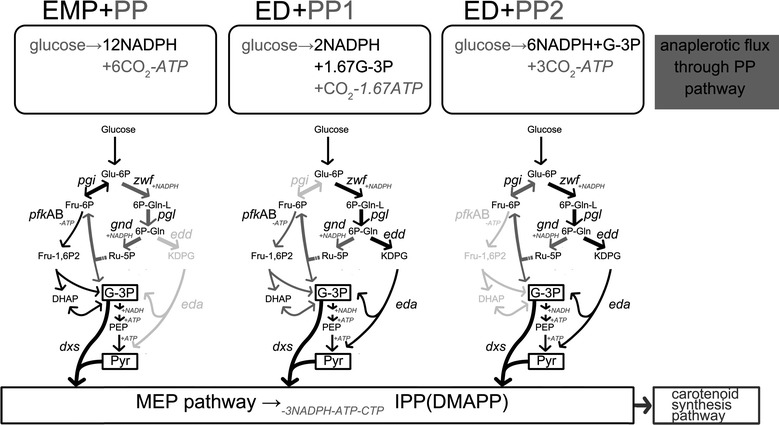
NADPH and G3P generation through different pathway routes. ED/PP pathways routes were created by either Δpgi or ΔpfkAB, resulting in ED + PP1 or ED + PP2. EMP or ED pathway is shown in black; PP pathway is shown in dark gray. NADPH and G3P generation through PP pathways in different routes is summarized in corresponding boxes.
The bioconversion efficiency of a given chemical is determined by both the stoichiometric yield of the pathway and the kinetic efficiency of pathway reactions. The metabolic route established by deleting pgi not only provided a balanced precursor ratio for efficient turnover in the MEP pathway, but also created a well-distributed metabolic flux between carotenoid biosynthesis and other pathways. Acetate overflow is always a major metabolic burden in E. coli K-12 strains because of the imbalance of central metabolic flux in high-glucose culture conditions. Surprisingly, the improvement of carotenoid biosynthesis in the redistribution route led to both greater cell growth and less acetate accumulation. This indicated that MEP-dependent carotenoid biosynthesis might relieve the metabolic burden by balancing the level of redox or metabolites, e.g., pyruvate. Indeed, redistribution of central metabolic flux always has complex effects. A global flux response could occur when MEP precursors are used efficiently or when redox conditions are balanced, resulting in flexible adaptation of flux distribution. Additional factors in central metabolic pathways, such as the level of the regulator intermediate fructose 6-bisphosphate, might be affected in the redistribution route, resulting in various responses and effects on the carotenoid biosynthesis pathway. Further research should focus on the effects of global regulation of central metabolic networks and their flux in carotenoid biosynthesis.
Methods
Bacterial strains and plasmids
Escherichia coli BW25113 was used as the parental strain for genetic modification and carotenoid production. Gene knock-out strains were obtained from the KEIO collection [23]. Escherichia coli DH5α was used for standard plasmid construction. The plasmids used for MEP gene expression and as reporter vectors were derived from expression vectors previously developed in our laboratory (unpublished). These vectors have the following features: a promoter (araBAD), multiple cloning sites, the rrnB terminator, origin of replication (p15A, RSF1030 or pSC101), and an antibiotic resistance gene (Kan, Cm, or Sm resistance). The plasmids pUKM (derivative of pKD4), pKD46, and pCP20 were used for gene integration. All E. coli strains and plasmids used in this study are described in Table 3.
Table 3.
Strains and plasmids used in this study
| Strain/plasmid | Description | References |
|---|---|---|
| Escherichia coli strains | ||
| BW25113 | Wild type K-12 strain | Our lab |
| DH5αa | For genetic manipulation | Invitrogen |
| TX | PT5-dxs K-12 strain, for construction of strains that harboring PT5-dxs phenotype | This study |
| TI | PT5-idi K-12 strain, for construction of strains that harboring PT5-idi phenotype | This study |
| GRK | galR::P119-glk K-12 strain, for construction of strains that harboring galR::P119-glk phenotype | This study |
| W036 | BW25113, pLY036 plasmid | This study |
| P036 | Δpgi, pLY036 plasmid | This study, KEIO |
| KA036 | ΔpfkA, pLY036 plasmid | This study, KEIO |
| KB036 | ΔpfkB, pLY036 plasmid | This study, KEIO |
| KAB036 | ΔpfkAΔpfkB, pLY036 plasmid | This study |
| F036 | ΔfbaB, pLY036 plasmid | This study, KEIO |
| W-AZ036 | BW25113, pBAD-zwf and pLY036 plasmids | This study |
| W-AE036 | BW25113, pBAD-eda and pLY036 plasmids | This study |
| W-AD036 | BW25113, pBAD-edd and pLY036 plasmids | This study |
| TX036 | PT5-dxs, pLY036 plasmid | This study |
| PTX036 | ΔpgiPT5-dxs, pLY036 plasmid | This study |
| W-SX036 | BW25113, pSB1s-dxs and pLY036 plasmids | This study |
| P-SX036 | Δpgi, pSB1s-dxs and pLY036 plasmids | This study, KEIO |
| TX-SX036 | PT5-dxs, pSB1s-dxs and pLY036 plasmids | This study |
| PTX-SX036 | Δpgi PT5-dxs, pSB1s-dxs and pLY036 plasmids | This study |
| P-AE036 | Δpgi, pBAD-eda and pLY036 plasmids | This study, KEIO |
| P-A-SEX036 | Δpgi, pBAD, pSB1s-eda-dxs and pLY036 plasmids | This study, KEIO |
| P-AE-SX036 | Δpgi, pBAD-eda, pSB1s- dxs and pLY036 plasmids | This study, KEIO |
| P-AS-SEX036 | Δpgi, pBAD-SS, pSB1s-eda-dxs and pLY036 plasmids | This study, KEIO |
| PD-SX036 | ΔpgiΔedd, pSB1s-dxs and pLY036 plasmids | This study |
| PG-SX036 | ΔpgiΔgnd, pSB1s-dxs and pLY036 plasmids | This study |
| P-AEG036 | Δpgi, pBAD-eda-gnd and pLY036 plasmids | This study, KEIO |
| P-AEGG036 | Δpgi, pBAD-eda-GCgnd and pLY036 plasmids | This study, KEIO |
| PG036 | ΔpgiΔgnd, pLY036 plasmid | This study |
| PG-AE036 | ΔpgiΔgnd, pBAD-eda and pLY036 plasmids | This study |
| PG-AEG036 | ΔpgiΔgnd, pBAD-eda-gnd and pLY036 plasmids | This study |
| PG-AEGG036 | ΔpgiΔgnd, pBAD-eda-GCgnd and pLY036 plasmids | This study |
| W-SXID036 | BW25113, pSB1s-dxs-idi-ispDF and pLY036 plasmids | This study |
| P-SXID036 | Δpgi, pSB1s-dxs-idi-ispDF and pLY036 plasmids | This study, KEIO |
| KA-SXID036 | ΔpfkA, pSB1s-dxs-idi-ispDF and pLY036 plasmids | This study |
| KAB-SXID036 | ΔpfkAΔpfkB, pSB1s-dxs-idi-ispDF and pLY036 plasmids | This study |
| PKAB-SXID036 | ΔpgiΔpfkAΔpfkB, pSB1s-dxs-idi-ispDF and pLY036 plasmids | This study |
| PF-SXID036 | ΔpgiΔfbaB, pSB1s-dxs-idi-ispDF and pLY036 plasmids | This study |
| PY-SXID036 | ΔpgiΔytjC, pSB1s-dxs-idi-ispDF and pLY036 plasmids | This study |
| PTXI036 | Δpgi PT5-dxs PT5-idi, pLY036 plasmids | This study |
| PTXI-SXID036 | Δpgi PT5-dxs PT5-idi, pSB1s-dx -idi-ispDF and pLY036 plasmids | This study |
| W10RK | BW25113, pLY10RK plasmid | This study |
| TXI-SXID10RK | PT5-dxs PT5-idi, pSB1s-dx -idi-ispDF and pLY10RK plasmids | This study |
| PTXI-SXID10RK | Δpgi PT5-dxs PT5-idi, pSB1s-dx -idi-ispDF and pLY10RK plasmids | This study, KEIO |
| PTXIGK-AEGG-SXID10RK | Δpgi, PT5-dxsPT5-idi, ΔptsGgalR::P119-glk, pBAD-eda-GCgnd, pSB1s-dxs-idi-ispDF and pLY10RK plasmids | This study, KEIO |
| Other bacterial strains | ||
| Anabaena sp. PCC7120 | crtE, crtB for pLY036 | Our lab |
| Rhodobacter sphaeroides 2.4.1 | crtI for pLY036 | Our lab |
| Pantoea ananatis | crtB. crtI for pLY10RK | Our lab |
| Pantoea agglomerans | crtE for pLY10RK | Our lab |
| Plasmids | ||
| pKD46 | For lambda-Red mediated recombination | [25] |
| pUKM | For lambda-Red mediated recombination, derived from pKD4 by inserting multiple cloning site closed to FRT | Our lab |
| pCP20 | For lambda-Red mediated recombination | [25] |
| pUKM-T5 | For construction of TX and TI, derived from pUKM by inserting T5 promoter at multiple cloning site | This study |
| pBAD-HisA | ColE1 origin, araBAD promoter, AmpR | Invitrogen |
| pSB1s | pSC101 origin, araBAD promoter, StrR | Our lab |
| pS95s | pSC101 origin, constitutive P119 promoter (derived from iGEM part BBa_J23119), StrR | Our lab |
| pS95s-glk | pSC101 origin, constitutive P119 promoter, StrR,E. coli glk, for construction of pUKM-glk | This study |
| pUKM-glk | For construction of GRK, derived from pUKM by inserting P119-glk fragment at multiple cloning site | This study |
| pLY036 | p15A origin, araBAD promoter, CmR, Anabaena sp crtEB, R. sphaeroides crtI, E. coli idi | This study |
| pLY10RK | RSF1030 origin, araBAD promoter, KanR, P. agglomerans crtE, P. ananatis crtBI, E. coli idi | This study |
| pSB1s-dxs | pSC101 origin, araBAD promoter, StrR,E. coli dxs | This study |
| pSB1s-eda-dxs | pSC101 origin, araBAD promoter, StrR, dockerin-fused E. coli eda, dockerin-fused E. coli dxs | This study |
| pSB1s-dxs-idi-ispDF | pSC101 origin, araBAD promoter, StrR, E. coli dxs,idi,ispDF | This study |
| pBAD-zwf | ColE1 origin, araBAD promoter, AmpR, E. coli zwf | This study |
| pBAD-edd | ColE1 origin, araBAD promoter, AmpR, E. coli edd | This study |
| pBAD-eda | ColE1 origin, araBAD promoter, AmpR, E. coli eda | This study |
| pBAD-eda-gnd | ColE1 origin, araBAD promoter, AmpR, E. coli eda,gnd | This study |
| pBAD-eda-GCgnd | ColE1 origin, araBAD promoter, AmpR, E. coli eda,GCtag-gnd | This study |
| pBAD-SS | ColE1 origin, araBAD promoter, AmpR, clostridium sp cellulose scaffold protein | This study |
Construction of plasmids and genomic integration
Plasmids were constructed using standard molecular biological protocols [24]. For chromosomal promoter replacement and gene integration, heterogeneous gene fragments were inserted at the MCS site downstream of the FRT-kanamycin resistance cassette in pUKM. The gene-FRT-kan-FRT fragment was then amplified by PCR. Lambda-Red-mediated recombination was performed as described elsewhere [25]. For chromosomal phenotype integration, P1 virus-mediated transfection was performed as described elsewhere [26]. The primers used in this study are listed in Additional file 5: Table S2.
Growth media
LB medium was used for all molecular construction experiments and strain cultures. Auto-inducing medium [27] was used to induce protein expression. Bioconversion medium was prepared by adding 4% (w/v) glucose to M9 medium or LB medium.
Culture conditions
All strains were stored at −80°C until use. Strains were pre-cultured in LB medium supplemented with appropriate antibiotics at 37°C overnight and then transferred into auto-inducing medium to induce expression of recombinant proteins. For neurosporene and lycopene production, cells were harvested by centrifugation after protein induction. Cells were then transferred to bioconversion medium at a starting biomass of 4 OD/L. For bioconversion experiments, cells were cultured in a flask at 37°C with shaking at 220 rpm for indicated times, and then harvested to quantify products and metabolites.
Measurement of carotenoid production and production of other metabolites
Cells were harvested by centrifugation at 12,000g for 5 min. The cell pellet was washed and then extracted in 1 mL acetone at 4°C for 1 h. The mixture was centrifuged at 12,000g, and the absorbance of the supernatant was measured at 470 nm. Neurosporene was quantified as described elsewhere [28]. Lycopene was quantified using a standard curve of pure standard lycopene (Sigma). The biomass was determined by measuring cell density (OD600). The acetate concentration was determined by HPLC using a C18 column (Agilent), and the glucose concentration was measured using a glucose monitor (SDBI).
Batch fermentation in a microbioreactor
For batch fermentation of carotenoids, the strains were pre-cultured in auto-inducing medium to induce the synthesis of recombinant proteins. Then, cells were transferred into bioconversion medium in a bioreactor (BioLector), with a beginning cell density of 4 OD. The cells were cultured at 37°C with shaking at 800 rpm.
Authors’ contributions
YT participated in the conception and design of the study and revised the manuscript. W-FL participated in data collection and analysis, and drafted the manuscript. CL and L-QY participated in data collection and analysis. S–SZ and NC provided original data for some sections. All authors read and approved the final manuscript.
Acknowledgements
The authors gratefully acknowledge funding from the following organizations: the National Basic Research Program of China (973 Program) (Grant No. 2012CB721105); the Key Research Program of the Chinese Academy of Sciences (Grant No. KSZD-EW-Z-016-1); and the Key Research Program of the Chinese Academy of Sciences (Grant No. KGZD-EW-606-2).
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Additional files
Table S1. Biomass and carotenoid production of strains.
Figure S1. SDS-PAGE analysis of recombinant proteins produced by Escherichia coli strains: (M) protein markers; (0) strain W036; (1) strain W-AZ036; (2) strain W-AE036; (3) strain W-AD036.
Figure S2. Results of His-tag pull-down assay. Briefly, strains were induced to express target genes, then cells were lysed by sonication. Cell lysates were centrifuged and supernatants were incubated with Ni–NTA beads for 6 h. Beads were washed to remove unbound proteins. Bound proteins were eluted and analyzed by SDS-PAGE. (M) Markers; (1) strain with pSB1s-eda-dxs (dockerin-fused); (2) strain with pBAD-SS (His-tag fused scaffold protein); (3) strain with pBAD-SS and pSB1s-dxs (dockerin-fused); (4) strain with pBAD-SS and pSB1s-eda-dxs.
Figure S3. SDS-PAGE analysis of recombinant proteins produced by Escherichia coli strains: (M) protein markers; (1) strain P036; (2) strain P-AE036; (3) strain P-AEG036; (4) strain P-AEGG036.
Table S2. Primers used in this study.
Footnotes
Chun Li and Lan-Qing Ying contributed equally to this work
Contributor Information
Chun Li, Email: cindy1982@gmail.com.
Lan-Qing Ying, Email: lainie521@yahoo.cn.
Sha-Sha Zhang, Email: zhss31@126.com.
Nan Chen, Email: xiao_zhi_er@yahoo.com.cn.
Wei-Feng Liu, Email: liuwfv@163.com.
Yong Tao, Email: taoyong@im.ac.cn.
References
- 1.Colin VL, Rodriguez A, Cristobal HA. The role of synthetic biology in the design of microbial cell factories for biofuel production. J Biomed Biotechnol. 2011;2011:601834. doi: 10.1155/2011/601834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carothers JM, Goler JA, Keasling JD. Chemical synthesis using synthetic biology. Curr Opin Biotechnol. 2009;20:498–503. doi: 10.1016/j.copbio.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Alper H, Jin YS, Moxley JF, Stephanopoulos G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng. 2005;7:155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Alper H, Stephanopoulos G. Uncovering the gene knockout landscape for improved lycopene production in E. coli. Appl Microbiol Biotechnol. 2008;78:801–810. doi: 10.1007/s00253-008-1373-x. [DOI] [PubMed] [Google Scholar]
- 5.Choi HS, Lee SY, Kim TY, Woo HM. In silico identification of gene amplification targets for improvement of lycopene production. Appl Environ Microbiol. 2010;76:3097–3105. doi: 10.1128/AEM.00115-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodwin TW. Metabolism, nutrition, and function of carotenoids. Annu Rev Nutr. 1986;6:273–297. doi: 10.1146/annurev.nu.06.070186.001421. [DOI] [PubMed] [Google Scholar]
- 7.Fraser ML, Lee AH, Binns CW. Lycopene and prostate cancer: emerging evidence. Expert Rev Anticancer Ther. 2005;5:847–854. doi: 10.1586/14737140.5.5.847. [DOI] [PubMed] [Google Scholar]
- 8.Hughes DA. Effects of carotenoids on human immune function. Proc Nutr Soc. 1999;58:713–718. doi: 10.1017/S0029665199000932. [DOI] [PubMed] [Google Scholar]
- 9.Farmer WR, Liao JC. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol. 2000;18:533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Villalon A, Perez-Gil J, Rodriguez-Concepcion M. Carotenoid accumulation in bacteria with enhanced supply of isoprenoid precursors by upregulation of exogenous or endogenous pathways. J Biotechnol. 2008;135:78–84. doi: 10.1016/j.jbiotec.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 11.Jin YS, Stephanopoulos G. Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab Eng. 2007;9:337–347. doi: 10.1016/j.ymben.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Kang MJ, Lee YM, Yoon SH, Kim JH, Ock SW, Jung KH, et al. Identification of genes affecting lycopene accumulation in Escherichia coli using a shot-gun method. Biotechnol Bioeng. 2005;91:636–642. doi: 10.1002/bit.20539. [DOI] [PubMed] [Google Scholar]
- 13.Farmer WR, Liao JC. Precursor balancing for metabolic engineering of lycopene production in Escherichia coli. Biotechnol Prog. 2001;17:57–61. doi: 10.1021/bp000137t. [DOI] [PubMed] [Google Scholar]
- 14.Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol. 2005;23:612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Li Q, Sun T, Zhu X, Xu H, Tang J, et al. Engineering central metabolic modules of Escherichia coli for improving beta-carotene production. Metab Eng. 2013;17:42–50. doi: 10.1016/j.ymben.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Sun Y, Ramos KR, Nisola GM, Valdehuesa KN, Lee WK, et al. Combination of Entner–Doudoroff pathway with MEP increases isoprene production in engineered Escherichia coli. PLoS One. 2013;8:e83290. doi: 10.1371/journal.pone.0083290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Nambou K, Wei L, Cao J, Imanaka T, Hua Q. Lycopene production in recombinant strains of Escherichia coli is improved by knockout of the central carbon metabolism gene coding for glucose-6-phosphate dehydrogenase. Biotechnol Lett. 2013;35:2137–2145. doi: 10.1007/s10529-013-1317-0. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, Wang Y, Tang Q, Kong W, Chung WJ, Lu T. MEP Pathway-mediated isopentenol production in metabolically engineered Escherichia coli. Microb Cell Fact. 2014;13:135. doi: 10.1186/s12934-014-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruger NJ, von Schaewen A. The oxidative pentose phosphate pathway: structure and organisation. Curr Opin Plant Biol. 2003;6:236–246. doi: 10.1016/S1369-5266(03)00039-6. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, San KY, Bennett GN. Improvement of NADPH bioavailability in Escherichia coli through the use of phosphofructokinase deficient strains. Appl Microbiol Biotechnol. 2013;97:6883–6893. doi: 10.1007/s00253-013-4859-0. [DOI] [PubMed] [Google Scholar]
- 21.Siedler S, Bringer S, Blank LM, Bott M. Engineering yield and rate of reductive biotransformation in Escherichia coli by partial cyclization of the pentose phosphate pathway and PTS-independent glucose transport. Appl Microbiol Biotechnol. 2012;93:1459–1467. doi: 10.1007/s00253-011-3626-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2006(2):0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green MR, Sambrook J, Sambrook J. Molecular cloning : a laboratory manual. 4. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- 25.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomason LC, Costantino N, Court DL (2007) E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol Chapter 1:1–17 [DOI] [PubMed]
- 27.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Farasat I, Kushwaha M, Collens J, Easterbrook M, Guido M, Salis HM. Efficient search, mapping, and optimization of multi-protein genetic systems in diverse bacteria. Mol Syst Biol. 2014;10:731. doi: 10.15252/msb.20134955. [DOI] [PMC free article] [PubMed] [Google Scholar]


