Abstract
Mass spectrometry (MS) is a powerful tool for determining the mass of biomolecules with high accuracy and sensitivity. MS performed under so-called “native conditions” (native MS) can be used to determine the mass of biomolecules that associate noncovalently. Here we review the application of native MS to the study of protein−ligand interactions and its emerging role in elucidating the structure of macromolecular assemblies, including soluble and membrane protein complexes. Moreover, we discuss strategies aimed at determining the stoichiometry and topology of subunits by inducing partial dissociation of the holo-complex. We also survey recent developments in "native top-down MS", an approach based on Fourier Transform MS, whereby covalent bonds are broken without disrupting non-covalent interactions. Given recent progress, native MS is anticipated to play an increasingly important role for researchers interested in the structure of macromolecular complexes.
Keywords: native mass spectrometry (MS), top-down MS, protein−ligand interactions, stoichiometry, 2D interaction map, macromolecular complexes
Introduction
Non-covalent protein interactions play a vital role in biology.1 Through noncovalent electrostatic, van der Waals, hydrophobic and hydrogen bonding interactions, proteins recognize nearly all their binding partners: other proteins, nucleic acids, lipids, carbohydrates, and small molecules. Recently, mass spectrometry (MS) has emerged as an important approach to study such interactions. Using conditions that preserve non-covalent interactions (i.e., native MS), one can determine the mass of intact assemblies, their precise stoichiometry, direct interactions between subunits, the relative position (core vs. periphery) of subunits within an assembly and the strength of inter-subunit interactions2–8 (Fig. 1). By mixing subunits in a stepwise manner, a hierarchy in the assembly pathway can be determined.9 When native MS is coupled with ion mobility (IM; approaches mentioned in this review are summarized in Table1), information regarding the shape of complexes can be obtained.10,11 Combined with bioinformatics, native MS has also provided important insights into the evolution of protein complexes.12,13
Figure 1.
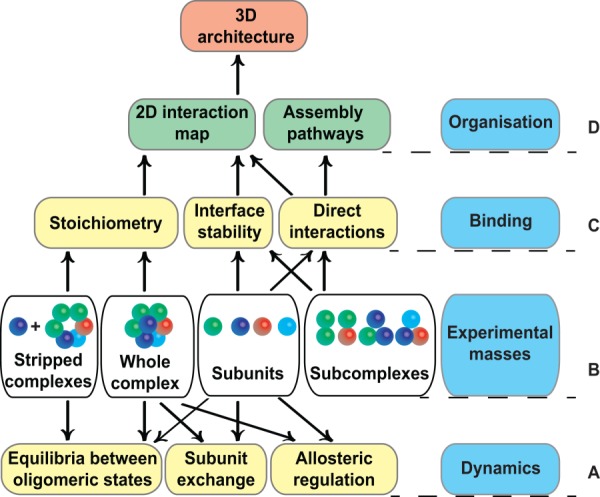
Native MS provides diverse information about macromolecular complexes. Primary data concerning the masses of the holo-complex, subcomplexes, and individual subunits (row B) provide access to a wealth of secondary information, such as binding interactions and dynamic properties (rows A, C), which in turn allows for higher inferences to be made concerning the organization of the complex (row D). Rows B-C: The experimental masses of individual subunits of a complex are determined following chromatographic separation under denaturing conditions.157 The use of MS conditions optimised for preserving noncovalent interactions (i.e., native MS) allows the mass of the intact complex to be measured. The combined subunit and holo-complex data reveal the stoichiometry of subunits, which can be further confirmed by a series of tandem MS spectra (i.e., “gas-phase dissociation” to generate stripped complexes32). The use of perturbing conditions or destabilizing agents (“in-solution dissociation”) allows one to generate overlapping subcomplexes (dimers, trimers, etc.), whose composition reveals direct interactions between subunits47 and allows interface stability to be assessed.13 Row D: The above data allow one to determine the two dimensional (2D) interaction network of subunits within a protein complex and, when combined with structural data (EM or SAXS envelopes), to deduce the 3D architecture of a macromolecular assembly (e.g., Refs.90 and91). In addition, individual subunits (or subcomplexes) can be mixed in solution and a mass shift can be detected if a (sub-)complex is generated (e.g., Ref.158), allowing the assembly pathway of a macromolecular complex to be identified. Row A: Native MS can provide information regarding the dynamic behavior of complexes. First, it can reveal the presence of different oligomeric states and monitor changes in equilibrium induced by different solution pH values and concentrations.5 Second, the subunit composition of intact complexes can be varied and monitored as a function of time by incubating light and heavy isoforms of a protein (e.g., labeled with 13C and 15N). The kinetics of subunit exchange can reveal distinct pathways for wild-type and mutant proteins (e.g., involved in amyloidosis,159 or assessing the effect of substrates and products.57) Third, native MS can relatively quantify the populations of macromolecular complexes containing different numbers of bound molecules, providing information about allostery.27
Table 1.
Summary of MS-Based Approaches Mentioned in the Text
| Acronym | Technique | Description |
|---|---|---|
| CID (or CAD) | Collision induced dissociation (Collisionally activated dissociation) | Technique for fragmenting ions in the gas phase, whereby ions are accelerated by an electrical potential and allowed to collide with neutral gas molecules such as argon or xenon. |
| ECD | Electron capture dissociation | Technique for fragmenting ions in the gas phase by causing them to interact directly with low-energy free electrons. |
| ESI | Electrospray ionisation | Soft ionisation technique whereby ions are produced in the gas phase by applying a voltage to the sample solution and creating an aerosol. |
| ETD | Electron transfer dissociation | Technique for fragmenting ions in the gas phase by transferring electrons to them from a radical anion such as anthracene or azobenzene. |
| FT MS | Fourier Transform based MS | Technique for determining the m/z ratio of ions through the frequency signals that the ions generate within the instrument. The mass spectrum is derived by applying a Fourier transform to the frequency data. |
| FT-ICR | Fourier Transform ion cyclotron resonance | Type of FT-based MS in which mass spectra are derived from the ion cyclotron resonance frequencies produced by ions as they rotate in a magnetic field. |
| HCD | Higher energy C-trap (or collisional) dissociation | A CID technique specific to Orbitrap instruments in which the fragmentation of gas phase ions takes place outside the orbitrap analyzer. |
| IM | Ion mobility | Technique used to separate molecular ions in the gas phase based on their mobility in a buffer gas under the influence of a weak electric field. |
| ISCID | In-source collision induced dissociation | Type of CID in which ions are fragmented in the source region of the mass spectrometer. |
| IRMPD | Infrared multiphoton dissociation | Technique for fragmenting ions in the gas phase by the absorption of multiple infrared photons. |
| MALDI | Matrix-assisted laser desorption/ionisation | Soft ionisation technique whereby macromolecules are embedded in a solid organic matrix and subsequently desorbed and ionised by a pulse of laser light. |
| MS/MS or MS2 | Tandem MS | Method of analysis involving two stages of MS selection. The first MS stage separates sample components according to their m/z. During the second MS stage, the selected ions are subsequently subjected to fragmentation and mass spectra of the fragmentation products are obtained. |
| - | Orbitrap | A FT-based analyzer composed of an outer barrel-like electrode and an inner spindle-like electrode. Ions are trapped in an orbital motion around the spindle and the frequency signals that arise from the resulting image current are used to calculate the mass spectrum. |
| NEMS | nanoelectromechanical systems | A nanoscale device which resonates at high frequency and can function as a highly sensitive mass sensor. Instead of measuring m/z ratio, NEMS sensors register jumps in frequencies that are directly proportional to the mass of the adsorbed species. |
| Q | Quadrupole | An analyzer composed of four parallel metal rods to which a radio frequency voltage and direct current voltage are applied. For a given ratio of voltages, ions travelling down the quadrupole having the appropriate m/z ratio will proceed through the analyzer, while others will have unstable trajectories and collide with the rods. |
| TOF | Time-of-flight | An analyzer in which ions are accelerated in an electric field and then allowed to drift through a field-free region to a detector; the m/z of the ions is calculated from the time taken to reach the detector. |
The focus of this review is the emerging role of native MS in characterizing the structure and dynamics of macromolecular assemblies, including protein-ligand interactions and soluble and membrane protein complexes (Table2). We illustrate different approaches used to establish the stoichiometry and topology of macromolecular complexes, including the partial dissociation of holo-complexes under controlled conditions in the gas phase and in solution. Moreover, we review recent progress in "native top-down MS," a technique based on Fourier Transform MS, whereby covalent bonds are broken and noncovalent interactions are preserved. Finally, we present a perspective on the future of macromolecular complex studies by native MS.
Table 2.
Examples of Soluble and Membrane Protein Complexes Analyzed by Native MS
| Complex | Mass (kDa) | Outcome of analysis | Prior atomic structure known | References |
|---|---|---|---|---|
| Soluble Complexes | ||||
| Cascade | 405 | Two dimensional map | N | 99 |
| Csy (CRISPR system yersinia) | 350 | “ “ “ | N | 100 |
| Eukaryotic translation factor eIF3 | 798 | “ “ “ | N | 46 |
| DNA-Central glycolytic genes Repressor (CggR) | 92−183 | Functional insights | N | 102 |
| EcoP15I, EcoPI, PstII | 63−311 | “ “ | N | 162 |
| Bacteriophage HK97 capsid | 18,000 | Assembly pathway | N | 34 |
| TrV virions (from Triatoma infestans) | 8,300 | “ “ | N | 105 |
| Norwalk virus-like particles | 10,100 | “ “ | N | 106 |
| Heat Shock Protein 90 chaperone | 441−138 | “ “ | Y | 95 |
| HK97 Prohead-1 particles | ∼21,400 | “ “ | Y | 98 |
| Trax–translin endonuclease | 206−246 | Three dimensional model of the full-length complex | Y | 90 |
| DegP oligomers | 143−575 | Model for the transition between the resting and active states of an enzyme | Y | 93 |
| Protruding (P) domain of the norovirus capsid protein oligomers | 72−1,361 | Discovery of multiple oligomeric states controlled by buffer conditions (e.g., pH) | Y | 97 |
| Membrane Complexes | ||||
| E. coli Translocon (ColE9-Im9 complex, BtuB, OmpF trimer, and TolB) | 296 | Functional insights | Y | 108 |
| MscL | 78 | “ “ | Y | 163 |
| ATPase and subcomplexes | 690−208 | “ “ | N | 107 |
| OmpA | 69 | Low-resolution model for the full-length dimer | N | 164 |
| DgkA, pSRII, LacY-GFP | 13−78 | Reconstitution in detergent, amphipols, bicelles and nanodiscs | Y | 110 |
| PagP and OmpT, Mhp1 and GalP | 20−54 | Same as above. | Y | 111 |
| PagP | 21 | Same as above | Y | 165 |
| ATP-Binding Cassette transporter P-glycoprotein (P-gp) | 141−147 | Ligand binding affinities | Y | 109 |
| K channel KcsA | 192 | “ “ “ | Y | 36 |
| EmrE, LmrP, MscL, BtuCD, LmrCD, MacB, MexB, ATP synthase | 12−344 | “ “ “ | Y | 40 |
| MscL, AqpZ and AmtB | 85−126 | “ “ “ | Y | 166 |
| B subunits of cholera and heat labile toxins | 58−62 | “ “ “ | Y | 112 |
| OmpF, AmtB and P-gp | 111−141 | Stabilization of membrane protein complexes by charge reduction | Y | 167 |
Advantages and limitations of native MS
MS-based approaches are useful for gaining important insights into the structure and dynamics of macromolecular complexes. Native MS has several advantages compared to other techniques. First, it is widely applicable to samples that vary greatly in mass, degree of flexibility, symmetry, and polydispersity. Second, multiple oligomeric states can be analyzed simultaneously, providing specific information about each individual species (i.e., without averaging the data over different species). This allows the dynamics of quaternary structure to be studied in real time. Third, native MS is highly sensitive. In several cases successful analyses required only a few microlitres of sample at relatively low (μM) concentration. Finally, native MS does not require samples to be chemically labeled or crosslinked.
However, native MS also has important limitations. MS is performed under vacuum conditions and the detection of macromolecular complexes takes place in the gas phase. Consequently, the relative abundance of detected complexes may deviate from that in solution because of the distinct ionization, transmission and detection probabilities of different complexes.5 Moreover, in the gas phase hydrophobic and electrostatic interactions become weaker and stronger, respectively, than in solution. This may render the detection of certain assemblies impossible without prior crosslinking.14 Nevertheless, computational and experimental evidence indicates that the transition from solution to the gas phase does not drastically alter biomolecules.15 For instance, lysozyme and trypsin retain their catalytic activity after their ionization, mass selection, and soft-landing onto surfaces.16 Also, when solution conditions such as pH and concentration change, the gas-phase spectra change accordingly5 (Fig. 2). Even so, the above limitations should be borne in mind when interpreting the results of a native MS experiment.
Figure 2.
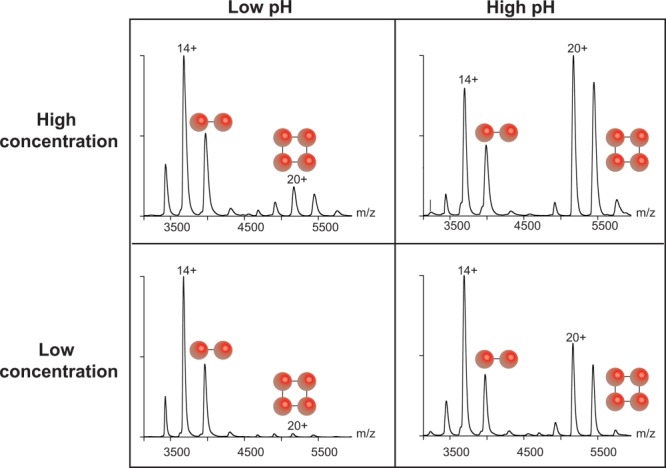
Solution conditions influence the species detected by native MS in the gas phase. The solution pH and sample concentration affect the relative abundance of dimers and tetramers of the protein concanavalin A. Spectra of intact concanavalin A are shown at the following concentrations and pH values (clockwise from top left): 18.3 μM and pH 3.4; 16.9 μM and pH 8.4; 2.0 μM and pH 8.4; and 2.1 μM and pH 3.4. Two spheres indicate the dimeric ions and four spheres indicate tetrameric ions. These spectra were reproduced with permission from Ref. 5, ©(2011) ACS publications.
Preserving noncovalent complexes in the gas phase
Native MS requires gentle ionization of the macromolecular complex being analyzed. Of the two soft ionization methods generally used in biological MS, electrospray ionization (ESI), and matrix-assisted laser desorption/ionization (MALDI),17 ESI is the preferred method for native MS because noncovalent interactions are preserved. The principles underlying ESI have been described previously,18–21 although how protein ions are produced and subjected to gas-phase transition remains poorly understood.22,23 Noncovalent interactions are primarily studied by nano-electrospray ionization (nano-ESI), which involves a spray orifice diameter (1–10 μm) smaller than that used for conventional ESI (∼100 μm).24,25 The small opening allows the use of reduced sample volumes (2−5 μL) at micromolar concentrations and at low flow rates (20−200 nL/min).26 ESI-MS analysis can be influenced by the nature of the intermolecular interactions, by the composition, ionic strength and pH of the sample buffer, and by the voltages and pressures within the mass spectrometer. Native MS is typically carried out using volatile buffers such as ammonium acetate2 or ethylenediammonium diacetate27 and hence usually necessitates exchanging the sample buffer prior to analysis. In some cases the biochemical steps involved in sample preparation may need to be optimized so that the native state of the complex is preserved during buffer exchange. Unlike other types of ESI-MS analysis, neither acidic conditions nor organic solvents are used.
Nano-ESI Q-TOF-based native MS
The study of noncovalent interactions requires the use of mass spectrometers specifically modified to transmit and detect large, fragile assemblies. In the early 1990s, a few groups demonstrated that relatively small-sized noncovalent assemblies could be analyzed by nano-ESI-MS using conventional spectrometers28–31 (see Fig. 3 for timeline). However, the analysis of larger complexes required customising the mass spectrometers, because protein complexes with molecular masses above 60 kDa typically form ions with mass-to-charge (m/z) ratios greater than 4000, which is above the detection limit of standard instruments. Major hardware modifications enabled the detection of larger complexes, leading to tandem MS experiments being performed on nano-ESI-quadrupole-time-of-flight (Q-TOF) mass spectrometers.32,33 The use of modified instruments has enabled the investigation of very large protein complexes (e.g., an intact 18 MDa viral capsid34) and of interactions involving membrane proteins,35–41 described further below.
Figure 3.
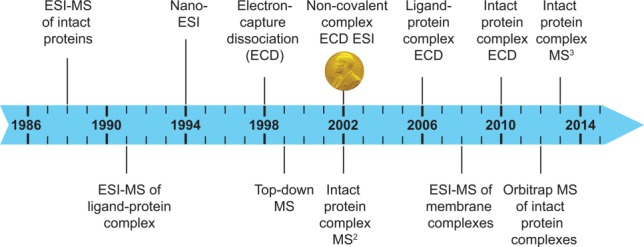
Timeline of MS-based analyses of macromolecular complexes. The advent of ESI127,160 and of nano-ESI161 laid the groundwork for native MS. In 1991 the first examples of the analysis of macromolecular complexes were published and in 1998 electron capture dissociation (ECD) was developed.74 In 2002 John Bennett Fenn was awarded a share of the Nobel Prize in Chemistry for his contribution to the development of electrospray ionization. In the same year noncovalent complexes were fragmented by ECD76 and tandem MS (i.e., MS2) experiments of protein complexes were performed.32 In 2006 ECD fragmentation of a ligand−protein complex was first reported.80 Subsequently, intact protein complexes were fragmented by ECD121 and in 2013 subunits ejected from intact protein complexes were broken down using MS3 experiments.131
Dissociation of macromolecular complexes in the gas phase and in solution
Tandem MS (also called MS2 or MS/MS) is based on several MS steps whereby selected ions are broken down in the gas phase and the product ions are analyzed.42 In particular, when protein complexes are studied, an ion population (characterized by a specific charge state, z) is selected according to its m/z ratio in the first mass analyzer (e.g., a quadrupole) and then accelerated toward a collision cell filled with a gas such as argon or xenon, where it undergoes collision induced dissociation (CID). Afterwards, the dissociation products are further analyzed (e.g., in a TOF region). With a few exceptions,43,44 monomers are ejected from macromolecular complexes one after the other. Moreover, the charge is distributed across the dissociated ions in a highly asymmetric manner. For example, a tetramer will generate highly charged monomers and lowly charged “stripped” trimers that can further dissociate into monomers and dimers. The ejection of monomers allows the assessment of stoichiometry with high accuracy. Tandem MS can also provide an insight into the location (core vs. periphery) of subunits within the complex,45 as peripheral subunits are expelled at lower energy compared to the core subunits.9 In certain cases, tandem MS can provide structural information on subunit interactions and their relative spatial arrangements.43
In addition to gas-phase dissociation, macromolecular complexes can also be dissociated in solution. “Destabilising agents” [e.g., dimethyl sulfoxide (DMSO) or methanol] are used to selectively break some interactions within the complex before introducing the sample into the mass spectrometer. One study exploited this “in-solution dissociation” approach to elucidate the assembly and disassembly pathways of certain homo-complexes, revealing these pathways to reflect the evolutionary pathways whereby different quaternary structures arise.13 In the case of hetero-complexes, in-solution dissociation allows one to establish the direct interactions between subunits by generating subcomplexes. In the case of the 13-subunit human eukaryotic translation factor eIF3, simply increasing the ionic strength of the ammonium acetate buffer was sufficient to perturb ionic and polar interactions at subunit interfaces, generating almost thirty different subcomplexes.46 The addition of DMSO or methanol was also used to partially disrupt the 10-subunit yeast exosome.47 The analysis of more than a dozen subcomplexes led to a 2D interaction map that was subsequently combined with a low-resolution electron microscopy (EM) envelope to deduce a 3D starting model of the yeast exosome. To summarise, the ability to dissociate assemblies either in the gas phase or in solution is of key importance for confirming stoichiometry and determining direct interactions between subunits.2
Protein-ligand interactions studied by native Q-TOF based MS
The use of native MS to analyze noncovalent interactions between proteins and small-molecule ligands has been reviewed previously.48–50 Here we focus on examples that illustrate the application of native MS and Q-TOF instruments to address diverse questions regarding protein−ligand interactions in the context of structural studies.
A study concerning the pore-forming leucotoxins secreted by S. aureus illustrates the use of native MS to evaluate the binding affinity of protein−ligand interactions.51 The leukotoxins studied were of two types, class S (LukS-PV, HlgA, and HlgC with a mass between 31 and 32 kDa) and class F (LukF-PV and HlgB; 34–35 kDa). Native MS and surface plasmon resonance were used to study the inhibition of these toxins by p-sulfonato-calix[n]arenes (SCns, including SC4, SC6, and SC8) and confirmed that the SCns only bind class S, not class F, leucotoxins.51 MS experiments determined the SCn-leucotoxin stoichiometry, ranked the affinity of the different SCns for the class S toxins (SC8>SC6>SC4) and ranked the leukotoxins according to their relative affinity for SCn (HlgA>HlgC>LukS-PV).
Native MS experiments characterized the binding of a recombinant protein to an unknown ligand that was present in the expression medium.52 The squid nerve protein ReP1-NCXSQ (Regulatory protein of the squid nerve sodium calcium exchanger) was studied by MS and X-ray crystallography.52 In native conditions, MS detected the presence of unknown ligands bound to ReP1-NCXSQ, subsequently identified by liquid chromatography and high-resolution MS as fatty acids. When the binding of palmitoleic, palmitic, and oleic acids to ReP1-NCXSQ was specifically evaluated, native MS revealed that the occupancy of the bound fatty acid was greatly reduced upon mutating a key residue within the phospholipid binding site.
Native MS was used to establish the stoichiometry of protein−ligand interactions in a study of the Kelch-like ECH-associated protein 1 (Keap1).53 Keap1 represses the activation of Nrf2, a transcription factor which mediates protective anti-oxidant and anti-inflammatory responses. The authors used chemical screening to identify compounds that block the interaction between Nrf2 and the C-terminal domain of Keap1 (Kelch-DC) and determined co-crystal structures for Kelch-DC in complex with two such inhibitors. Interestingly, these structures exhibited a difference in inhibitor:protein binding stoichiometry. The authors used native MS to confirm the 2:1 binding stoichiometry for one of these compounds and to rule out a crystal packing artefact. By contrast, native MS was used to demonstrate that crystal packing environment alters the relative binding affinity of aldose reductase for two different inhibitors.54 Native MS has also been successfully used to investigate the effect of inhibitors on a protein:RNA complex.55 Another study that combined native MS with X-ray crystallography concerned two nickel import proteins of Staphylococcus aureus.56 One of these was successfully crystallized bound to a Ni-(L-His)2 ligand. The second protein could not be crystallized, but native MS indicated that it binds Ni(II) ions via a His-dependent chelator different from Ni-(L-His)2 (Fig. 4).
Figure 4.
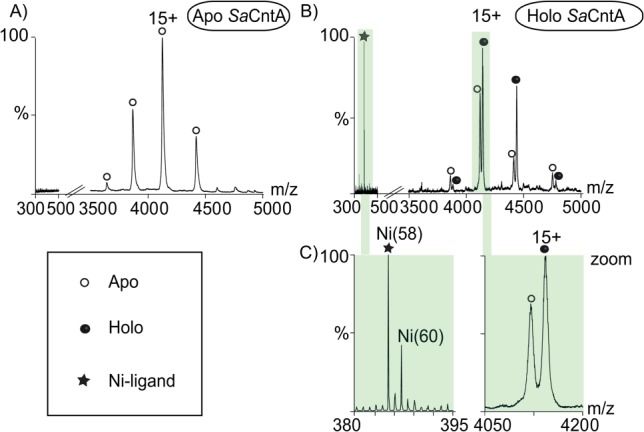
Native mass spectra of Staphylococcus aureus (Sa) cobalt and nickel transporter (Cnt) A protein. CntA is an extracytoplasmic nickel-binding subunit of an ABC-importer. A: Apo form of SaCntA analyzed at high collision energy (i.e., E: 140 Vs).56 The 300–500 and 3500–5000 m/z ranges are shown. B: SaCntA, incubated with culture supernatant, was studied at the collision energy of 140 Vs. C: Expanded views of the 380–395 and 4050–4200 m/z ranges. The left view illustrates the isotopic pattern of the nickel-binding ligand. The right view shows the presence of two peaks corresponding to the apo and liganded SaCntA. Empty circles, apo SaCntA; filled circles, liganded SaCntA; asterisk, Ni(II)-containing ligand.
Another remarkable example used native MS to investigate the allosteric mechanism of ATP binding to the chaperonin GroEL, a 14-mer formed by two heptameric subunit rings.27 Allosteric models were considered in which subunits exist in either a low-affinity (T) or high-affinity (R) state. The MS data allowed determination of the relative coexisting populations of GroEL with different numbers of bound ligand, thereby yielding individual binding affinities for all 14 ATP molecules. These data confirmed positive cooperativity within each heptameric ring and negative cooperativity between the two rings. Specifically, when three ATP molecules were bound to the first heptamer, this ring shifted from a low- to high-affinity condition (T to R state transition), whereas five ligands were required to shift the second ring to the high-affinity state.
Finally, native MS was used to investigate the dynamics of dimer formation in a study of E. coli glucosamine-6-phosphate synthase (GlmS).57 X-ray crystallography had previously localized the substrate binding site within the dimer interface. The kinetics of GlmS subunit exchange were examined by mixing 14N and 15N labeled proteins in the presence of various substrates and products and the resulting dimers were analyzed by native MS in a time-resolved manner. The rate constants of subunit exchange determined from these experiments revealed that certain ligands accelerated, while others prevented, subunit exchange.
The above examples illustrate that native MS carried out with nano-ESI-Q-TOF instruments represents a versatile approach for studying protein−ligand interactions.
Protein−ligand interactions studied by Fourier transform based MS and electron-based dissociation
Besides nano-ESI-Q-TOF instruments, Fourier transform (FT) based mass spectrometers have also successfully been used to investigate non−covalent interactions. These studies exploit the high accuracy and resolution of two types of mass analyzer, the FT ion cyclotron resonance (ICR)58 and Orbitrap59–62 spectrometers. For example, in early studies the high resolution offered by FT-ICR enabled the detection of toxin-oligosaccharide complexes and the estimation of their association constants,63 and was exploited to analyze the competitive binding of inhibitors to bovine carbonic anhydrase II.64 Orbitrap instruments, introduced in the year 2000,59 are comparable in resolution and accuracy to FT-ICR mass spectrometers without the need for a superconducting magnet.60–62 Several reviews illustrate the features and general applications of FT based MS.58,65–68 Here we focus on the study of protein−ligand interactions by native ESI coupled with FT instruments.
A key feature of FT-based MS is the fragmentation induced by electrons, namely, electron capture dissociation (ECD)69–71 and electron transfer dissociation (ETD).72,73 In ECD, first described in 1998,74 trapped multiply protonated molecules in the gas phase capture low energy electrons, resulting in the formation and subsequent fragmentation of odd-electron ions.69,74 This process is highly bond specific; for instance, the breaking of disulfide interactions and N–Cα bonds is favored by ECD. Like ECD, ETD is a fragmentation method based on the transfer of electrons originating from radical anions.72 Both ECD and ETD allow the preservation of non-covalent bonds and of PTMs,73,75 whereas covalent interactions such as peptide bonds are broken.
One of the first examples of noncovalent binding maintained intact by ECD was described in 2002.76 Haselmann et al. investigated the interactions mediating homodimer formation for a series of peptides as well as those mediating the binding of two glycopeptide antibiotics (vancomycin and eremomycin) to their bacterial tripeptide target.76 In this study, only strong covalent bonds were broken by ECD, while weak bonding within a noncovalent complex was preserved, allowing the gas-phase structure of the noncovalent complexes, position of the binding site and primary sequence information to be determined. ECD was also used to investigate the direct interactions of a neuropeptide, Substance P, with divalent metals,77 to identify the copper binding site of the chloroplastic protein CP1278 and to map the primary binding sites of the anticancer drugs cisplatin, transplatin, and oxaliplatin on ubiquitin.79
An FT-ICR mass spectrometer was utilised to study the noncovalent complex formed by the small disordered protein α-synuclein and the polycationic amine spermine, allowing the spermine binding site to be mapped to a C-terminal fragment of α-synuclein.80 CID and ECD were used to investigate the interaction between chicken adenylate kinase and ATP, allowing localization of the ATP-binding pocket.81 Similarly, the combination of these two fragmentation techniques was used to map the zinc binding site on bovine carbonic anhydrase II and the ATP binding site on chicken muscle adenylate kinase.82 CID and ECD were also used to characterize nanoscale bilayers (nanodiscs) formed by the binding of phospholipids to a scaffold protein.83 Both gas-phase dissociative approaches were utilized to study metallodrug-peptide complexes and characterize the amino acid binding sites.84,85 CID, ETD, and higher-energy C-trap dissociation (HCD) were also applied to study a metallodrug-protein complex (i.e., the oxaliplatin-ubiquitin assembly).86 Among the three approaches, ETD was the most useful for identifying the metal binding sites. Another interesting example compared ETD with ECD to study the interactions between basic and acidic peptides, concluding that the two approaches yielded a similar abundance and type of intramolecular fragments.87 In summary, FT-based MS combined with electron-based dissociation is a powerful method for investigating protein−ligand interactions and characterizing binding sites.
The role of native MS in the structure elucidation of soluble protein complexes
Numerous reports illustrate the power of native MS using nano-ESI-Q-TOF instruments and CID to advance structural studies of soluble protein assemblies (reviewed in3,4,8–10,45,88,89). The examples below are grouped to illustrate how native MS (i) can be combined with other structural approaches, (ii) provides useful information in the absence of high-resolution structural data, and (iii) sheds light on the assembly pathway of macromolecular complexes.
(i) Native MS provides complementary information to other structural methods. For example, MS was instrumental in elucidating the structure of the Trax-Translin endonuclease complex involved in the RNA interference pathway.90 Whereas the crystal structure of the truncated proteins indicated the formation of a hexameric assembly, native MS revealed that the full-length proteins assembled as ∼240 kDa octamers.90 Combining data from crystallography, EM and MS yielded a three dimensional model of the full-length octameric complex. Most recently, a computational method was developed for combining native MS data with other MS-based information (IM-MS and chemical crosslinking) to generate pseudo-atomic models of protein complexes.91,92
Native MS was also used to study the E. coli protease-chaperone DegP, which exists in different oligomeric states.93 Structures of the DegP hexameric resting state and the 12meric and 24meric active states had previously been observed by X-ray crystallography.94 Native MS experiments suggested the mechanism whereby substrate-mimicking peptides induce the transition from the resting to the active state. Similarly, native MS was utilized to study the dynamics of Hsp90-containing complexes formed in the presence of co-chaperones.95 In this case, the assembly of ten different complexes was monitored in real time, KD values were calculated and a kinetic model for the Hsp90 assemblies was proposed to illustrate the most favorable reaction pathways.95
Several viruses have been successfully analyzed by native MS (reviewed in Refs. 88, 96). The major structural capsid protein of noroviruses is VP1, whose protruding (P) domain forms antigenic assemblies that might be useful for vaccine development. In addition to the P assemblies (dimer, 12mer, and 24mer) previously identified by structural studies, native and IM-MS identified additional P particles (18mer and 36mer), whose abundance depended on the buffer conditions (pH and DTT concentration) used.97 In a separate example involving the bacteriophage HK97, single-particle cryo-EM, X-ray crystallography, label-free quantitative proteomics and native and hydrogen/deuterium exchange MS were combined to better understand the role of the HK97 protease in maturation of the bacteriophage capsid.98 In particular, native MS allowed the mass determination of protease-free and protease-containing procapsids and of the mature capsid (17.9, 21.4, and 12.9 MDa, respectively). These results allowed the number of protease molecules in the procapsid to be estimated and the efficiency of procapsid cleavage to be assessed, shedding light on the capsid assembly mechanism.
(ii) Native MS provides useful structural information when no high-resolution structure is available. For example, native MS was used to investigate Cascade, a 405 kDa ribonucleoprotein (RNP) complex required for CRISPR-mediated immunity in E. coli.99 Native MS provided a 2D interaction map of Cascade, complementing the low-resolution structures obtained by negative-stain EM and small-angle X-ray scattering (SAXS). The combined data led to a structural model for how double-stranded DNA targets are recognized by Cascade. Another CRISPR-associated RNP studied by native MS is the Csy complex (350 kDa) from P. aeruginosa.100 This approach provided the stoichiometry of the complex and revealed the peripheral location of certain subunits, complementing EM and SAXS data which revealed the overall shape of the Csy complex. The Type III-A CRISPR-Cas Csm complex (427 kDa) from T. thermophilus was also studied by proteomics, native MS and EM.101
An additional example is a study of the human translation initiation factor eIF3, a 13-subunit complex of 798 kDa.46 Its stoichiometry, overall subunit architecture and the existence of three submodules were established by native MS. These results, in combination with EM data, allowed the identification of subunits located near the decoding center of the ribosomal 40S subunit.
Native MS was also combined with SAXS and fluorescence cross-correlation spectroscopy to study the Central glycolytic genes Repressor (CggR), a transcription factor that regulates carbohydrate metabolic pathways in B. subtilis.102 Crystal structures of the CggR effector-binding domain indicated structural changes upon the binding of glycolytic intermediates,103 but the DNA-bound structure of CggR was unknown. Native MS was used to analyze CggR bound to a full-length operator DNA or to a half-site DNA sequence in the presence and absence of the inducer metabolite, fructose 1,6-bisphosphate (FBP). These experiments revealed that CggR forms a compact tetramer upon binding either DNA sequences and that FBP causes the tetramer to dissociate into two physically independent dimers. Further support for these conclusions was obtained using IM-MS.104
(iii) Native MS allows the investigation of assembly pathways. An example is the characterization of Prohead-1, an icosahedral intermediate in the assembly of the bacteriophage HK97 with a mass of 18 MDa and formed by 420 identical subunits.34 In another case, MS was combined with atomic force microscopy (AFM) to study virion assembly and viral genome release (uncoating) of the picorna-like Triatoma virus.105 Native MS revealed the composition of a labile intermediate in the uncoating pathway, providing key information unattainable by AFM. Both native MS and AFM were also used to assess the influence of solution pH, ionic strength and subunit concentration on the assembly pathway of Norwalk virus-like particles.106
Investigation of membrane protein complexes
In the last decade, interactions involving membrane proteins have been studied by MS, including protein-drug interactions,35 protein−lipid complexes,36 and multiprotein assemblies.37,38 More recently, heteromeric membrane protein complexes containing both soluble and transmembrane subunits were successfully analyzed by native MS39,40,107 (Table2). A few of the most recent such studies are described below.
Native MS was instrumental for deciphering the structure of the E. coli translocon complex that mediates the translocation of colicin E9 (ColE9) across the bacterial outer membrane.108 This complex is composed of two membrane proteins (the trimeric porin OmpF and the vitamin B12 receptor BtuB) and three soluble proteins (ColE9, the immunity protein Im9 and the periplasmic protein TolB). By native MS the heptameric complex was detected, its mass determined and the presence of an additional lipopolysaccharide bound to BtuB was assessed (Fig. 5). Native MS also identified a stable subcomplex obtained by limited proteolysis, corresponding to the intact OmpF trimer, TolB and a ColE9 fragment. In addition, native MS and electrophysiological measurements in which OmpF was exposed to increasing concentrations of ColE9 peptides suggested a novel model for the sequential binding of ColE9 to OmpF. These MS results were combined with a negative-stain EM envelope and previous crystal structures to deduce a 3D model of the translocon.
Figure 5.
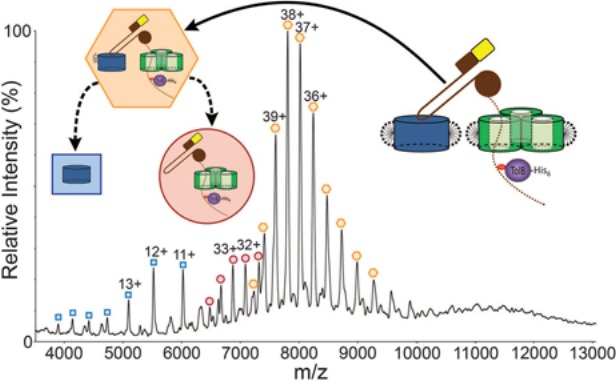
Native mass spectrum of the ColE9 translocon. The inserts represent the observed species: orange hexagon, intact complex bound to a lipopolysaccharide; red circle, translocon lacking BtuB and the lipopolysaccharide; blue square, BtuB. See text for further details. This spectrum was reproduced with permission from Ref.108 © (2013) AAAS.
Native MS was also used to study the ATP-Binding Cassette transporter P-glycoprotein (P-gp) and its binding to lipids, nucleotides, and drugs in real time.109 Lipid binding rates and apparent Kd values were determined, indicating that P-gp preferentially binds anionic (versus zwitterionic) phospholipids and short-chain (versus long-chain) cardiolipins. The interaction between P-gp and cyclosporin A (CsA), a P-gp inhibitor, was also assessed and was shown to increase the subsequent interaction of P-gp with cardiolipin. IM-MS indicated that open and closed P-gp conformations (revealed by crystallography) were readily interconverted by ligand binding.
The above MS studies employed detergents such as β-octylglucoside and dodecylmaltoside to maintain membrane protein complexes in their native oligomeric state in the gas phase. Efforts to develop detergent-free alternatives revealed that the native state of proteins and macromolecular complexes can also be preserved when certain lipid bicelles, amphipols, or nanodiscs are used.110,111 Nanodiscs were also exploited in a native MS study of the interaction between glycosphingolipids (GSL) and the B subunit homopentamers of cholera toxin and heat labile toxin.112 By screening different nanodisc-incorporated glycosphingolipids against the proteins, specific protein−ligand interactions could be identified and their relative affinities ranked.
Analysis of soluble protein complexes by native top-down MS
The introduction of ECD as a technique for fragmenting biomolecules enabled the study of intact proteins by so-called “top-down MS”.113 In top-down MS, samples are not proteolytically digested as in “bottom-up” proteomics,114 but are directly analyzed as intact proteins and fragmented inside FT-based mass spectrometers.115 Top-down MS has allowed for the characterization of different proteoforms,116 protein conformations, PTMs and protein–ligand binding site locations.117–120
The mass and complexity of samples that can be analyzed by top-down MS are steadily increasing. In early studies, small noncovalent complexes76 and a protein−ligand complex80 were analyzed by “native top-down MS” using ECD fragmentation (as detailed above). The approach has more recently been extended to large noncovalent assemblies.26,121 Notably, in 2010 Gross et al. analyzed the tetrameric yeast alcohol dehydrogenase (ADH, 147 kDa) using an ECD-enabled ESI-FT-ICR instrument.121 They reported the fragmentation of ADH subunits (breaking covalent bonds within monomers) without disrupting noncovalent interactions, thus demonstrating for the first time the ability to obtain information on both the primary and quaternary structures in the same analysis. The same group later extended their analyses to include concanavalin A (103 kDa) and the Fenna-Matthews-Olson antenna protein complex (140 kDa).122 They used different types of dissociative approaches to extract primary sequence information from highly flexible terminal regions and less flexible internal regions. The unfolding of ADH subunits was monitored by combining “in-source dissociation” (ISCID), CID and ECD, revealing that gas-phase protein unfolding initiated at the N-terminus and then moved towards the complex core.122 Interestingly, the extent of fragmentation observed by MS correlated well with the atomic displacement factor determined by crystallography (the B-factor, which reflects the relative degree of motion of protein atoms), suggesting that the ECD fragmentation pattern may be a useful indicator of protein flexibility. Moreover, the ADH from horse liver and from yeast were further studied by FT-ICR MS, infrared multiphoton dissociation (IRMPD), ISCID, CID and ECD to characterize the binding sites of zinc atoms and NAD/NADH ligand and to quantify different proteoforms.123 These examples highlight the ability of FT-ICR MS to provide information not only on inter-subunit interactions within protein assemblies, but also on subunit primary sequence and structural flexibility in the gas phase.
ECD and ETD are mainly used in FT instruments, but can also be incorporated in ion traps70 and Q-TOF spectrometers.71,124 Recently, ETD was utilized to sequence fragments of a noncovalent protein complex (ADH) using a Q-TOF instrument.125
In 2012, Heck et al. first demonstrated the use of an Orbitrap mass spectrometer (Exactive Plus™, Thermo Fisher Scientific) for the analysis of intact protein complexes ranging from 150 kDa to 800 kDa.126–129 They subsequently extended this range to include virus-like nanoparticles with a mass up to 4.5 MDa130 (Fig. 6). To carry out these analyses, an Orbitrap instrument was modified to extend its m/z range to 40000 and to transmit “heavy ions”.88,126 This modified mass spectrometer reached a very high sensitivity, enabling the analysis of IgG at 1 nM concentration, and a remarkable resolution at high m/z, allowing for the characterization of different glycosylation forms of IgG.127 The high resolution of this instrument also made it possible to determine the type (ADP or ATP) and number of nucleotides bound to such a large assembly as GroEL (800 kDa).
Figure 6.
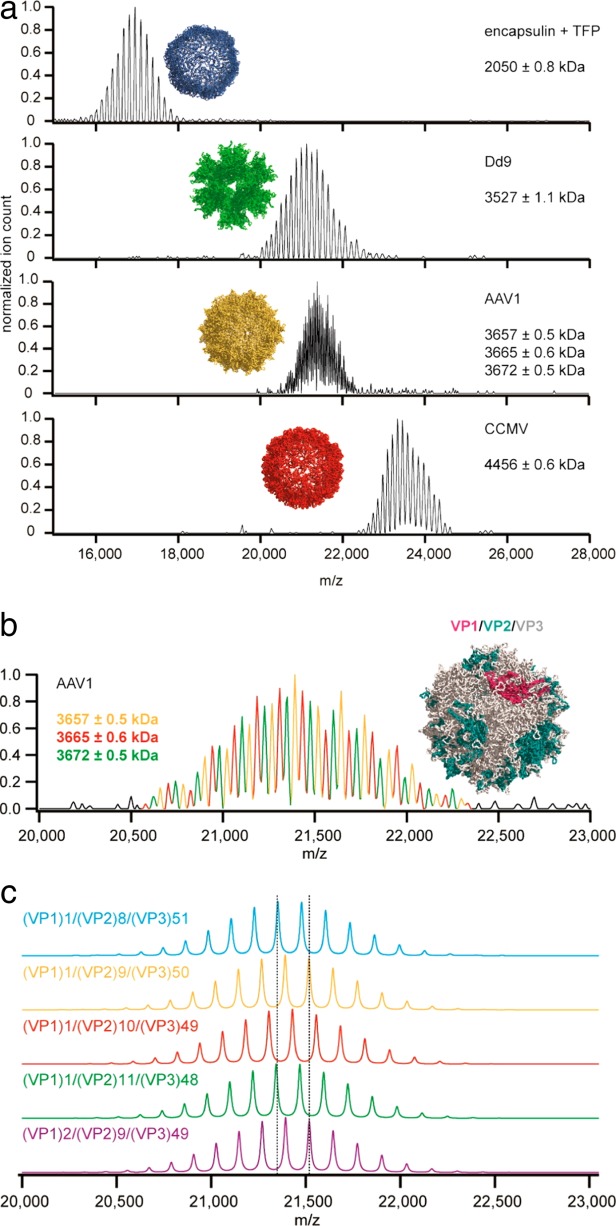
Native mass spectra of bacterial and viral nanoparticles. A: The native spectra of encapsulin, adenovirus dodecahedron (Dd9), adeno-associated virus serotype 1 (AAV1) and cowpea chlorotic mottle virus (CCMV) were acquired using a modified Orbitrap instrument. Even though the macromolecular complexes were larger than 2 MDa, the exceptional mass resolving power of the mass spectrometer allowed the determination of the particle masses with high accuracy. B: Native spectrum of the AAV1 capsid. The remarkable resolution of the spectra allowed Heck and colleagues to determine the stoichiometry of three different proteoforms (VP1, VP2, VP3) of the AAV1 capsid viral protein. C: Simulated spectra of AAV1 capsids showing different VP1:VP2:VP3 ratios. These spectra were reproduced with permission from Ref.130 © (2014) American Chemical Society.
Kelleher's group further modified an Exactive Orbitrap mass spectrometer to perform MS3 experiments on intact protein complexes.131 This means that protein assemblies were dissociated (in MS2 experiments) and then the ejected monomers were further fragmented in the HCD cell (in MS3 experiments). Finally, the fragment ions were transferred into the C-trap and Orbitrap to record spectra at high resolution and accuracy. The MS2 step allowed the stoichiometry of the protein complex to be confirmed while the MS3 step allowed the primary sequence of the ejected subunits to be characterized.
Concluding Remarks and Outlook
Twenty years ago there was great excitement about the emerging role of MS in biology, especially in the field of proteomics.132–134 At that time, MALDI- and ESI-MS had already exhibited excellent sensitivity, accuracy of mass measurement and speed of data acquisition.135,136 Without doubt, MS has more than met the expectations and predictions of two decades ago. Currently, MS-based proteomics allows one to quantify the intracellular changes in protein abundance in a few hours137 as well as to profile PTMs and protein interactions on a large scale.138 Not surprisingly, MS-based proteomics was recently ranked among the 10 methods with the highest impact on biological investigations.139
Although the first MS studies of non-covalent interactions also date from over two decades ago,28–31 native MS has had less of an impact than MS-based proteomics. Nevertheless, it is clearly gaining momentum in opening new scenarios in structural biology. Native MS has already proved itself to be a valuable technique in hybrid structural studies,140 especially because of its sensitivity, wide applicability, speed of analysis and selectivity (i.e., several species with different masses can be simultaneously analyzed and separated). This latter feature (mass selectivity) was recently exploited to separate the apo and holo forms of ferritin, a 24-meric complex, for subsequent visualization by negative-stain EM.141 The recorded images confirmed the preservation of the soft-landed complexes, showing specific features such as the large central cavity and iron core characteristic of apoferritin and holoferritin, respectively. This study epitomizes the use of MS-based separation to selectively purify heterogeneous protein complexes prior to structural investigation.
A key advantage of native MS is its ability to determine the composition and relative abundance of different macromolecular complexes in real time. Consequently, native MS is destined to play an increasingly important role in deciphering the functional regulation of PTMs, cofactors, and transient complexes assembled in a cellular milieu. In years to come, the routine analysis of intact macromolecular assemblies may be possible directly from cells without the need for recombinant over-expression. Indeed, the literature already contains a few examples of endogenous particles studied by native MS.47,142,143 For instance, the comparison between an endogenous human and recombinant form of the small nuclear ribonucleoprotein U1, a component of the spliceosome, revealed the effect of specific PTMs on the stability of U1 sub-complexes.143 However, the broader application of native MS to endogenous complexes will require technical advances that increase the yield of such complexes purified from cells at their natural expression level.144,145 We will also need instrumentation with increased sensitivity, resolution, and ionization efficiency. Orbitrap instruments already represent an improvement for their sensitivity and resolution.126,127,129 Novel ionization approaches could increase the sample fraction introduced in mass spectrometers.146–151 Moreover, new detectors will have a positive impact on the applicability of native MS. For instance, nanoelectromechanical systems (NEMS) can detect very high masses with unprecedented sensitivity,152 requiring only a few hundred single-molecule adsorption events to detect megadalton molecules. Another example is charge detection mass spectrometry (CDMS), whereby the m/z and z are simultaneously measured for each ion.153 CDMS combined with cryo-EM was recently utilized to characterize the oligomeric states of large intermediates (>3.5 MDa) in the assembly of the hepatitis B virus capsid.154
Another area where there is room for improvement is in data analysis. Currently, a few MS software packages are available for the characterization of macromolecular complexes (e.g., Refs.155–156), but manual evaluation remains unavoidable. To carry out high-throughput studies of macromolecular complexes, software as effective as that available for bottom-up proteomics will need to be developed.
In conclusion, given the growing trend towards the use of multiple, complementary techniques for elucidating the structure of macromolecular complexes, native MS will clearly play an increasingly important role in addressing biological problems of ever greater complexity.
References
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, Babu M, Craig SA, Hu P, Wan C, Vlasblom J, Dar VU, Bezginov A, Clark GW, Wu GC, Wodak SJ, Tillier ER, Paccanaro A, Marcotte EM, Emili A. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nature Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- Heck AJ. Native mass spectrometry: a bridge between interactomics and structural biology. Nature Meth. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- Benesch JL, Ruotolo BT. Mass spectrometry: come of age for structural and dynamical biology. Curr Opin Struct Biol. 2011;21:641–649. doi: 10.1016/j.sbi.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeri Erba E, Barylyuk K, Yang Y, Zenobi R. Quantifying protein-protein interactions within noncovalent complexes using electrospray ionization mass spectrometry. Analyt Chem. 2011;83:9251–9259. doi: 10.1021/ac201576e. [DOI] [PubMed] [Google Scholar]
- Morgner N, Robinson CV. Linking structural change with functional regulation-insights from mass spectrometry. Curr Opin Struct Biol. 2012;22:44–51. doi: 10.1016/j.sbi.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Laganowsky A, Reading E, Hopper JT, Robinson CV. Mass spectrometry of intact membrane protein complexes. Nature Protoc. 2013;8:639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon M. Biochemistry. Structural MS pulls its weight. Science. 2013;340:1059–1060. doi: 10.1126/science.1236303. [DOI] [PubMed] [Google Scholar]
- Sharon M. How far can we go with structural mass spectrometry of protein complexes? J Am Soc Mass Spectrom. 2010;21:487–500. doi: 10.1016/j.jasms.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Konijnenberg A, Butterer A, Sobott F. Native ion mobility-mass spectrometry and related methods in structural biology. Biochim Biophys Acta. 2013;1834:1239–1256. doi: 10.1016/j.bbapap.2012.11.013. [DOI] [PubMed] [Google Scholar]
- Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJ. Ion mobility mass spectrometry of proteins and protein assemblies. Chem Soc Rev. 2010;39:1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- Marsh JA, Hernandez H, Hall Z, Ahnert SE, Perica T, Robinson CV, Teichmann SA. Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell. 2013;153:461–470. doi: 10.1016/j.cell.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bich C, Baer S, Jecklin MC, Zenobi R. Probing the hydrophobic effect of noncovalent complexes by mass spectrometry. J Am Soc Mass Spectrom. 2010;21:286–289. doi: 10.1016/j.jasms.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Meyer T, de la Cruz X, Orozco M. An atomistic view to the gas phase proteome. Structure. 2009;17:88–95. doi: 10.1016/j.str.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Takats Z, Blake TA, Gologan B, Guymon AJ, Wiseman JM, Oliver JC, Davisson VJ, Cooks RG. Preparing protein microarrays by soft-landing of mass-selected ions. Science. 2003;301:1351–1354. doi: 10.1126/science.1088776. [DOI] [PubMed] [Google Scholar]
- Madler S, Boeri Erba E, Zenobi R. MALDI-ToF mass spectrometry for studying noncovalent complexes of biomolecules. Topics Curr Chem. 2013;331:1–36. doi: 10.1007/128_2011_311. [DOI] [PubMed] [Google Scholar]
- Wilm M. Principles of electrospray ionization. Mol Cell Proteom. 2011;10:M111 009407. doi: 10.1074/mcp.M111.009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebarle P, Verkerk UH. Electrospray: from ions in solution to ions in the gas phase, what we know now. Mass Spectrom Rev. 2009;28:898–917. doi: 10.1002/mas.20247. [DOI] [PubMed] [Google Scholar]
- Fernandez de la Mora J. Electrospray ionization of large multiply charged species proceeds via Dole's charged residue mechanism. Analyt Chim Acta. 2000;406:93–104. [Google Scholar]
- Kebarle P. Brief overview of the present status of the mechanisms involved in electrospray mass spectrometry. J Mass Spectrom. 2000;35:804–817. doi: 10.1002/1096-9888(200007)35:7<804::AID-JMS22>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Ogorzalek Loo RR, Lakshmanan R, Loo JA. What protein charging (and supercharging) reveal about the mechanism of electrospray ionization. J Am Soc Mass Spectrom. 2014;25:1675–1693. doi: 10.1007/s13361-014-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Vahidi S, Konermann L. Insights into the mechanism of protein electrospray ionization from salt adduction measurements. J Am Soc Mass Spectrom. 2014;25:1322–1331. doi: 10.1007/s13361-014-0905-0. [DOI] [PubMed] [Google Scholar]
- Wilm M, Mann M. Electrospray and Taylor-Cone theory, Dole's beam of macromolecules at last. Int J Mass Spectrom Ion Proc. 1994;136:167–180. [Google Scholar]
- Wilm M, Mann M. Analytical properties of the nanoelectrospray ion source. Analyt Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cui W, Gross ML, Blankenship RE. Native mass spectrometry of photosynthetic pigment-protein complexes. FEBS Lett. 2013;587:1012–1020. doi: 10.1016/j.febslet.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyachenko A, Gruber R, Shimon L, Horovitz A, Sharon M. Allosteric mechanisms can be distinguished using structural mass spectrometry. Proc Natl Acad Sci USA. 2013;110:7235–7239. doi: 10.1073/pnas.1302395110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, Light-Wahl KJ. The observation of non-covalent interactions in solution by electrospray ionization mass spectrometry: promise, pitfalls and prognosis. Biol Mass Spectrom. 1993;22:493–501. [Google Scholar]
- Katta V, Chait BT. Observation of the heme-globin complex in native myoglobin by electrospray-ionization mass spectrometry. J Am Chem Soc. 1991;113:8535–8537. [Google Scholar]
- Ganem B, Li YT, Henion JD. Detection of noncovalent receptor ligand complexes by mass spectrometry. J Am Chem Soc. 1991;113:6294–6296. [Google Scholar]
- Ganem B, Li YT, Henion JD. Observation of noncovalent enzyme-substrate and enzyme-product complexes by ion-spray mass spectrometry. J Am Chem Soc. 1991;113:7818–7819. [Google Scholar]
- Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Analyt Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- van den Heuvel RH, van Duijn E, Mazon H, Synowsky SA, Lorenzen K, Versluis C, Brouns SJ, Langridge D, van der Oost J, Hoyes J, Heck AJ. Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Analyt Chem. 2006;78:7473–7483. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- Snijder J, Rose RJ, Veesler D, Johnson JE, Heck AJ. Studying 18 MDa virus assemblies with native mass spectrometry. Angew Chem Int Ed Engl. 2013;52:4020–4023. doi: 10.1002/anie.201210197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilag LL, Ubarretxena-Belandia I, Tate CG, Robinson CV. Drug binding revealed by tandem mass spectrometry of a protein-micelle complex. J Am Chem Soc. 2004;126:14362–14363. doi: 10.1021/ja0450307. [DOI] [PubMed] [Google Scholar]
- Demmers JA, van Dalen A, de Kruijff B, Heck AJ, Killian JA. Interaction of the K+ channel KcsA with membrane phospholipids as studied by ESI mass spectrometry. FEBS Lett. 2003;541:28–32. doi: 10.1016/s0014-5793(03)00282-5. [DOI] [PubMed] [Google Scholar]
- Lengqvist J, Svensson R, Evergren E, Morgenstern R, Griffiths WJ. Observation of an intact noncovalent homotrimer of detergent-solubilized rat microsomal glutathione transferase-1 by electrospray mass spectrometry. J Biol Chem. 2004;279:13311–13316. doi: 10.1074/jbc.M310958200. [DOI] [PubMed] [Google Scholar]
- Esteban O, Bernal RA, Donohoe M, Videler H, Sharon M, Robinson CV, Stock D. Stoichiometry and localization of the stator subunits E and G in Thermus thermophilus H+-ATPase/synthase. J Biol Chem. 2008;283:2595–2603. doi: 10.1074/jbc.M704941200. [DOI] [PubMed] [Google Scholar]
- Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321:243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- Barrera NP, Isaacson SC, Zhou M, Bavro VN, Welch A, Schaedler TA, Seeger MA, Miguel RN, Korkhov VM, van Veen HW, Venter H, Walmsley AR, Tate CG, Robinson CV. Mass spectrometry of membrane transporters reveals subunit stoichiometry and interactions. Nature Meth. 2009;6:585–587. doi: 10.1038/nmeth.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Morgner N, Barrera NP, Politis A, Isaacson SC, Matak-Vinkovic D, Murata T, Bernal RA, Stock D, Robinson CV. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 2011;334:380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesch JL, Aquilina JA, Ruotolo BT, Sobott F, Robinson CV. Tandem mass spectrometry reveals the quaternary organization of macromolecular assemblies. Chem Biol. 2006;13:597–605. doi: 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Boeri Erba E, Ruotolo BT, Barsky D, Robinson CV. Ion mobility-mass spectrometry reveals the influence of subunit packing and charge on the dissociation of multiprotein complexes. Analyt Chem. 2010;82:9702–9710. doi: 10.1021/ac101778e. [DOI] [PubMed] [Google Scholar]
- Pagel K, Hyung SJ, Ruotolo BT, Robinson CV. Alternate dissociation pathways identified in charge-reduced protein complex ions. Analyt Chem. 2010;82:5363–5372. doi: 10.1021/ac101121r. [DOI] [PubMed] [Google Scholar]
- Lorenzen K, van Duijn E. Native mass spectrometry as a tool in structural biology. Curr Protoc Prot Sci Chapter. 2010;17 doi: 10.1002/0471140864.ps1712s62. :Unit17 12. [DOI] [PubMed] [Google Scholar]
- Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, Doudna JA, Robinson CV. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci USA. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez H, Dziembowski A, Taverner T, Seraphin B, Robinson CV. Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep. 2006;7:605–610. doi: 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivat Hannah V, Atmanene C, Zeyer D, Van Dorsselaer A, Sanglier-Cianferani S. Native MS: an 'ESI' way to support structure- and fragment-based drug discovery. Future Med Chem. 2010;2:35–50. doi: 10.4155/fmc.09.141. [DOI] [PubMed] [Google Scholar]
- Niu S, Rabuck JN, Ruotolo BT. Ion mobility-mass spectrometry of intact protein—ligand complexes for pharmaceutical drug discovery and development. Curr Opin Chem Biol. 2013;17:809–817. doi: 10.1016/j.cbpa.2013.06.019. [DOI] [PubMed] [Google Scholar]
- Schmidt C, Robinson CV. Dynamic protein ligand interactions—insights from MS. FEBS J. 2014;281:1950–1964. doi: 10.1111/febs.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laventie BJ, Potrich C, Atmanene C, Saleh M, Joubert O, Viero G, Bachmeyer C, Antonini V, Mancini I, Cianferani-Sanglier S, Keller D, Colin DA, Bourcier T, Anderluh G, van Dorsselaer A, Dalla Serra M, Prevost G. p-Sulfonato-calix[n]arenes inhibit staphylococcal bicomponent leukotoxins by supramolecular interactions. Biochem J. 2013;450:559–571. doi: 10.1042/BJ20121628. [DOI] [PubMed] [Google Scholar]
- Cousido-Siah A, Ayoub D, Berberian G, Bollo M, Van Dorsselaer A, Debaene F, DiPolo R, Petrova T, Schulze-Briese C, Olieric V, Esteves A, Mitschler A, Sanglier-Cianferani S, Beauge L, Podjarny A. Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger. Acta Cryst. 2012;D68:1098–1107. doi: 10.1107/S090744491202094X. [DOI] [PubMed] [Google Scholar]
- Marcotte D, Zeng W, Hus JC, McKenzie A, Hession C, Jin P, Bergeron C, Lugovskoy A, Enyedy I, Cuervo H, Wang D, Atmanene C, Roecklin D, Vecchi M, Vivat V, Kraemer J, Winkler D, Hong V, Chao J, Lukashev M, Silvian L. Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism. Bioorgan Med Chem. 2013;21:4011–4019. doi: 10.1016/j.bmc.2013.04.019. [DOI] [PubMed] [Google Scholar]
- Cousido-Siah A, Petrova T, Hazemann I, Mitschler A, Ruiz FX, Howard E, Ginell S, Atmanene C, Van Dorsselaer A, Sanglier-Cianferani S, Joachimiak A, Podjarny A. Crystal packing modifies ligand binding affinity: the case of aldose reductase. Proteins. 2012;80:2552–2561. doi: 10.1002/prot.24136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritschel T, Atmanene C, Reuter K, Van Dorsselaer A, Sanglier-Cianferani S, Klebe G. An integrative approach combining noncovalent mass spectrometry, enzyme kinetics and X-ray crystallography to decipher Tgt protein-protein and protein-RNA interaction. J Mol Biol. 2009;393:833–847. doi: 10.1016/j.jmb.2009.07.040. [DOI] [PubMed] [Google Scholar]
- Lebrette H, Borezee-Durant E, Martin L, Richaud P, Boeri Erba E, Cavazza C. Novel insights into nickel import in Staphylococcus aureus: the positive role of free histidine and structural characterization of a new thiazolidine-type nickel chelator. Metallomics. 2015 doi: 10.1039/c4mt00295d. in press. [DOI] [PubMed] [Google Scholar]
- Chevreux G, Atmanene C, Lopez P, Ouazzani J, Van Dorsselaer A, Badet B, Badet-Denisot MA, Sanglier-Cianferani S. Monitoring the dynamics of monomer exchange using electrospray mass spectrometry: the case of the dimeric glucosamine-6-phosphate synthase. J Am Soc Mass Spectrom. 2011;22:431–439. doi: 10.1007/s13361-010-0054-z. [DOI] [PubMed] [Google Scholar]
- Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom Rev. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Makarov A. Electrostatic axially harmonic orbital trapping: a high-performance technique of mass analysis. Analyt Chem. 2000;72:1156–1162. doi: 10.1021/ac991131p. [DOI] [PubMed] [Google Scholar]
- Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Analyt Chem. 2006;78:2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- Makarov A, Denisov E, Lange O, Horning S. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J Am Soc Mass Spectrom. 2006;17:977–982. doi: 10.1016/j.jasms.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Mohr J, Swart R, Samonig M, Bohm G, Huber CG. High-efficiency nano- and micro-HPLC–high-resolution Orbitrap-MS platform for top-down proteomics. Proteomics. 2010;10:3598–3609. doi: 10.1002/pmic.201000341. [DOI] [PubMed] [Google Scholar]
- Kitova EN, Kitov PI, Bundle DR, Klassen JS. The observation of multivalent complexes of Shiga-like toxin with globotriaoside and the determination of their stoichiometry by nanoelectrospray Fourier-transform ion cyclotron resonance mass spectrometry. Glycobiology. 2001;11:605–611. doi: 10.1093/glycob/11.7.605. [DOI] [PubMed] [Google Scholar]
- Cheng X, Chen R, Bruce JE, Schwartz BL, Anderson GA, Hofstadler SA, Gale DC, Smith RD. Using electrospray ionization FTICR mass spectrometry to study competitive binding of inhibitors to carbonic anhydrase. J Am Chem Soc. 1995;117:8859–8860. [Google Scholar]
- Scigelova M, Hornshaw M, Giannakopulos A, Makarov A. Fourier transform mass spectrometry. Mol Cell Proteom. 2011;10:M111.009431. doi: 10.1074/mcp.M111.009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubarev RA, Makarov A. Orbitrap mass spectrometry. Anal Chem. 2013;85:5288–5296. doi: 10.1021/ac4001223. [DOI] [PubMed] [Google Scholar]
- Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Graham Cooks R. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- Marshall AG, Hendrickson CL. High-resolution mass spectrometers. Annu Rev Anal Chem. 2008;1:579–599. doi: 10.1146/annurev.anchem.1.031207.112945. [DOI] [PubMed] [Google Scholar]
- Zubarev RA. Electron-capture dissociation tandem mass spectrometry. Curr Opin Biotechnol. 2004;15:12–16. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Ding L, Brancia FL. Electron capture dissociation in a digital ion trap mass spectrometer. Analyt Chem. 2006;78:1995–2000. doi: 10.1021/ac0519007. [DOI] [PubMed] [Google Scholar]
- Voinov VG, Deinzer ML, Beckman JS, Barofsky DF. Electron capture, collision-induced, and electron capture-collision induced dissociation in Q-TOF. J Am Soc Mass Spectrom. 2011;22:607–611. doi: 10.1007/s13361-010-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Nat Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- Kjeldsen F, Giessing AM, Ingrell CR, Jensen ON. Peptide sequencing and characterization of post-translational modifications by enhanced ion-charging and liquid chromatography electron-transfer dissociation tandem mass spectrometry. Analyt Chem. 2007;79:9243–9252. doi: 10.1021/ac701700g. [DOI] [PubMed] [Google Scholar]
- Haselmann KF, Jorgensen TJ, Budnik BA, Jensen F, Zubarev RA. Electron capture dissociation of weakly bound polypeptide polycationic complexes. Rapid Commun Mass Spectrom. 2002;16:2260–2265. doi: 10.1002/rcm.853. [DOI] [PubMed] [Google Scholar]
- Liu H, Hakansson K. Divalent metal ion-peptide interactions probed by electron capture dissociation of trications. J Am Soc Mass Spectrom. 2006;17:1731–1741. doi: 10.1016/j.jasms.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Erales J, Gontero B, Whitelegge J, Halgand F. Mapping of a copper-binding site on the small CP12 chloroplastic protein of Chlamydomonas reinhardtii using top-down mass spectrometry and site-directed mutagenesis. Biochem J. 2009;419:75–82. doi: 10.1042/BJ20082004. [DOI] [PubMed] [Google Scholar]
- Hartinger CG, Tsybin YO, Fuchser J, Dyson PJ. Characterization of platinum anticancer drug protein-binding sites using a top-down mass spectrometric approach. Inorgan Chem. 2008;47:17–19. doi: 10.1021/ic702236m. [DOI] [PubMed] [Google Scholar]
- Xie Y, Zhang J, Yin S, Loo JA. Top-down ESI-ECD-FT-ICR mass spectrometry localizes noncovalent protein-ligand binding sites. J Am Chem Soc. 2006;128:14432–14433. doi: 10.1021/ja063197p. [DOI] [PubMed] [Google Scholar]
- Yin S, Loo JA. Elucidating the site of protein-ATP binding by top-down mass spectrometry. J Am Soc Mass Spectrom. 2010;21:899–907. doi: 10.1016/j.jasms.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Yin S, Loo JA. Top-down mass spectrometry of supercharged native protein-ligand complexes. Intl J Mass Spectrom. 2011;300:118–122. doi: 10.1016/j.ijms.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty MT, Zhang H, Cui W, Blankenship RE, Gross ML, Sligar SG. Native mass spectrometry characterization of intact nanodisc lipoprotein complexes. Analyt Chem. 2012;84:8957–8960. doi: 10.1021/ac302663f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills RH, Habtemariam A, Lopez-Clavijo AF, Barrow MP, Sadler PJ, O'Connor PB. Insights into the binding sites of organometallic ruthenium anticancer compounds on peptides using ultra-high resolution mass spectrometry. J Am Soc Mass Spectrom. 2014;25:662–672. doi: 10.1007/s13361-013-0819-2. [DOI] [PubMed] [Google Scholar]
- Li H, Snelling JR, Barrow MP, Scrivens JH, Sadler PJ, O'Connor PB. Mass spectrometric strategies to improve the identification of Pt(II)-modification sites on peptides and proteins. J Am Soc Mass Spectrom. 2014;25:1217–1227. doi: 10.1007/s13361-014-0877-0. [DOI] [PubMed] [Google Scholar]
- Meier SM, Tsybin YO, Dyson PJ, Keppler BK, Hartinger CG. Fragmentation methods on the balance: unambiguous top-down mass spectrometric characterization of oxaliplatin-ubiquitin binding sites. Analyt Bioanalyt Chem. 2012;402:2655–2662. doi: 10.1007/s00216-011-5523-0. [DOI] [PubMed] [Google Scholar]
- Jackson SN, Dutta S, Woods AS. The use of ECD/ETD to identify the site of electrostatic interaction in noncovalent complexes. J Am Soc Mass Spectrom. 2009;20:176–179. doi: 10.1016/j.jasms.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder J, Heck AJ. Analytical approaches for size and mass analysis of large protein assemblies. Annu Rev Anal Chem. 2014;7:43–64. doi: 10.1146/annurev-anchem-071213-020015. [DOI] [PubMed] [Google Scholar]
- Gulbakan B, Barylyuk K, Zenobi R. Determination of thermodynamic and kinetic properties of biomolecules by mass spectrometry. Curr Opin Biotechnol. 2014;31C:65–72. doi: 10.1016/j.copbio.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ascano M, Diaz-Avalos R, Park AY, Juranek SA, Rice WJ, Yin Q, Robinson CV, Tuschl T, Patel DJ. Multimeric assembly and biochemical characterization of the Trax-translin endonuclease complex. Nature Struct Mol Biol. 2011;18:658–664. doi: 10.1038/nsmb.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis A, Stengel F, Hall Z, Hernandez H, Leitner A, Walzthoeni T, Robinson CV, Aebersold R. A mass spectrometry-based hybrid method for structural modeling of protein complexes. Nature Meth. 2014;11:403–406. doi: 10.1038/nmeth.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis A, Schmidt C, Tjioe E, Sandercock AM, Lasker K, Gordiyenko Y, Russel D, Sali A, Robinson CV. Topological models of heteromeric protein assemblies from mass spectrometry: application to the yeast eIF3:eIF5 complex. Chem Biol. 2015;22:117–128. doi: 10.1016/j.chembiol.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson NJ, Merdanovic M, Ehrmann M, van Duijn E, Heck AJ. Substrate occupancy at the onset of oligomeric transitions of DegP. Structure. 2014;22:281–290. doi: 10.1016/j.str.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature. 2002;416:455–459. doi: 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- Ebong IO, Morgner N, Zhou M, Saraiva MA, Daturpalli S, Jackson SE, Robinson CV. Heterogeneity and dynamics in the assembly of the heat shock protein 90 chaperone complexes. Proc Natl Acad Sci USA. 2011;108:17939–17944. doi: 10.1073/pnas.1106261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetrecht C, Heck AJ. Modern biomolecular mass spectrometry and its role in studying virus structure, dynamics, and assembly. Angew Chem Int Ed Engl. 2011;50:8248–8262. doi: 10.1002/anie.201008120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereszczak JZ, Barbu IM, Tan M, Xia M, Jiang X, van Duijn E, Heck AJ. Structure, stability and dynamics of norovirus P domain derived protein complexes studied by native mass spectrometry. J Struct Biol. 2012;177:273–282. doi: 10.1016/j.jsb.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Veesler D, Khayat R, Krishnamurthy S, Snijder J, Huang RK, Heck AJ, Anand GS, Johnson JE. Architecture of a dsDNA viral capsid in complex with its maturation protease. Structure. 2014;22:230–237. doi: 10.1016/j.str.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, Beijer MR, Barendregt A, Zhou K, Snijders AP, Dickman MJ, Doudna JA, Boekema EJ, Heck AJ, van der Oost J, Brouns SJ. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nature Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, Doudna JA. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, Sakamoto K, Suzuki T, Dohmae N, Yokoyama S, Schaap PJ, Urlaub H, Heck AJ, Nogales E, Doudna JA, Shinkai A, van der Oost J. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaix D, Ferguson ML, Atmanene C, Van Dorsselaer A, Sanglier-Cianferani S, Royer CA, Declerck N. Physical basis of the inducer-dependent cooperativity of the Central glycolytic genes Repressor/DNA complex. Nucleic Acids Res. 2010;38:5944–5957. doi: 10.1093/nar/gkq334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezacova P, Kozisek M, Moy SF, Sieglova I, Joachimiak A, Machius M, Otwinowski Z. Crystal structures of the effector-binding domain of repressor Central glycolytic gene Regulator from Bacillus subtilis reveal ligand-induced structural changes upon binding of several glycolytic intermediates. Mol Microbiol. 2008;69:895–910. doi: 10.1111/j.1365-2958.2008.06318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmanene C, Chaix D, Bessin Y, Declerck N, Van Dorsselaer A, Sanglier-Cianferani S. Combination of noncovalent mass spectrometry and traveling wave ion mobility spectrometry reveals sugar-induced conformational changes of central glycolytic genes repressor/DNA complex. Analyt Chem. 2010;82:3597–3605. doi: 10.1021/ac902784n. [DOI] [PubMed] [Google Scholar]
- Snijder J, Uetrecht C, Rose RJ, Sanchez-Eugenia R, Marti GA, Agirre J, Guerin DM, Wuite GJ, Heck AJ, Roos WH. Probing the biophysical interplay between a viral genome and its capsid. Nature Chem. 2013;5:502–509. doi: 10.1038/nchem.1627. [DOI] [PubMed] [Google Scholar]
- Shoemaker GK, van Duijn E, Crawford SE, Uetrecht C, Baclayon M, Roos WH, Wuite GJ, Estes MK, Prasad BV, Heck AJ. Norwalk virus assembly and stability monitored by mass spectrometry. Mol Cell Proteom. 2010;9:1742–1751. doi: 10.1074/mcp.M900620-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Politis A, Davies RB, Liko I, Wu KJ, Stewart AG, Stock D, Robinson CV. Ion mobility-mass spectrometry of a rotary ATPase reveals ATP-induced reduction in conformational flexibility. Nature Chem. 2014;6:208–215. doi: 10.1038/nchem.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden NG, Hopper JT, Lukoyanova N, Rodriguez-Larrea D, Wojdyla JA, Klein A, Kaminska R, Bayley H, Saibil HR, Robinson CV, Kleanthous C. Intrinsically disordered protein threads through the bacterial outer-membrane porin OmpF. Science. 2013;340:1570–1574. doi: 10.1126/science.1237864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoux J, Wang SC, Politis A, Reading E, Ma J, Biggin PC, Zhou M, Tao H, Zhang Q, Chang G, Morgner N, Robinson CV. Mass spectrometry reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump. Proc Natl Acad Sci USA. 2013;110:9704–9709. doi: 10.1073/pnas.1303888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper JT, Yu YT, Li D, Raymond A, Bostock M, Liko I, Mikhailov V, Laganowsky A, Benesch JL, Caffrey M, Nietlispach D, Robinson CV. Detergent-free mass spectrometry of membrane protein complexes. Nature Meth. 2013;10:1206–1208. doi: 10.1038/nmeth.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese AN, Watkinson TG, Henderson PJ, Radford SE, Ashcroft AE. Amphipols outperform dodecylmaltoside micelles in stabilizing membrane protein structure in the gas phase. Anal Chem. 2015;87:1118–1126. doi: 10.1021/ac5037022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leney AC, Fan X, Kitova EN, Klassen JS. Nanodiscs and electrospray ionization mass spectrometry: a tool for screening glycolipids against proteins. Anal Chem. 2014;86:5271–5277. doi: 10.1021/ac4041179. [DOI] [PubMed] [Google Scholar]
- Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top down versus bottom up protein characterization by tandem high-resolution mass spectrometry. J Am Chem Soc. 1999:806–812. [Google Scholar]
- Chait BT. Chemistry. Mass spectrometry: bottom-up or top-down? Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- Kelleher NL. Top-down proteomics. Analyt Chem. 2004;76:197A–203A. [PubMed] [Google Scholar]
- Smith LM, Kelleher NL. Proteoform: a single term describing protein complexity. Nature Meth. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armirotti A, Damonte G. Achievements and perspectives of top-down proteomics. Proteomics. 2010;10:3566–3576. doi: 10.1002/pmic.201000245. [DOI] [PubMed] [Google Scholar]
- Cui W, Rohrs HW, Gross ML. Top-down mass spectrometry: recent developments, applications and perspectives. Analyst. 2011;136:3854–3864. doi: 10.1039/c1an15286f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia BA. What does the future hold for top down mass spectrometry? J Am Soc Mass Spectrom. 2010;21:193–202. doi: 10.1016/j.jasms.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nature Meth. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Native electrospray and electron-capture dissociation in FTICR mass spectrometry provide top-down sequencing of a protein component in an intact protein assembly. J Am Soc Mass Spectrom. 2010;21:1966–1968. doi: 10.1016/j.jasms.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Native electrospray and electron-capture dissociation FTICR mass spectrometry for top-down studies of protein assemblies. Analyt Chem. 2011;83:5598–5606. doi: 10.1021/ac200695d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wongkongkathep P, Van Orden SL, Ogorzalek Loo RR, Loo JA. Revealing ligand binding sites and quantifying subunit variants of noncovalent protein complexes in a single native top-down FTICR MS experiment. J Am Soc Mass Spectrom. 2014;25:2060–2068. doi: 10.1007/s13361-014-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsybin YO, Fornelli L, Stoermer C, Luebeck M, Parra J, Nallet S, Wurm FM, Hartmer R. Structural analysis of intact monoclonal antibodies by electron transfer dissociation mass spectrometry. Analyt Chem. 2011;83:8919–8927. doi: 10.1021/ac201293m. [DOI] [PubMed] [Google Scholar]
- Lermyte F, Konijnenberg A, Williams JP, Brown JM, Valkenborg D, Sobott F. ETD allows for native surface mapping of a 150 kDa noncovalent complex on a commercial Q-TWIMS-TOF instrument. J Am Soc Mass Spectrom. 2014;25:343–350. doi: 10.1007/s13361-013-0798-3. [DOI] [PubMed] [Google Scholar]
- Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJ. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nature Meth. 2012;9:1084–1086. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- Rosati S, Rose RJ, Thompson NJ, van Duijn E, Damoc E, Denisov E, Makarov A, Heck AJ. Exploring an orbitrap analyzer for the characterization of intact antibodies by native mass spectrometry. Angew Chem Int Ed Engl. 2012;51:12992–12996. doi: 10.1002/anie.201206745. [DOI] [PubMed] [Google Scholar]
- Compton PD, Kelleher NL. Spinning up mass spectrometry for whole protein complexes. Nature Meth. 2012;9:1065–1066. doi: 10.1038/nmeth.2216. [DOI] [PubMed] [Google Scholar]
- Whitelegge J. Intact protein mass spectrometry and top-down proteomics. Expert Rev Proteom. 2013;10:127–129. doi: 10.1586/epr.13.10. [DOI] [PubMed] [Google Scholar]
- Snijder J, van de Waterbeemd M, Damoc E, Denisov E, Grinfeld D, Bennett A, Agbandje-McKenna M, Makarov A, Heck AJ. Defining the stoichiometry and cargo load of viral and bacterial nanoparticles by Orbitrap mass spectrometry. J Am Chem Soc. 2014;136:7295–7299. doi: 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov ME, Damoc E, Denisov E, Compton PD, Horning S, Makarov AA, Kelleher NL. From protein complexes to subunit backbone fragments: a multi-stage approach to native mass spectrometry. Analyt Chem. 2013;85:11163–11173. doi: 10.1021/ac4029328. [DOI] [PubMed] [Google Scholar]
- Chait BT. Mass spectrometry—a useful tool for the protein X-ray crystallographer and NMR spectroscopist. Structure. 1994;2:465–467. doi: 10.1016/s0969-2126(00)00047-2. [DOI] [PubMed] [Google Scholar]
- Wang R, Chait BT. High-accuracy mass measurement as a tool for studying proteins. Curr Opin Biotechnol. 1994;5:77–84. doi: 10.1016/s0958-1669(05)80074-6. [DOI] [PubMed] [Google Scholar]
- Chait BT, Kent SB. Weighing naked proteins: practical, high-accuracy mass measurement of peptides and proteins. Science. 1992;257:1885–1894. doi: 10.1126/science.1411504. [DOI] [PubMed] [Google Scholar]
- Tsarbopoulos A, Karas M, Strupat K, Pramanik BN, Nagabhushan TL, Hillenkamp F. Comparative mapping of recombinant proteins and glycoproteins by plasma desorption and matrix-assisted laser desorption/ionization mass spectrometry. Analyt Chem. 1994;66:2062–2070. doi: 10.1021/ac00085a022. [DOI] [PubMed] [Google Scholar]
- Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- Mann M, Kulak NA, Nagaraj N, Cox J. The coming age of complete, accurate, and ubiquitous proteomes. Mol Cell. 2013;49:583–590. doi: 10.1016/j.molcel.2013.01.029. [DOI] [PubMed] [Google Scholar]
- Bruce JE. In vivo protein complex topologies: sights through a cross-linking lens. Proteomics. 2012;12:1565–1575. doi: 10.1002/pmic.201100516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten years of methods. Nature Meth. 2014;11:1000–1001. doi: 10.1038/nmeth1014-1000. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Musacchio A, Cusack S, Petosa C. Investigating a macromolecular complex: the toolkit of methods. J Struct Biol. 2011;175:106–112. doi: 10.1016/j.jsb.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Mikhailov VA, Mize TH, Benesch JL, Robinson CV. Mass-selective soft-landing of protein assemblies with controlled landing energies. Analyt Chem. 2014;86:8321–8328. doi: 10.1021/ac5018327. [DOI] [PubMed] [Google Scholar]
- Chorev DS, Moscovitz O, Geiger B, Sharon M. Regulation of focal adhesion formation by a vinculin-Arp2/3 hybrid complex. Nature Commun. 2014;5:3758. doi: 10.1038/ncomms4758. [DOI] [PubMed] [Google Scholar]
- Hernandez H, Makarova OV, Makarov EM, Morgner N, Muto Y, Krummel DP, Robinson CV. Isoforms of U1–70k control subunit dynamics in the human spliceosomal U1 snRNP. PloS One. 2009;4:e7202. doi: 10.1371/journal.pone.0007202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Maeda K, Kuhner S. Recent advances in charting protein-protein interaction: mass spectrometry-based approaches. Curr Opin Biotechnol. 2011;22:42–49. doi: 10.1016/j.copbio.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Domanski M, Molloy K, Jiang H, Chait BT, Rout MP, Jensen TH, LaCava J. Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels. BioTechniques. 2012;0:1–6. doi: 10.2144/000113864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimpin S, Inutan ED. New ionization method for analysis on atmospheric pressure ionization mass spectrometers requiring only vacuum and matrix assistance. Analyt Chem. 2013;85:2005–2009. doi: 10.1021/ac303717j. [DOI] [PubMed] [Google Scholar]
- Wang B, Tisdale E, Trimpin S, Wilkins CL. Matrix-assisted ionization vacuum for high-resolution Fourier transform ion cyclotron resonance mass spectrometers. Analyt Chem. 2014;86:6792–6796. doi: 10.1021/ac500511g. [DOI] [PubMed] [Google Scholar]
- Yoon SH, Huang Y, Edgar JS, Ting YS, Heron SR, Kao Y, Li Y, Masselon CD, Ernst RK, Goodlett DR. Surface acoustic wave nebulization facilitating lipid mass spectrometric analysis. Analyt Chem. 2012;84:6530–6537. doi: 10.1021/ac300807p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yoon SH, Heron SR, Masselon CD, Edgar JS, Turecek F, Goodlett DR. Surface acoustic wave nebulization produces ions with lower internal energy than electrospray ionization. J Am Soc Mass Spectrom. 2012;23:1062–1070. doi: 10.1007/s13361-012-0352-8. [DOI] [PubMed] [Google Scholar]
- Heron SR, Wilson R, Shaffer SA, Goodlett DR, Cooper JM. Surface acoustic wave nebulization of peptides as a microfluidic interface for mass spectrometry. Analyt Chem. 2010;82:3985–3989. doi: 10.1021/ac100372c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik A, Hauf W, Hoffmann J, Cernescu M, Brutschy B, Braun V. Oligomeric structure of ExbB and ExbB-ExbD isolated from Escherichia coli as revealed by LILBID mass spectrometry. Biochemistry. 2011;50:8950–8956. doi: 10.1021/bi2008195. [DOI] [PubMed] [Google Scholar]
- Hanay MS, Kelber S, Naik AK, Chi D, Hentz S, Bullard EC, Colinet E, Duraffourg L, Roukes ML. Single-protein nanomechanical mass spectrometry in real time. Nature Nanotechnol. 2012;7:602–608. doi: 10.1038/nnano.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contino NC, Pierson EE, Keifer DZ, Jarrold MF. Charge detection mass spectrometry with resolved charge states. J Am Soc Mass Spectrom. 2013;24:101–108. doi: 10.1007/s13361-012-0525-5. [DOI] [PubMed] [Google Scholar]
- Pierson EE, Keifer DZ, Selzer L, Lee LS, Contino NC, Wang JC, Zlotnick A, Jarrold MF. Detection of late intermediates in virus capsid assembly by charge detection mass spectrometry. J Am Chem Soc. 2014;136:3536–3541. doi: 10.1021/ja411460w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgner N, Robinson CV. Massign: an assignment strategy for maximizing information from the mass spectra of heterogeneous protein assemblies. Analyt Chem. 2012;84:2939–2948. doi: 10.1021/ac300056a. [DOI] [PubMed] [Google Scholar]
- Tseng YH, Uetrecht C, Heck AJ, Peng WP. Interpreting the charge state assignment in electrospray mass spectra of bioparticles. Analyt Chem. 2011;83:1960–1968. doi: 10.1021/ac102676z. [DOI] [PubMed] [Google Scholar]
- Rozen S, Tieri A, Ridner G, Stark AK, Schmaler T, Ben-Nissan G, Dubiel W, Sharon M. Exposing the subunit diversity within protein complexes: a mass spectrometry approach. Methods. 2013;59:270–277. doi: 10.1016/j.ymeth.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Sharon M, Mao H, Boeri Erba E, Stephens E, Zheng N, Robinson CV. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009;17:31–40. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Keetch CA, Bromley EH, McCammon MG, Wang N, Christodoulou J, Robinson CV. L55P transthyretin accelerates subunit exchange and leads to rapid formation of hybrid tetramers. J Biol Chem. 2005;280:41667–41674. doi: 10.1074/jbc.M508753200. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Fenn JB. Electrospray ion source. Another variation on the free-jet theme. J Phys Chem. 1984;88:4451–4459. [Google Scholar]
- Wilm M, Mann M. Electrospray and Taylor-Cone theory, Dole's beam of macromolecules at last? Intl J Mass Spectrom Ion Proc. 1994;136:167–180. [Google Scholar]
- Butterer A, Pernstich C, Smith RM, Sobott F, Szczelkun MD, Toth J. Type III restriction endonucleases are heterotrimeric: comprising one helicase-nuclease subunit and a dimeric methyltransferase that binds only one specific DNA. Nucleic Acids Res. 2014;42:5139–5150. doi: 10.1093/nar/gku122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konijnenberg A, Yilmaz D, Ingolfsson HI, Dimitrova A, Marrink SJ, Li Z, Venien-Bryan C, Sobott F, Kocer A. Global structural changes of an ion channel during its gating are followed by ion mobility mass spectrometry. Proc Natl Acad Sci USA. 2014;111:17170–17175. doi: 10.1073/pnas.1413118111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoux J, Politis A, Rinehart D, Marshall DP, Wallace MI, Tamm LK, Robinson CV. Mass spectrometry defines the C-terminal dimerization domain and enables modeling of the structure of full-length OmpA. Structure. 2014;22:781–790. doi: 10.1016/j.str.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borysik AJ, Hewitt DJ, Robinson CV. Detergent release prolongs the lifetime of native-like membrane protein conformations in the gas-phase. J Am Chem Soc. 2013;135:6078–6083. doi: 10.1021/ja401736v. [DOI] [PubMed] [Google Scholar]
- Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood S, Marcoux J, Hopper JT, Allison TM, Liko I, Borysik AJ, Robinson CV. Charge reduction stabilizes intact membrane protein complexes for mass spectrometry. J Am Chem Soc. 2014;136:17010–17012. doi: 10.1021/ja510283g. [DOI] [PMC free article] [PubMed] [Google Scholar]


