Abstract
Nanoelectrospray ionization-mass spectrometry and ion mobility-mass spectrometry have been used to study the interactions of the large, multidomain, and conformationally flexible deubiquitinating enzyme ubiquitin specific protease 5 (USP5) with mono- and poly-ubiquitin (Ub) substrates. Employing a C335A active site mutant, mass spectrometry was able to detect the stable and cooperative binding of two mono-Ub molecules at the Zinc-finger ubiquitin binding protein (ZnF-UBP) and catalytic site domains of USP5. Tetra-ubiquitin, in contrast, bound to USP5 with a stoichiometry of 1 : 1, and formed additional interactions with USP5's two ubiquitin associated domains (UBAs). Charge-state distribution and ion mobility analysis revealed that both mono- and tetra-ubiquitin bound to the compact conformation of USP5 only, and that tetra-ubiquitin binding was able to shift the conformational distribution of USP5 from a mixture of extended and compact forms to a completely compact conformation.
Keywords: ubiquitin specific protease 5, deubiquitinase, native electrospray ionization-mass spectrometry, traveling wave ion mobility spectrometry, noncovalent interactions, structural proteomics
Introduction
Mass spectrometry (MS) and ion mobility spectrometry (IMS), particularly when combined with nano electrospray ionization (nESI), are rapidly becoming indispensable tools in protein structural studies.1–4 Providing that the protein is electrosprayed from aqueous, volatile buffers, such as ammonium acetate, at physiological pH, elements of higher protein structure can be maintained to a sufficient extent to gain useful structural information.5 It is well established that protein-small molecule, and protein− protein interactions can survive the ESI process to be detected, and quantified, by MS.6–10 Moreover, it is becoming clear that, despite some collapse upon desolvation, biologically relevant information regarding protein structure and conformation is available from MS and IMS measurement.11–15 These approaches have been used to study, for example, polydisperse proteins such as αB-crystallin, which are difficult to probe by other techniques.16
Recently, we examined the conformationally flexible enzyme ubiquitin specific protease 5 (USP5) using MS and IMS-MS, and found evidence for two distinct conformational forms based on charge state distributions (CSDs) and collisional cross section (CCS) measurement.17 USP5, also known as isopeptidase T (IsoT), is part of a family of deubiquitinating (DUB) enzymes, whose role is to regulate the ubiquitin system.18 Ubiquitination, a protein PTM, is involved in many cellular regulatory mechanisms, such as proteasomal degradation, and signaling.19 Classically the C-terminus of ubiquitin (Ub) is attached to lysine residues on selected target proteins. Furthermore, poly-Ub chains can be assembled on target proteins by the successive linkage of additional Ub moieties to one of seven internal K-residues, or the N-terminal amino group. By utilizing different K-residues to construct poly-Ub chains, a range of poly-Ub linkage types, or topologies, is produced, which results in the creation of a “ubiquitin code”.20 Labeled target proteins are then recognized by interaction of (poly)Ub modifications with ubiquitin binding domains (UBDs) found within receptor proteins, which may result in degradation of the target, or a particular signaling output. This process is negatively regulated by DUBs, which disassemble poly-Ub chains. USP5 specifically has a role in the degradation of unanchored poly-Ub chains. These unanchored chains, which have a free C-terminus, have been demonstrated to be a physiologically relevant species, and perform a number of regulatory roles.21–23
USP5 possesses five folded domains linked by flexible loops.24 The catalytic domain is flanked by two Zinc finger (ZnF) Ub-binding protein (UBP) domains, and two Ub-associated (UBA) domains. Wilkinson and coworkers have studied the role of each domain in binding poly-Ub substrates using a combination of mutagenesis, isothermal titration calorimetry (ITC), and activity assays.18 The C-terminal ZnF-UBP is known to possess a deep binding pocket to accommodate the free C-terminus of unanchored poly-Ub chains, which is used as the principal means of selectivity for this type of poly-Ub.25 In contrast, no direct evidence was found for direct interaction between Ub and the (cryptic) N-terminal ZnF-UBP, but the domain appears essential for catalytic function. The catalytic domain itself is of the papain-type, and shows significant interaction with Ub, as evidenced by the X-ray crystal structure of USP5 with a covalently attached, modified Ub suicide inhibitor.24 Mutation to the two UBA domains only affected interactions with tri- and tetra-Ub, indicating that binding to the UBAs occurs only with tri-Ub and longer chains, and that these domains occupy a position distal to the C-terminal ZnF-UBP. The recent crystal structure of USP5 confirms the relative positions of the five domains, but raises a number of important questions concerning the structure and dynamics of the enzyme; in particular the orientation of the C-terminal ZnF-UBP and the two UBAs with respect to the catalytic domain.24
To provide further insights into substrate binding of USP5, and its effect on the structure of the enzyme, we have examined the interactions between catalytically inactive C335A-USP5 and mono- and tetra-Ub using native MS and IMS. These techniques were able to successfully reveal (i) cooperative binding of two mono-Ub molecules to C335A-USP5, (ii) increased affinity for poly-Ub chains, and—most significantly— (iii) conformational change in C335A-USP5 induced by binding of tetra-Ub.
Results
In order to examine the binding of poly-Ub chains to USP5 we expressed and purified a C335A mutant of the enzyme as described previously.17 This active site mutant is unable to catalyze the hydrolysis of poly-Ub chains, and thus forms stable complexes with the Ub substrates. Additional mutations were introduced to knock out binding at the C-terminal ZnF-UBP (R221A/C335A-USP5), or the two UBA (M643E/M711E/C335A-USP5) domains. To examine the role of the key C-terminal region of mono-Ub on interactions with C335A-USP5, ΔG75/G76 mutant mono-Ub construct was also successfully expressed and purified.
USP5 binding to mono-Ub
Addition of WT mono-Ub (2 μM) to C335A-USP5 (10 μM) resulted in the binding of one and two Ub monomers (Fig. 1). It was readily apparent from the spectrum that C335A-USP5•mono-Ub and C335A-USP5•mono-Ub•mono-Ub complexes were observed for the lower CSD only, and that the intensity of the signals for the binding of two Ubs were significantly greater than those for one Ub. Calculation of the apparent dissociation constant (Kd), based on the populations of one Ub bound-, two Ub bound-, and unbound-C335A-USP5 taken from the signal intensities seen in the mass spectrum yielded an approximate apparent Kd value of 2.5 (±0.3) μM for the first binding event. This value represented an average for the addition of 2, 3, and 4 μM mono-Ub to C335-USP5 (10 μM) (see Materials and Methods). The Kd value for the second binding event was too low to be quantified using the concentrations of binding partners required for detection by MS, which indicated a high affinity interaction. In contrast to WT mono-Ub, the ΔG75/G76 mutant mono-Ub bound only once to C335A-USP5 (Fig. 2), with an apparent approximate Kd value of 26.7 (±0.8) μM. As binding with this mutant can only take place at the catalytic site, it is evident that binding at the ZnF-UBP domain is required for optimal interaction at the catalytic site. It was very clear from these data that positive cooperativity was exhibited during binding of mono-Ub to C335A-USP5. To further support this, the use of a ZnF-UBP mutant of USP5 (Supporting Information Fig. S1) showed binding to one mono-Ub moiety. Interestingly, the double UBA mutant of C335A-USP5 bound WT-Ub in a manner identical to C335A-USP5, which demonstrates that these domains are not involved in high affinity interactions with mono-Ub (Supporting Information Fig. S1).
Figure 1.
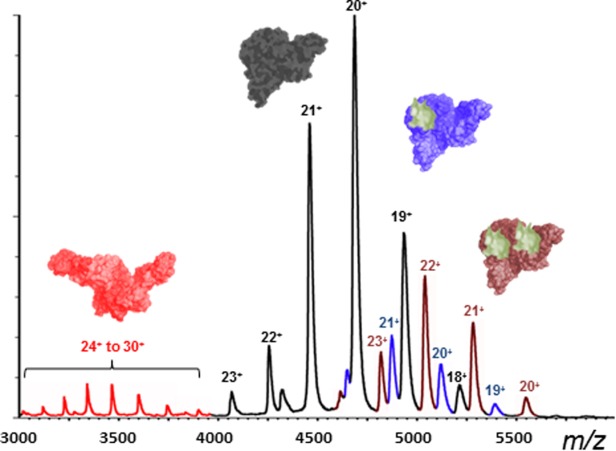
nESI-MS of C335A-USP5 (10 µm) in complex with mono-Ub (2 µm). Highlighted are the extended USP5 conformer (red), compact USP5 conformer (black), and the USP5•mono-Ub complex (1 Ub moiety bound in blue, 2 Ub moieties bound in brown).
Figure 2.
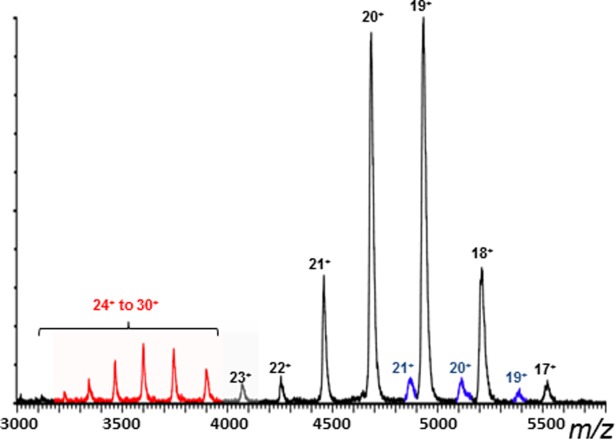
nESI-MS spectrum of C335A-USP5 (10 µm) in complex with ΔG75/G76 mutant mono-Ub (2 µm). Extended USP5 conformer (red), compact USP5 conformer (black) and the USP5.mono-Ub complex (1 Ub moiety bound in blue).
We subjected the C335A-USP5•mono-Ub and C335A-USP5•mono-Ub•mono-Ub complexes to IM-MS analysis using traveling wave ion mobility spectrometry (TWIMS). The resulting CCS measurements, summarized in Table1 (see also Supporting Information Figs. S3-S5), show that each bound Ub monomer contributed 4.4 (±0.02) nm2 to the overall CCS of each complex. These findings were rather surprising as the two high affinity sites for mono-Ub, namely the ZnF-UBP and the catalytic site, occupy quite different locations on the USP5 structure, which might be expected to produce complexes with correspondingly different CCSs. Models of the two C335A-USP5•mono-Ub complexes were constructed to investigate this phenomenon further. Using as a template the output of a 2 ns gas-phase MD simulation of USP5 reported earlier,17 mono-Ub was docked either onto the ZnF-UBP or the catalytic site domain by alignment with partial structures 2G45 and 3IHP from the Protein Data Bank, respectively. Due to the presence of multiple clashes it was found that gas-phase collapse following 2 ns of MD was incompatible with mono-Ub binding in the active site. The output following only 100 ps of MD simulation, however, allowed mono-Ub to be accommodated by the active site domain without clashes, and was utilized for the C335A-USP5 structure in this case [Fig. 3(A)]. These findings indicated that structural collapse of C335A-USP5 around the proximity of the catalytic site may have been prevented by the presence of mono-Ub in that location. Due to its peripheral position, no such stabilization was expected when mono-Ub bound the ZnF-UBP domain, and the 2 ns MD structure was retained for this model [Fig. 3(B)]. Calculation of the theoretical CCS for the two mono-Ub binding modes yielded values of 57.2 nm2 and 56.9 nm2 for the ZnF-UBP and catalytic site domains, respectively. The two CCSs are similar in magnitude, and ∼4 nm2 larger than the theoretical CCS of unbound, thus closely mimicking the experimental results described above. Moreover, the experimental CCS for the complex between USP5 and ΔG75/G76-mono-Ub was determined to be 55 (±0.7) nm2. This mono-Ub mutant should bind at the catalytic site domain only, and the fact that it produces a complex with a very similar CCS to that of WT-mono-Ub also supports the notion that the two binding events lead to similar size complexes (at least within the limits of the relatively modest IM resolution available).
Table 1.
The Molecular Masses, and Measured and Theoretical Collisional Cross Sections (CCS) for USP5 and Its Ubiquitin Complexes
| Protein complex | Molecular Mass/Da | Mean CCS/nm2 | |
|---|---|---|---|
| Experimentala | Theoretical | ||
| USP5 | 93,792 | 50.2 (±1.9) | 53.5 |
| USP5•mono-Ub | 102,357 | 54.7 (±0.9) | 57.2,b 56.9c |
| USP5•mono-Ub•mono-Ub | 110,922 | 59.0 (±0.7) | — |
| USP5•ΔG75/G76-mono-Ub | 102,243 | 55.2 (±0.7) | — |
| USP5•tetra-Ub | 127,980 | 66.1 (±1.4) | — |
Mean CCS are derived from the IM drift-trace peak maxima for each charge state detected (see Supporting Information Fig. S7-9 for drift trace plots). Errors are ± standard deviation.
ZnF-UBP-bound.
catalytic site-bound.
- indicates not calculated.
Figure 3.
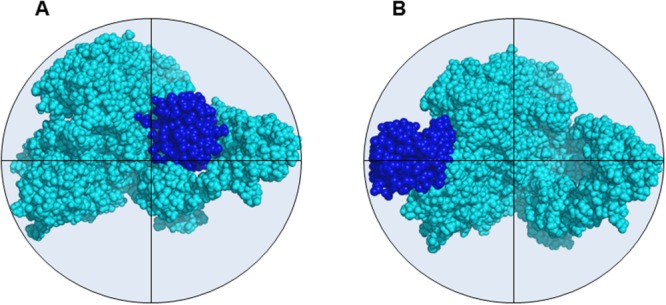
Molecular models of USP5 with mono-Ub bound at (A) the active site domain and (B) the ZnF-UBP domain showing the similar dimensions of the two complexes (calculated CCSs = 56.9 nm2 and 57.2 nm2, respectively).
USP5 binding to tetra-Ub
To investigate the binding of a poly-Ub substrate to USP5, we utilized K63-linked tetra-Ub. Addition of this poly-Ub chain to C335A-USP5 resulted in a complex with an apparently high affinity interaction (Fig. 4). In the presence of 0.5 molar equivalents of tetra-Ub, a 1 : 1 ratio of bound to unbound C335A-USP5 was observed, and when 1 equivalent of tetra-Ub was added, a single set of signals for the C335A-USP5•Ub4 complex was seen with very little uncomplexed protein [Fig. 4(C)]. As with the second binding event of mono-Ub, the affinity of the interaction was too high to quantify by MS. Interestingly, it was clear from the spectra in Figure 4 that the binding of tetra-Ub had a dramatic effect on the CSD of C335A-USP5. Previously we had shown that two distinct CSDs were visible for the unbound enzyme, as seen in Figure 4(A). With the addition of tetra-Ub, the higher CSD became increasingly depleted [Fig. 4(B,C)], which demonstrates that the structural forms responsible for the low and high CSDs seen in the MS are in equilibrium, and that the position of the equilibrium can be shifted by binding to tetra-Ub.
Figure 4.
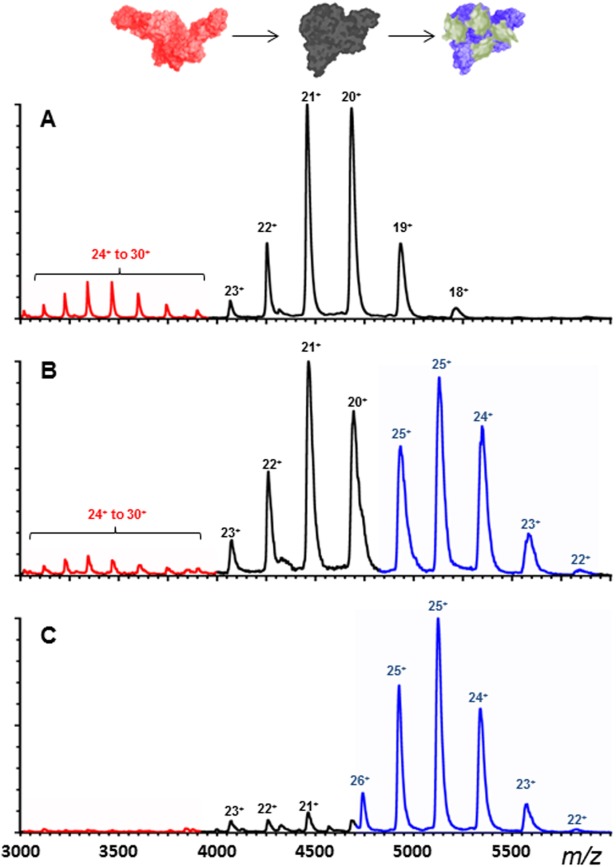
nESI-MS spectrum of C335A-USP5 (10 µm) (A) alone, (B) with 5 µm, and (C) with 10 µm K-63 linked tetra-Ub. Highlighted are the extended USP5 conformer (red), the compact USP5 conformer (black) and the USP5•tetra-Ub complex (blue).
In contrast to that seen with mono-Ub, the complex between tetra-Ub and C335A-USP5 was affected by UBA domain mutations, which confirms that longer poly-Ub chains interact, presumably by avidity effects, with the UBA domains (Supporting Information Fig. S2).
IM-MS measurement of the C335A-USP5•tetra-Ub complex showed a significant increase in CCS for the bound species over the unbound (low CSD in both cases), as was to be expected for the addition of a large protein such as tetra-Ub (see Table1). The CCS of the complex was 16 nm2 larger than that C335A-USP5 alone. Interestingly, this corresponded to 4 nm2 per Ub unit, which is relatively similar to the 4.4 nm2 increase seen for mono-Ub.
Discussion
nESI-MS measurements for the interaction of C335A-USP5 with mono-Ub showed a clear stoichiometry of 1 : 1 and 1 : 2 for C335A-USP5:mono-Ub. A relatively high affinity complex (2.5 μM) for the first mono-Ub binding event was seen, and a very high affinity for the second binding event detected. Although this second mono-Ub interaction was too strong to quantify, based on previous experience10 and the MS signal intensities observed for USP5, we can place an upper limit on the associated Kd of 100 nM. This > 20-fold increase in affinity reveals strong positive cooperativity in the binding of mono-Ub to USP5. A similar result was obtained by Wilkinson et al. using ITC.18 They determined Kd values of ∼600 nM and 20−30 nM for the first and second binding events, respectively. The highest affinity site for mono-Ub binding to USP5 is believed to be at the ZnF-UBP domain, which—as an isolated domain—has been found to possess Kd values for mono-Ub of 2.3 and 2.8 μM, based on two independent measurements using different biophysical techniques.10,25 It is noteworthy that these numbers are in excellent agreement with our estimate for the single mono-Ub binding to full-length C335A-USP5. Interaction between the C335A-USP5 catalytic site domain and mono-Ub is believed to be of lower inherent affinity, but clearly binding here is greatly enhanced by the presence of mono-Ub at the ZnF-UBP site. As expected the ΔG75/G76 mono-Ub mutant exhibited reduced affinity for C335A-USP5, as it is unable to bind the ZnF-UBP domain.
IM-MS measurement of the C335A-USP5•Ub complexes showed that each mono-Ub binding event contributed ∼4.4 nm2 to the CCS. Although the ZnF-UBP binding site is in a more peripheral position on the USP5 structure, and a Ub unit in that position might, therefore, be expected to make a greater contribution to the CCS, modelling indicates that mono-Ub binding at the active site prevents partial structural collapse. The result is two structures with very similar CCSs, despite the different locations of the bound Ub monomer.
Addition of K63-linked tetra-Ub to C335A-USP5 resulted in a clear, very high affinity, 1 : 1 binding as judged by nESI-MS. No free tetra-Ub signals were present in the spectrum, indicating that 100% bound to C335A-USP5. ITC measurements by Wilkinson et al. reveal a Kd ≤ 2 nM for this interaction, which is consistent with such behavior. The most notable result was the depletion of high CSD C335A-USP5 ions as tetra-Ub was titrated into the enzyme solution (see Fig. 4). Previously we have shown that C335A-USP5 exhibits two well-defined CSDs: a major one between 17+ and 21+, and a minor distribution between 24+ and 29+.17 These were attributed to compact and extended conformations of C335A-USP5. It is clear from Figure 4 that the USP5•tetra-Ub complex only exhibits relatively low charge states, and that when one equivalent on the poly-Ub is added, no high CSD is visible in the spectrum. This is a significant result, as it indicates that the high and low CSDs represent species that are capable of inter-conversion in solution, and that tetra-Ub binding is able to shift this equilibrium distribution in favor of structures that give rise to the low CSD. USP5 contains a number of highly flexible loop regions between its folded domains, which are not seen by X-ray crystallography. It may be that the high CSDs seen in the MS are derived from structural states with these loops in a more extended conformation, which gives rise to greater separation between domains, and a resulting higher CSD and CCS. Upon binding tetra-Ub the USP5 domains appear to occupy a more compact structure, as they make contact with the various units of the poly-Ub. Functionally, such structural flexibility may serve to accommodate the various possible topologies of poly-Ub that arise due to different chain linkages and may also be important in the catalytic process itself. It should also be pointed out that mono-Ub, in binding to the lower charge states of C335A-USP5 only, may also be capable of altering the conformation of the enzyme. The relatively low affinity of this interaction prevented us from saturating C335A-USP5 with mono-Ub and observing any significant shift in the overall charge state distribution.
In summary, we have demonstrated that nESI-MS can be used to detect the complexes of C335A-USP5 with mono- and tetra-Ub. For the lower affinity interactions, we have been able to quantify these interactions. MS clearly revealed the stoichiometry of USP5•Ub binding, and allowed detection of cooperativity. Changes in CSD upon tetra-Ub binding provided insights into the equilibrium between extended and compact forms of USP5.
Materials and Methods
Protein expression and purification
The protein coding region of full length short human USP5 (residues 1-835) was cloned from PCR amplified human U20S cDNA and ligated into the BamHI/XhoI sites of pGEX-4T-1 (GE Healthcare, Buckinghamshire, UK). Mutations to this construct (C335A, R221A, and M643E/M711E) were introduced by site-directed mutagenesis (QuikChange kit, Stratagene, Agilent, Stockport, UK) and confirmed by DNA sequencing. The expression and purification of USP5 was carried out as described previously.17 Subsequently USP5 samples were desalted into aqueous ammonium acetate (200 mM, pH 7) using Viva-Spin ultrafilters (10 kDa MWCO, 0.5 mL, Sartorius Stedim Biotech, Epsom, UK), and quantified by nanodrop. Concentrate solutions were diluted into ammonium acetate (200 mM, pH 7) to the desired concentration.
Wild type and ΔG75/G76 mutant mono-Ub were expressed and purified as previously described.26 K-63 linked tetra-Ub was purchased (Boston Biochem, MA).
Mass spectrometry
Nanoelectrospray ionisation mass spectrometry experiments were performed on a Waters (Altrincham, UK) Synapt G1 High Definition Mass Spectrometer (HDMS)—a hybrid quadrupole/ion mobility/orthogonal acceleration time of flight (oa-TOF) instrument, equipped with the standard Waters nanospray source. Home-made nanospray tips were prepared as previously described.17
The instrument was operated in positive-ion mode, under the following optimized nESI-MS parameters: Capillary voltage, 1.3 kV, sample cone voltage, 30V, extraction cone voltage, 5V, backing pressure 4.0−4.2 mBar. The “trap” and “transfer” T-wave collision cells, containing argon gas held at a pressure of 2.5 × 10−2 mBar, were operated at a collision energy of 10 V. The oa-TOF-MS was operated over the scanning range of m/z 500−8000 at a pressure of 1.8 × 10−6 mBar. The instrument was controlled and data viewed using MassLynx 4.1 software (Waters).
Using nESI-MS, Kd values for Ub•C335A-USP5 complexes were determined by titration of USP5, fixed at 10 µM with 2/3/4 µM monoubiquitin. nESI-MS analysis of the resulting solutions was used to determine ratios of signal intensities attributed to free C335A-USP5 and C335A-USP5•Ub ions. Providing that the electrospray response factors for the bound and unbound USP5 ions are similar and that minimal dissociation of the complex occurs post-desolvation, then measured ratios can represent those present in solution. See Supporting Information for equation derivitization.
Ion mobility-mass spectrometry
The TWIMS ion mobility cell, which contained nitrogen gas at 0.45 mBar, was operated at ambient temperature with a wave-height of 10 V traveling at 300 m/s. Travelling wave parameters for the “trap” and “transfer” were: Trap 300 m/s, 0.5 V and transfer 248 m/s, 4 V. Other instrument parameters were as described above. TWIMS measurements on USP5 and USP5-ubiquitin complexes were calibrated, using identical instrument parameters, against a set of standard CCS values for beta-lactoglobulin, bovine serum albumin (BSA), and alcohol dehydrogenase (ADH) (Sigma-Aldrich, Dorset, UK) taken from Bush et al.27 using the method of Ruotolo et al.28 (see Supporting Information Figs. S6-S6). IM drift traces for each ion of USP5 and USP5-ubiquitin complexes were exported into Excel (Microsoft) and the arrival time data points converted onto the CCS scale using the calibration parameters obtained. This allowed the drift traces for each ion to be plotted on a CCS scale, with charge state correction. The combined calibration plot obtained displayed excellent linearity (R2 = 0.999; see Supporting Information Fig. S9).
Models of USP5•mono-Ub with binding at the catalytic site and ZnF-UBP domains were constructed from the USP5 gas-phase MD outputs obtained previously.17 To represent occupancy of the catalytic site, PDB file 3IHP was aligned to the 100 ps MD output of USP5 using Pymol. This structure shows mono-Ub covalently bound to the active site. The USP5 atoms from 3IHP were deleted, leaving the MD output and mono-Ub. A similar process was performed to generate the USP5 ZnF-UBP-bound complex, but using PDB file 2G45 to align to the 2 ns MD output of USP5. The 2G45 structure shows mono-Ub bound to the isolated ZnF-UBP domain. The theoretical CCS of each structure was obtained using the Waters projection approximation (PA) CCS calculator (DriftScope 2.1 CCSCalc program), and scaled by the established empirical correction factor 1.14 to produce a more realistic measure of CCS.29 The PA method of calculating CCSs, which has the advantage of computational speed, is known to underestimate the size of large biomolecules by a constant factor (ibid).
Acknowledgments
The authors are grateful to the Jed Long for the expression and purification of WT and ΔG75/G76 mono-Ub, as well as the University of Nottingham for a studentship to DS.
Supporting Information
Additional Supporting Information may be found in the online version of this article
Supporting Information
References
- Zhong Y, Hyung SJ, Ruotolo BT. Ion mobility-mass spectrometry for structural proteomics. Expert Rev Proteomics. 2012;1:47–58. doi: 10.1586/epr.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJ. Ion mobility mass spectrometry of proteins and protein assemblies. Chem Soc Rev. 2010;39:1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat Chem. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- Breuker K, McLafferty FW. Stepwise evolution of protein native structure with electrospray into the gas phase, 10(-12) to 10(2) s. Proc Natl Acad Sci USA. 2008;105:18145–18152. doi: 10.1073/pnas.0807005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JM, Friess SD, Rajagopalan S, Wendt S, Zenobi R. Quantitative determination of noncovalent binding interactions using soft ionization mass spectrometry. Int J Mass Spectrom. 2002;216:1–27. [Google Scholar]
- Hopper JTS, Robinson CV. Mass spectrometry quantifies protein interactions-from chaperones to membrane proteins. Angew Chem Int Ed. 2014;53:14002–14015. doi: 10.1002/anie.201403741. [DOI] [PubMed] [Google Scholar]
- Yin S, Loo JA. Mass spectrometry detection and characterization of noncovalent protein complexes. Methods Mol Biol. 2009;492:273–282. doi: 10.1007/978-1-59745-493-3_16. [DOI] [PubMed] [Google Scholar]
- El-Hawiet A, Kitova EN, Arutyunov D, Simpson DJ, Szymanski, Klassen JS. Quantifying ligand binding to large protein complexes using electrospray ionization mass spectrometry. Anal Chem. 2012;84:2867–3870. doi: 10.1021/ac3005082. [DOI] [PubMed] [Google Scholar]
- Sokratous K, Roach LV, Channing D, Strachan J, Long J, Searle MS, Layfield R, Oldham NJ. Probing affinity and ubiquitin linkage selectivity of ubiquitin-binding domains using mass spectrometry. J Am Chem Soc. 2012;134:6416–6424. doi: 10.1021/ja300749d. [DOI] [PubMed] [Google Scholar]
- Jurneczko E, Barran PE. How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collisional cross sections in the gas phase. Analyst. 2011;136:20–28. doi: 10.1039/c0an00373e. [DOI] [PubMed] [Google Scholar]
- Scarff CA, Thalassinos K, Hilton GR, Scrivens JH. Travelling wave ion mobility mass spectrometry studies of protein structure: biological significance and comparison with X-ray crystallography and nuclear magnetic resonance spectroscopy measurements. Rapid Commun Mass Spectrom. 2008;22:3296–3304. doi: 10.1002/rcm.3737. [DOI] [PubMed] [Google Scholar]
- Bleiholder C, Dupui NF, Wyttenbach T, Bowers MT. Ion mobility-mass spectrometry reveals a conformational conversion from random assembly to β -sheet in amyloid fibril formation. Nat Chem. 2011;3:172–177. doi: 10.1038/nchem.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapman TW, Warriner SL, Valette NM, Ashcroft AE. Ion mobility spectrometry-mass spectrometry of intrinsically unfolded proteins: trying to put order into disorder. Curr Anal Chem. 2013;9:181–191. doi: 10.2174/1573411011309020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner M, Ellis J, Huang WC, Raven EL, Roberts GCK, Oldham NJ. Detection of a protein conformational equilibrium by electrospray ionisation-ion mobility-mass spectrometry. Angew Chem Int Ed. 2011;50:8291–8294. doi: 10.1002/anie.201101077. [DOI] [PubMed] [Google Scholar]
- Baldwin AJ, Lioe H, Robinson CV, Kay LE, Benesch JLP. αB-crystallin polydispersity is a consequence of unbiased quaternary dynamics. J Mol Biol. 2011;413:297–309. doi: 10.1016/j.jmb.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Scott D, Layfield R, Oldham NJ. Ion mobility-mass spectrometry reveals conformational flexibility in the deubiquitinating enzyme USP5. Proteomics. 2015 doi: 10.1002/pmic.201400457. DOI: 10.1002/pmic.201400457. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008;283:19581–19592. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Dayal S, Sparks A, Jacob J, Allende-Vega N, Lane DP, Saville MK. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem. 2009;284:5030–5041. doi: 10.1074/jbc.M805871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Allali-Hassani A, Asinas A, Nair UB, Fang X, Zuo X, Wang Y-X, Wilkinson KD, Dhe-Paganon S. Two ZnF-UBP Domains in Isopeptidase T (USP5) Biochemistry. 2012;51:1188–1198. doi: 10.1021/bi200854q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Long J, Gallagher TRA, Cavey JR, Sheppard PW, Ralston SH, Layfield R, Searle MS. Ubiquitin Recognition by the Ubiquitin-associated Domain of p62 Involves a Novel Conformational Switch. J. Biol. Chem. 2008;283:5427–5440. doi: 10.1074/jbc.M704973200. [DOI] [PubMed] [Google Scholar]
- Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal. Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- Ruotolo BT, Benesch JLP, Sandercock AM, Hyung SJ, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- Benesch JLP, Ruotolo BT. Mass spectrometry: come of age for structural and dynamical biology. Cur. Opin. Struct. Biol. 2011;21:641–649. doi: 10.1016/j.sbi.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


