Significance
Development of preimplantation embryos entails global DNA demethylation on the zygotic genome. The original thought was that TET-deficient embryos would be unlikely to survive early embryogenesis because they would be unable to mediate genome-wide demethylation in the zygote and preimplantation embryo. However, mice lacking the individual TET proteins Tet1, Tet2, or Tet3 have survived until birth and beyond, suggesting redundancy among TET proteins in the early embryogenesis. Here we report that preimplantation embryos doubly disrupted for Tet1 and Tet3 show abnormal embryonic phenotypes, whose incomplete penetrance correlates with a high variability of transcriptional profiles and DNA methylation status. Our data suggest that in addition to facilitating DNA demethylation, TET proteins and oxidized methylcytosines may regulate the consistency of gene transcription during embryogenesis.
Keywords: DNA methylation, cholesterol biosynthesis, TET methylcytosine oxidases, 5-hydroxymethylcytosine, 5hmC
Abstract
Dioxygenases of the TET (Ten-Eleven Translocation) family produce oxidized methylcytosines, intermediates in DNA demethylation, as well as new epigenetic marks. Here we show data suggesting that TET proteins maintain the consistency of gene transcription. Embryos lacking Tet1 and Tet3 (Tet1/3 DKO) displayed a strong loss of 5-hydroxymethylcytosine (5hmC) and a concurrent increase in 5-methylcytosine (5mC) at the eight-cell stage. Single cells from eight-cell embryos and individual embryonic day 3.5 blastocysts showed unexpectedly variable gene expression compared with controls, and this variability correlated in blastocysts with variably increased 5mC/5hmC in gene bodies and repetitive elements. Despite the variability, genes encoding regulators of cholesterol biosynthesis were reproducibly down-regulated in Tet1/3 DKO blastocysts, resulting in a characteristic phenotype of holoprosencephaly in the few embryos that survived to later stages. Thus, TET enzymes and DNA cytosine modifications could directly or indirectly modulate transcriptional noise, resulting in the selective susceptibility of certain intracellular pathways to regulation by TET proteins.
The three mammalian TET proteins TET1, TET2, and TET3 are Fe(II) and 2-oxoglutarate-dependent dioxygenases that alter the modification status of cytosines in DNA by successively oxidizing 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (1–3). All three oxidized methylcytosines (oxi-mCs) are intermediates in DNA demethylation, the complete conversion of 5mC to C (reviewed in refs. 4, 5). At least two mechanisms appear to be involved: inhibition of the maintenance methyltransferase activity of the DNMT1/UHRF1 complex (6) and excision of 5caC and 5fC by thymine DNA glycosylase (TDG) (3, 7, 8). Because DNA demethylation occurs in a genome-wide fashion during embryonic development, there has been considerable interest in the role of TET proteins in early embryogenesis.
Genome-wide DNA demethylation is observed at two stages of embryonic development, in the fertilized zygote and during the specification of primordial germ cells (PGCs) (9, 10). In PGCs, Tet1 and Tet2 contribute to DNA demethylation through a replication-dependent mechanism (9). In fertilized zygotes, Tet3 was originally thought to oxidize 5mC preferentially in the paternally inherited genome (11–13); more recently, however, reduced-representation bisulfite sequencing (RRBS) has been used to suggest that demethylation of the maternal genome is also catalyzed by Tet3 (14). RRBS measures the sum of 5mC and 5hmC (vs. C, 5fC, and 5caC) at a fraction of cytosines in the genome, and the data show that loss of 5mC+5hmC in both maternal and paternal pronuclei occurs primarily through a passive, replication-dependent process (14, 15). Despite the high expression of Tet3 in oocytes and zygotes (11), Tet3-deficient zygotes display only a marginal increase in 5mC+5hmC in either paternal or maternal pronuclei (14), suggesting the redundant involvement of other TET proteins.
Further attesting to the redundant functions of TET-family proteins, mice lacking individual TET proteins develop relatively normally until birth and beyond (reviewed in ref. 16). Specifically, adult Tet1 and Tet2 KO mice are viable and fertile, displaying relatively mild behavioral and hematopoietic phenotypes, respectively (17–19), whereas Tet3 KO mice die perinatally for unknown reasons (11). Tet3-deficient early embryos show no difference in gene expression compared with WT embryos (14), suggesting compensation from Tet1, Tet2, or both. Even the double deletion of Tet1 and Tet2 has only minor consequences: a significant fraction of Tet1/Tet2 doubly deficient mice survive to adulthood, whereas the remainder succumb late in embryogenesis or shortly after birth (20). This relatively mild embryonic phenotype could reflect the potential involvement of Tet3, which is up-regulated compared with control (CTL) in embryonic stem (ES) cells, embryonic day 13.5 (E13.5) embryos, and adult brain and lung of Tet1/2 DKO mice (20). Tet3 is likely to be an important player, because triple Tet1/2/3-deficient ES cells show significant defects in differentiation (21) and triple TET-deficient mouse embryonic fibroblasts cannot be reprogrammed to induced pluripotent stem cells (22). Thus, a complete characterization of the roles of TET proteins and oxi-mC modifications in early embryogenesis will require significant interference with TET function, through analysis of embryos deficient for Tet1/Tet3, Tet2/Tet3, or all of the three TET proteins.
In this study, we investigated the roles of Tet1 and Tet3 in early embryonic development. We show that single cells of Tet1/3 DKO eight-cell embryos display a global loss of 5hmC and gain of 5mC, indicating that Tet1 and Tet3 are the primary contributors to oxi-mC production at this developmental stage. RNA sequencing of single blastomeres from these eight-cell embryos revealed an unexpected degree of transcriptome variability compared with controls, with a global effect on the majority of expressed genes irrespective of expression level. A similar global variability was observed at a slightly later stage of embryonic development, by RNA sequencing (RNA-seq) of single blastocysts collected at E3.5. RRBS analysis of individual blastocysts showed that the variability of gene transcription correlated with variably increased 5mC/5hmC in gene bodies and repetitive elements, which in turn correlated with pronounced phenotypic variability at both early and late stages of embryonic development. However, a small number of genes were reproducibly down-regulated in Tet1/3 DKO blastocysts compared with controls; many of these encoded enzymes and transcription factors involved in lipid and cholesterol biosynthesis, potentially explaining a phenotype resembling holoprosencephaly observed in the few embryos that survived to later developmental stages (E10.5).
Results
Embryonic Lethality of Tet1/3 Double-Deficient Mice.
Consistent with previous reports (23, 24), we detected Tet1 protein at the two-cell, eight-cell, and blastocyst stages by immunocytochemistry, and Tet3 protein at the two-cell but not the eight-cell stage (SI Appendix, Fig. S1 A–C; for specificity of the Tet1 and Tet antisera see SI Appendix, Fig. S1 D and E). Thus, Tet3 and Tet1 likely act in succession during early embryonic development, leading us to examine mice conditionally disrupted for both the Tet1 and the Tet3 genes (SI Appendix, Fig. S2 A and B).
Unlike Tet1-disrupted mice on a mixed genetic background (129Sv, C57BL/6) (17, 25), Tet1-disrupted C57BL/6 mice showed partial embryonic lethality—only 30% of the expected number of Tet1 KO pups survived to birth (SI Appendix, Fig. S3A and Table S1). Like the mixed background mice, however, C57BL/6-background Tet1-disrupted mice were runted (SI Appendix, Fig. S3 B and C); they showed hallmarks of hydrocephaly (SI Appendix, Fig. S3B), a phenomenon that may underlie some of the reported learning defects of the mixed background mice (17, 18); and the females were infertile with degenerated ovaries (SI Appendix, Fig. S3D), recalling the meiotic abnormality of Tet1 gene-trap mice (26). Tet3-disrupted mice on the C57BL/6 background showed perinatal lethality, as observed for 129Sv-background Tet3−/− mice (11). Intercrosses of Tet1+/−Tet3+/− double heterozygous parents yielded significantly smaller litters (3.4 pups per litter) than those from Tet1+/− or Tet3+/− crosses (4.5 and 7.2 pups per litter, respectively), and Tet1−/−Tet3−/− pups did not survive to birth (SI Appendix, Fig. S3E and Table S2).
To generate Tet1−/−Tet3−/− progeny efficiently, we bred Zp3-Cre or Stra8-Cre mice to Tet1fl/fl, Tet3fl/fl mice (hereafter termed Zp3DKO and Stra8DKO mice). As control breeders, we used Tet1fl/fl, Tet3fl/fl littermates of the Zp3DKO females and Stra8DKO males. None of the resulting embryos (Tet1fl/fl, Tet3fl/fl, Zp3mat, or Stra8pat, hereafter termed Tet1/3 DKO) was viable beyond day 10.5: the majority of embryos (45/58 at day E10.5–11; 11/15 at E12–12.5) were resorbed and the remainder showed abnormal development (SI Appendix, Fig. S3F). Genotyping of Tet1/3 DKO E10.5 embryos from two litters confirmed complete excision of the floxed Tet1 and Tet3 alleles (SI Appendix, Fig. S3G).
Early Developmental Abnormalities in Tet1/3 DKO Eight-Cell Embryos.
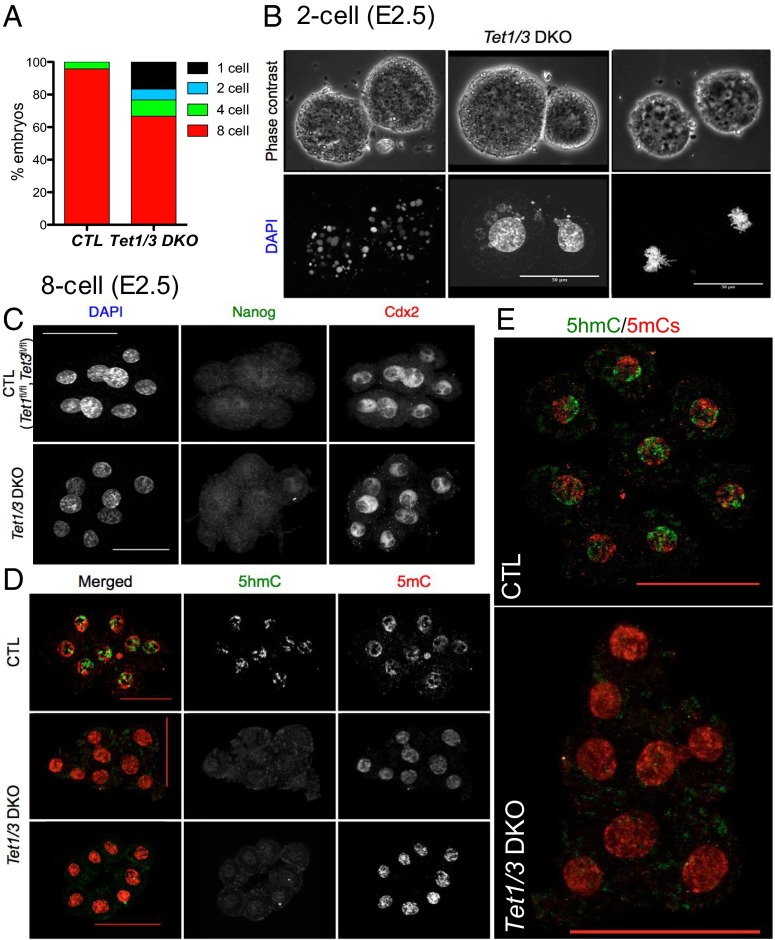
Focusing on early embryonic stages, we found that Tet1/3 DKO embryos showed variably delayed or aborted development relative to Tet1fl/fl, Tet3fl/fl (CTL) embryos: at E2.5, more than 95% of control embryos had reached the eight-cell stage, whereas ∼25% of Tet1/3 DKO embryos were one- and two-cell embryos displaying various abnormal phenotypes including mitotic defects, apoptosis, and/or nuclear blebbing or fragmentation (Fig. 1 A and B). The Tet1/3 DKO embryos (∼70%) that achieved the eight-cell stage by E2.5 were similar to control embryos, as judged by staining with antibodies to Nanog and Cdx2, lineage markers for primitive ectoderm and trophoectoderm, respectively (Fig. 1C). In control eight-cell embryos, 5hmC and 5mC occupied distinct areas of the nucleus (Fig. 1 D and E; the distinct staining patterns of 5mC and 5hmC confirm good antibody specificity), but in Tet1/3 DKO eight-cell embryos, 5hmC was undetectable and instead there was uniform 5mC staining throughout the nucleus (Fig. 1 D and E). Thus, Tet1 and Tet3 are the major catalysts of 5mC oxidation at early embryonic stages; in their absence, 5mC is not converted to 5hmC.
Fig. 1.
Analyses of control and Tet1/3 DKO two-cell and eight-cell embryos. (A) Bar graph showing the percentage of one-cell, two-cell, four-cell, and eight-cell embryos in control (CTL) and Tet1/3 DKO embryos harvested at E2.5. Almost all CTL embryos have reached the eight-cell stage, whereas a substantial fraction of Tet1/3 DKO embryos show aborted or delayed development. (B) Apoptosis (Left), nuclear fragmentation (Middle), and mitotic defects (Right) in representative nonviable two-cell embryos from Tet1/3 DKO mice revealed by DAPI staining. (C) Immunocytochemistry for the lineage markers Nanog and Cdx2 does not reveal any pronounced differences between CTL and Tet1/3 DKO eight-cell embryos. Cdx2 was expressed in all blastomeres, whereas Nanog expression was typically not detected in eight-cell embryos (or occasionally detected in one or a very few blastomeres). (D) Immunocytochemistry using antibodies to 5mC and 5hmC shows global loss of 5hmC and concomitant increase of 5mC in nuclei of Tet1/3 DKO eight-cell embryos compared with CTL. (Scale bar, 50 μm.) (E, Top) Merged and enlarged images of a second control embryo. (Bottom) Tet1/3 DKO embryo from D, Middle, stained with antibodies to 5mC and 5hmC.
Increased Transcriptome Variability in Tet1/3 DKO Eight-Cell Blastomeres Compared with Controls.
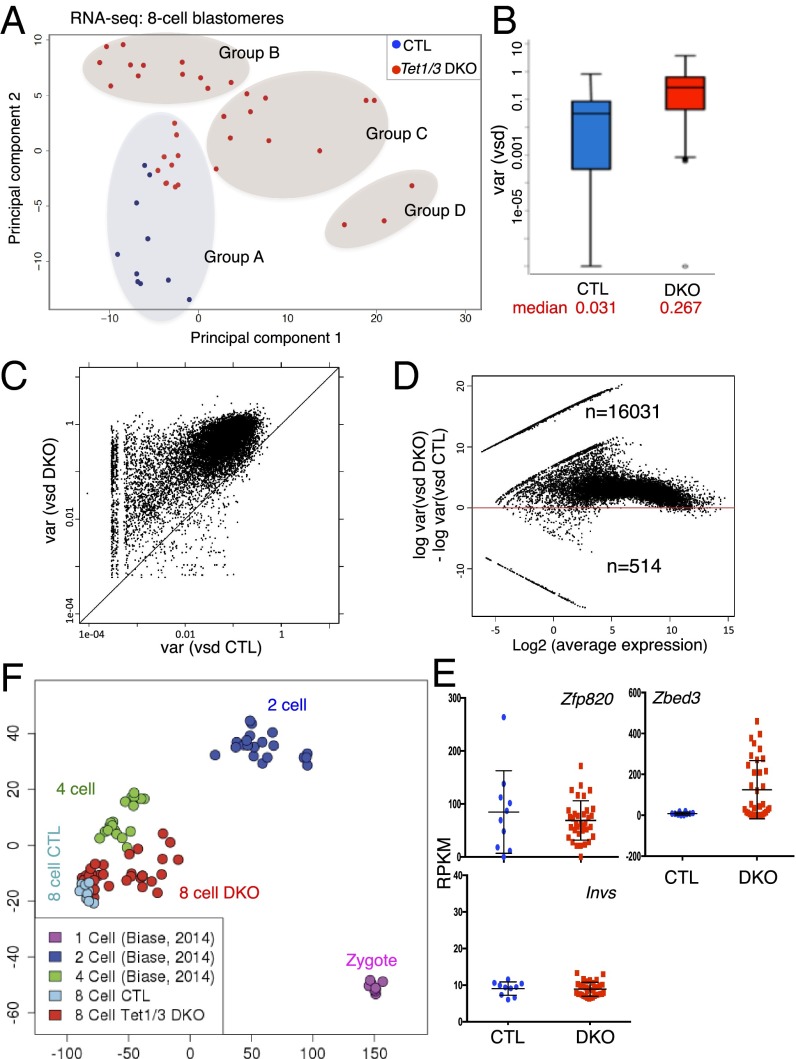
To investigate how the global loss of 5hmC and increase of 5mC in Tet1/3 DKO embryos affected gene expression profiles at the eight-cell stage, we performed RNA-seq on single cells (blastomeres) taken from eight-cell DKO and control embryos, a stage at which individual blastomeres are relatively, although not completely, homogeneous (27, 28). We collected six eight-cell embryos from control or Tet1/3 DKO timed matings and dissociated them into two pools of 48 single blastomeres each, then randomly chose 10 control and 35 Tet1/3 DKO single blastomeres and used them to generate single-cell RNA-seq libraries (SI Appendix, Table S3; the variability of the mutant phenotypes led us to collect more mutant than control blastomeres). When single-cell transcriptomes of all control and Tet1/3 DKO blastomeres were analyzed with respect to spike-in controls (red dots), overall gene expression levels were similar between control and Tet1/3 DKO blastomeres (SI Appendix, Fig. S4A), but the variability of gene expression was different (Fig. 2). Unsupervised clustering distinguished groups A–D (Fig. 2A and SI Appendix, Fig. S4B); principal component analysis showed considerably greater variability in the transcriptomes of single blastomeres from Tet1/3 DKO eight-cell embryos compared with controls (Fig. 2A). The group A cluster contained both control and DKO blastomeres, indicating that some Tet1/3 DKO blastomeres in group A possessed relatively normal transcriptomes (Fig. 2A and SI Appendix, Fig. S4B); however, even in group A, the expression of a subset of genes was different between control and Tet1/3 DKO single cells (SI Appendix, Fig. S4C). Gene ontology (GO) analysis of genes differentially expressed in groups B, C, and D compared with group A identified distinct categories (SI Appendix, Fig. S4D), with the GO terms “cell cycle” in group C and “programmed cell death” in group D (SI Appendix, Fig. S4D) recalling the abnormal phenotypes observed in two-cell embryos (Fig. 1B) and suggesting that the same failures occur in some blastomeres at the eight-cell stage.
Fig. 2.
Transcriptional variability associated with Tet1/3 deficiency revealed by single-cell RNA sequencing of individual blastomeres from eight-cell embryos. (A) Principal component analysis of the transcriptomes of 10 CTL and 35 Tet1/3 DKO with the sex chromosome information removed, showing that the 35 Tet1/3 DKO blastomeres are more widely separated than the 10 CTL blastomeres for both principal component 1 (PC1) (absolute gap, 37 vs. 8) and PC2 (17 vs. 13). Groups A–D identified by the clustering analysis of A are indicated. (B) The box-and-whisker plot depicts the distribution of stabilized variances (gene expression variances across samples independent of their expression strength; see Materials and Methods) in eight-cell embryos. Median variances are shown at Bottom. Upper and Lower whiskers represent >75% and <25%, respectively. (C) Correlation plot of expression variances of all genes expressed in single cells of CTL and Tet1/3 DKO eight-cell embryos. Each gene is represented by a dot. Most genes fall above the diagonal, indicating greater variability of expression in Tet1/3 DKO blastomeres compared with controls. (D) The ratios of gene expression variance between CTL and Tet1/3 DKO blastomeres are plotted against the average expression level for each gene. The increased gene expression variance observed in blastomeres from Tet1/3 DKO eight-cell embryos is independent of the level of gene expression. (E) The genes expressed with highest variance in CTL and Tet1/3 DKO blastomeres, Zfp820 (Left) and Zbed3 (Right), respectively, and a gene that shows low variance in both samples, Invs (Bottom), are indicated (also see SI Appendix, Fig. S4E). (F) Principal component analysis of the combined RNA-seq data from our study and the single-cell analysis of zygotes, two-cell, and four-cell embryos performed by Biase et al. (28).
To quantify the variability of gene expression between single blastomeres from control and Tet1/3 DKO eight-cell embryos we applied a variance-stabilizing transformation that normalizes variability to the level of gene expression (Fig. 2 B–D). The data revealed a global increase in the overall variance of gene expression in Tet1/3 DKO blastomeres compared with controls (Fig. 2 B–D). A correlation plot for the variance of gene expression among the 10 control and 35 DKO blastomeres showed that essentially all genes in DKO blastomeres were expressed more variably than in controls (Fig. 2C; each dot represents one gene, and the majority of dots fall above the diagonal), in a manner independent of the level of gene expression (Fig. 2D). To test if the greater number of Tet1/3 DKO single cells (n = 35) of the blastomeres compared with the number of control single cells (n = 10), had an influence on the observed difference of variances, we randomly selected 10 DKO samples and calculated gene expression variances based on the variance-stabilized data in 20 iterations. We observed increased variation of gene expression in all groups of 10 randomly chosen Tet1/3 DKO samples compared with the 10 control samples, demonstrating that the effect of increased variation is not dependent on the number of samples. Zfp820 and Zbed3 were the top variable genes in single blastomeres from control and Tet1/3 DKO eight-cell embryos, respectively, whereas Invs is an example of one of the few genes that was relatively stably expressed in both sets of blastomeres (Fig. 2E and SI Appendix, Fig. S4E).
We compared our RNA-seq data for single blastomeres from eight-cell embryos with data from two previous reports: one study profiled gene expression by RNA-seq in zygotes and single cells from two-cell and four-cell embryos (28), whereas the second examined the expression of 48 selected genes associated with embryonic development in single cells from the one-cell (zygote) until the 64-cell stage (27). Principal component analysis of the combined data from our study and that of Biase et al. (28) showed that the transcriptional profiles of single cells from control eight-cell embryos clustered tightly into a single group distinct from four-cell embryos, whereas single cells from Tet1/3 DKO eight-cell embryos showed a more variable distribution spread between the four-cell and 8 cell stages (Fig. 2F). Comparison with the data of Guo et al. (27) yielded a similar result (SI Appendix, Fig. S4F).
Together, these results emphasize the striking increase in the variability but not the level of gene expression in single blastomeres from eight-cell embryos lacking Tet1 and Tet3. This variability was maintained at later embryonic stages, as discussed for E3.5 blastocysts below.
Developmental Abnormalities in Tet1/3 DKO Preimplantation and Periimplantation Embryos.
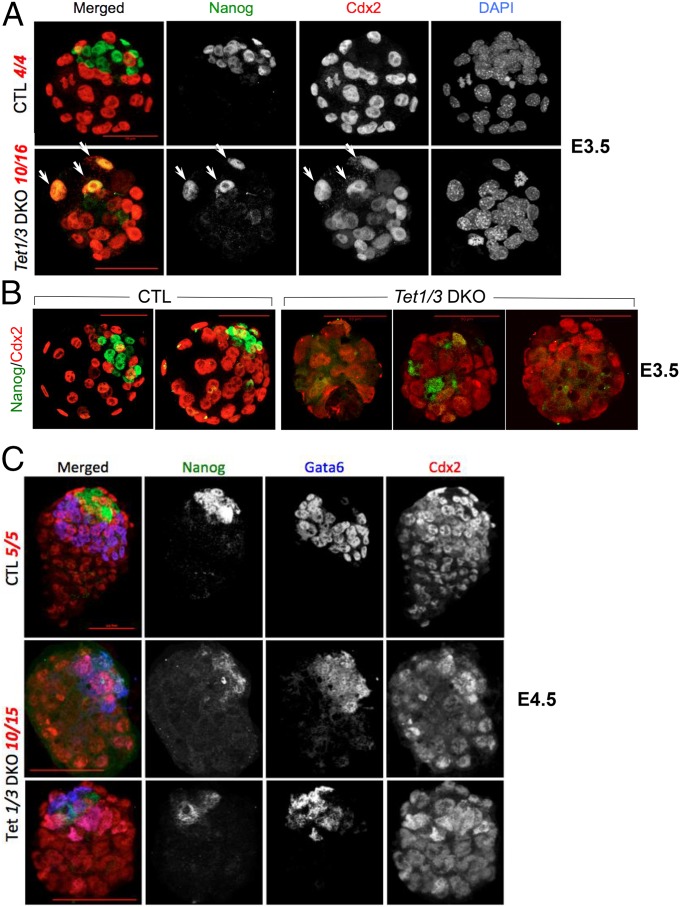
We examined slightly later stages of embryonic development by immunocytochemistry (Fig. 3 and SI Appendix, Fig. S5). All CTL E3.5 preimplantation blastocysts showed normal specification of the inner cell mass and trophectoderm, as judged by distinct expression of Nanog and Cdx2 (Fig. 3 A, Upper and B, Left). In contrast, more than 60% (10/16) of E3.5 Tet1/3 DKO embryos displayed either a severe loss of Nanog expression or aberrant coexpression of Nanog and Cdx2 (arrows); in many cases, the Nanog-expressing cells failed to form a well-defined inner cell mass (Fig. 3 A, Lower and B, Right and SI Appendix, Fig. S5A). A fraction of Tet1/3 DKO embryos (6/16) had a relatively normal appearance (e.g., SI Appendix, Fig. S5B, Left), but several of these showed variable coexpression of Nanog and Cdx2 (SI Appendix, Fig. S5B, Right).
Fig. 3.
Expression of the lineage markers Nanog, Cdx2, and Gata6 in control and Tet1/3 DKO E3.5 and E4.5 embryos. (A) Immunocytochemistry for Nanog and Cdx2 expression shows dysregulation of the first step of lineage commitment in Tet1/3 DKO E3.5 embryos compared with control (CTL). Arrows indicate aberrant coexpression of Nanog and Cdx2. (B) Two more control blastocysts (Left) and three Tet1/3 DKO blastocysts with poor or failed Nanog expression (Right). (C) Immunocytochemistry for Nanog, Cdx2, and Gata6 expression shows dysregulation of the second step of lineage commitment in Tet1/3 DKO E4.5 embryos. (Scale bar, 50 μm.) Listed in red are the numbers of embryos displaying the indicated normal or abnormal phenotype, out of the total number of embryos examined. For more examples see SI Appendix, Fig. S5.
A similar failure of early lineage specification was observed at the periimplantation blastocyst stage (E4.5). In control embryos, we observed well-developed layers of primitive ectoderm expressing Nanog, primitive endoderm expressing Gata6, and trophectoderm expressing Cdx2 (Fig. 3C and SI Appendix, Fig. S5C, Top), whereas Tet1/3 DKO embryos showed strong dysregulation of these three lineage markers, with aberrant coexpression of the markers, poor separation of germ cell layers, and poor or failed development of primitive ectoderm and primitive endoderm as judged by Nanog and Gata6 staining (Fig. 3C and SI Appendix, Fig. S5C, Bottom). Again, some Tet1/3 DKO embryos (5/15) had a relatively normal appearance at E4.5, with high Nanog and Gata6 expression (SI Appendix, Fig. S5D), but most embryos showed variably delayed development as judged by size (Fig. 3C and SI Appendix, Fig. S5 C and D; compare lengths of the 50-μm scale bars in each panel). Overall, there was marked phenotypic variability in Tet1/3 DKO embryos at each of the three stages profiled (E2.5, 3.5, and 4.5) and a steadily increasing fraction of abnormal embryos at later times.
Increased Transcriptome Variability in Tet1/3 DKO Blastocysts Compared with Controls.
To investigate the variable phenotypes of Tet1/3 DKO blastocysts at a molecular level, we performed RNA-seq on single blastocysts (SI Appendix, Table S4). At day 3.5 after timed matings, we randomly chose and sequenced three control blastocysts from a litter of seven, and five Tet1/3 DKO blastocysts from a litter of eight (again, the highly variable phenotypes of mutant embryos led us to sequence more mutant than control embryos). Despite low levels of Tet3 transcripts in control blastocysts, RNA-sequencing confirmed excision of the floxed Tet1 and Tet3 exons in Tet1/3 DKO blastocysts (SI Appendix, Fig. S6A and Dataset S1); Tet2 mRNA was expressed at considerably higher levels and showed no compensatory up-regulation in Tet1/3 DKO embryos relative to controls (SI Appendix, Fig. S6B). IDAX transcripts were not detected by RNA-seq (SI Appendix, Dataset S1), suggesting that Tet2 protein expression was unlikely to be controlled by IDAX (29) at this developmental stage. Expression of Eif2s3y, Zfy1, Ddx3y, Usp9y, and Uty on the Y chromosome and Xist on the X chromosome showed that the three control embryos were female, whereas the Tet1/3 DKO group had three male and two female embryos (SI Appendix, Fig. S6C). The expression of imprinted genes in the Tet1/3 DKO blastocysts was normal compared with controls.
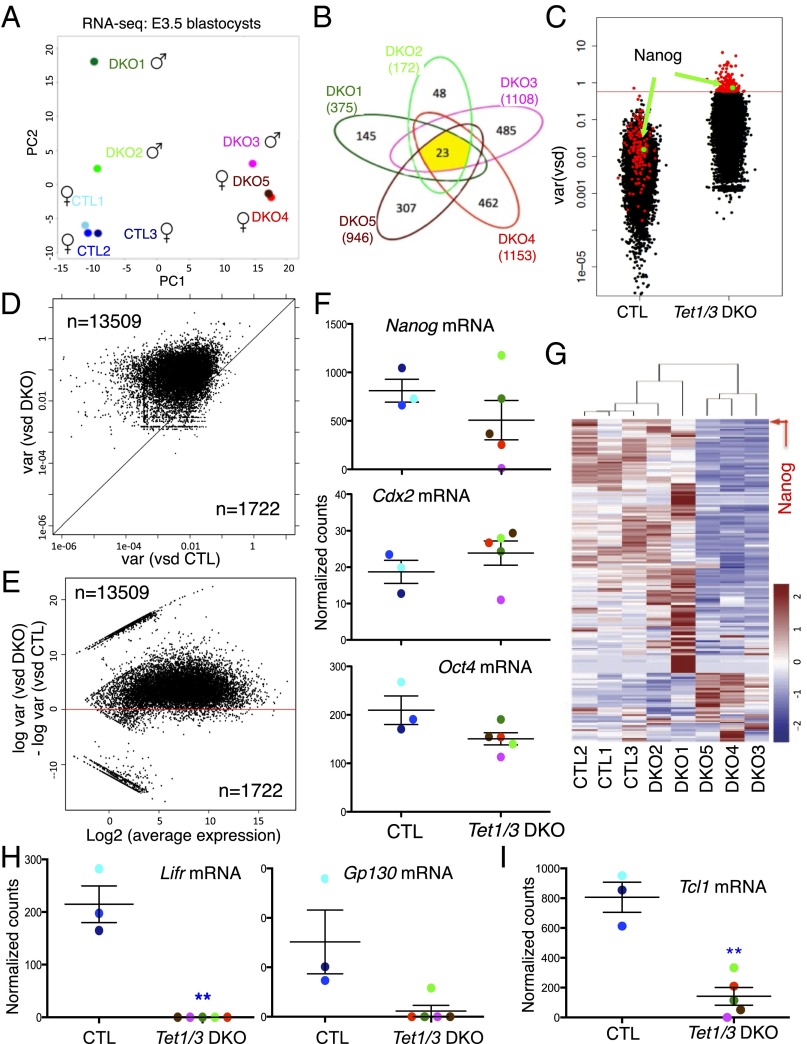
RNA sequencing revealed that Tet1/3 DKO blastocysts displayed considerably more transcriptome variability than the corresponding controls. Unsupervised clustering analysis based on similarities of the transcriptomes showed that control embryos clustered together, whereas Tet1/3 DKO embryos formed two clusters (SI Appendix, Fig. S6D). The clustering was not influenced by the sex of the embryos: DKO 1 and 2 (male) clustered with the three control embryos (female), whereas the other three embryos (DKO 3, male; DKO 4 and 5, female) formed a distinct group (SI Appendix, Fig. S6D). Principal component analysis based on the expression profiles (with sex chromosome data removed) confirmed that control embryos clustered together, whereas Tet1/3 DKO embryos showed a high degree of variance (Fig. 4A). Emphasizing this variability, the number of genes differentially expressed in each of the five Tet1/3 DKO blastocysts compared with the average of the three control blastocysts ranged from 172 differentially expressed genes in DKO 2 to 1,153 in DKO 4 (adjusted P value <0.05), with only 23 differentially expressed genes common to all five DKO blastocysts relative to controls (Fig. 4B).
Fig. 4.
Transcriptional variability associated with Tet1/3 deficiency revealed by RNA sequencing of single E3.5 blastocysts. (A) Principal component analysis (PCA) of three CTL and five Tet1/3 DKO transcriptomes with the sex chromosome information removed, showing that the three control blastocysts cluster together, whereas the five Tet1/3 DKO blastocysts are widely separated. (B) Numbers of genes differentially expressed in each of the Tet1/3 DKO blastocysts compared with the average of the CTL blastocysts. Only 23 genes are differentially expressed in all five Tet1/3 DKO blastocysts relative to control blastocysts. (C) Overall increase in the variance of gene expression in Tet1/3 DKO compared with CTL blastocysts. Each gene is represented by a dot, with Nanog indicated in green. Genes shown in red are those with highest variance (top 1%, n = 178) in Tet1/3 DKO blastocysts. (D) Correlation plot of expression variances of all genes from CTL and Tet1/3 DKO embryos. Most genes fall above the diagonal, indicating greater variability of expression in Tet1/3 DKO blastocysts compared with controls. (E) The ratios of gene expression variance between CTL and Tet1/3 DKO blastocysts are plotted against the average expression level for each gene. The increased gene expression variance observed in Tet1/3 DKO blastocysts is independent of the level of gene expression. (F) Nanog, Cdx2, and Oct4 mRNA expression in control (CTL) and Tet1/3 DKO E3.5 blastocysts quantified by RNA-seq. (G) Heatmap showing expression of the top 1% of variable genes in Tet1/3 DKO blastocysts compared with controls. Nanog is indicated by an arrow. (H) Lifr, Gp130, and (I) Tcl1 expression levels in control (CTL) and Tet1/3 DKO blastocysts assessed by RNA-seq (**adjusted P value <0.01).
Even in the averaged transcriptomes of E3.5 blastocysts, the vast majority of expressed genes showed a much greater variability of expression in Tet1/3 DKO blastocysts relative to controls (Fig. 4 C–E; each gene is represented by a dot). As in eight-cell blastomeres, a correlation plot for the variance of gene expression showed that essentially all genes in the five DKO blastocysts were expressed more variably than in the three controls (Fig. 4D), again in a manner independent of the level of gene expression (Fig. 4E). Consistent with our immunocytochemistry data, Nanog was one of the most variably expressed genes in DKO blastocysts compared with controls (Fig. 4 C, F, and G and SI Appendix, Dataset S1). Other genes, such as Cdx2 and Oct4, showed less variability: Cdx2 mRNA expression was similar between the two groups (Fig. 4F, Middle), whereas Oct4 mRNA expression showed a slight decrease in DKO embryos relative to control (Fig. 4F, Bottom). A heatmap comparing the expression of the top 1% of variable genes (n = 178; indicated by red dots in Fig. 4C) in DKO embryos relative to controls showed that the most common pattern for the highly variable genes was expression comparable to controls in DKO embryos 1 and 2, vs. poor expression relative to controls in DKO embryos 3, 4, and 5 (Fig. 4G).
Together, these results show that in E3.5 blastocysts as well as in single blastomeres from eight-cell embryos, deletion of Tet1 and Tet3 results in a profound and global increase in the variability of gene expression. We asked whether this variability was reflected in a corresponding global increase in the variability of DNA cytosine modification at different annotated regions of the genome (see section below).
A Few Genes Are Reproducibly Down-Regulated in Tet1/3 DKO Blastocysts.
Comparing the group of five Tet1/3 DKO blastocysts to the group of three control blastocysts, and applying an adjusted P value of <0.01, we identified only 40 genes as consistently differentially expressed in DKO blastocysts compared with controls (SI Appendix, Dataset S1). Among these 40 genes, the gene ranked at third place by P value was Lifr, the receptor for the cytokine LIF (Fig. 4H, Left and SI Appendix, Dataset S1). The active LIF receptor complex contains LIFR and its obligate binding partner, Gp130 (30), and Gp130 expression was also reproducibly low in Tet1/3 DKO blastocysts (Fig. 4H, Right). Mice deficient in LIFR subunits show variable phenotypes ranging from impaired implantation to perinatal lethality (31), potentially partly explaining the variable onset of embryonic lethality in Tet1/3 DKO embryos both before and after gastrulation. Embryonic stem cells (ESCs) cannot be derived from Lifr-deficient blastocysts (31), and indeed we consistently failed to derive ESCs from Tet1/3 DKO blastocysts (18 embryos from three litters), despite successful establishment of five different ESC lines from 11 control embryos. We also observed consistently reduced expression of several pluripotency-associated genes in Tet1/3 DKO embryos, including Tcl1 (T-cell leukemia-1, a gene involved in translocations to the TCR locus in T-cell leukemias and also a target of Tet1 in ES cells) (32) (Fig. 4I and SI Appendix, Fig. S6E and Dataset S1). Sca1/2 expression was diminished in Tet1/3 DKOs embryos (SI Appendix, Fig. S6F), potentially explaining the hematopoietic defects observed in some embryos at later developmental stages (SI Appendix, Fig. S3F). Genes involved in lipid and cholesterol biosynthesis were also reproducibly down-regulated, as discussed below.
Increased Levels and Variability of DNA Modification in Tet1/3 DKO Blastocysts Compared with Controls.
To investigate whether transcriptional variability reflected a corresponding variability in DNA modification status, we used targeted bisulfite sequencing to assess DNA cytosine modification at the Nanog locus, for which variable expression was strongly apparent both by immunocytochemistry and by RNA-seq. Because 5mC is not distinguished from 5hmC, nor unmodified C from 5fC and 5caC, in bisulfite sequencing (33, 34), and because Tet2 expressed in Tet1/3 DKO embryos can generate oxi-mC, we use the term “DNA modification” in preference to “DNA methylation,” and specify the modified bases actually examined in each case.
We first examined Nanog regulatory regions using bisulfite treatment followed by targeted PCR. At the eight-cell stage, RRBS still shows evidence of widespread 5mC/5hmC (35). However, we did not observe any substantial difference in the extent or variability of DNA modification at the Nanog promoter/transcription start site (TSS) or a distal (−5 kb) regulatory region (36) at either the E3.5 blastocyst or the E2.5 eight-cell stage (SI Appendix, Fig. S7 A and B).
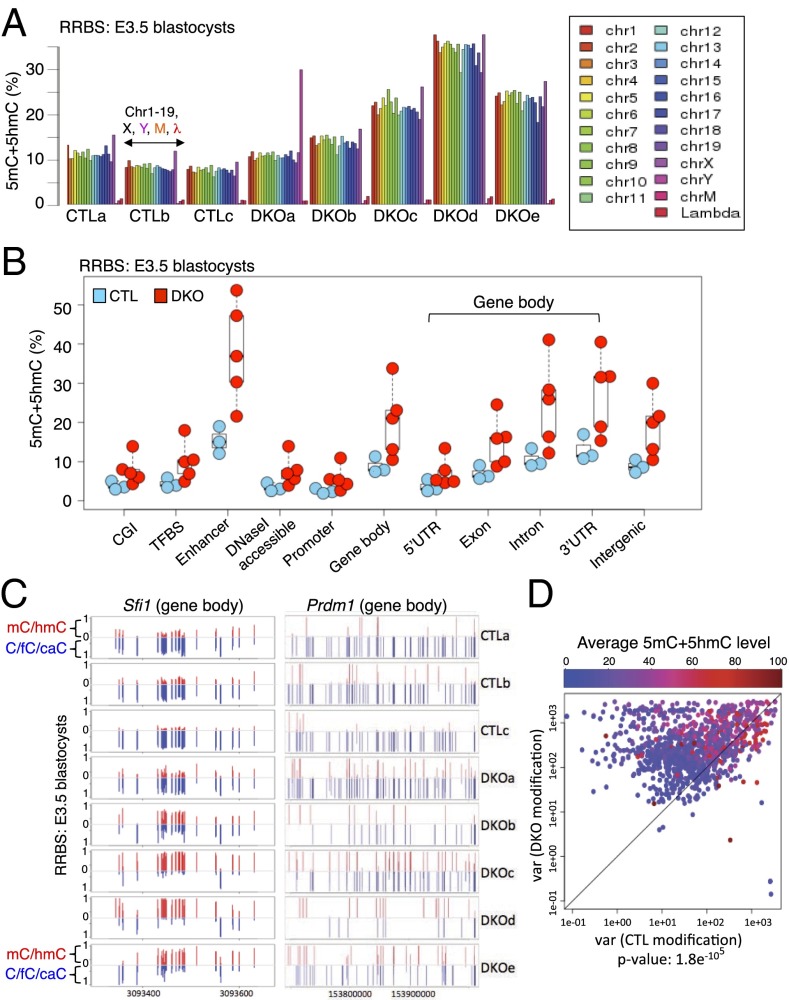
To examine DNA modification more globally, we prepared and sequenced RRBS libraries from three control blastocysts (from a litter of six), and five Tet1/3 DKO blastocysts (from a litter of six, SI Appendix, Table S5). As expected, control blastocysts were substantially demethylated on all chromosomes (7–10% 5mC/5hmC; Fig. 5A). In contrast, the overall levels of 5mC/5hmC in the five different Tet1/3 DKO embryos were highly variable, with two blastocysts (DKOa and DKOb) resembling controls, and the three others showing variably increased levels of 5mC/5hmC (Fig. 5A). Detailed analysis of DNA modification levels at specific genomic regions showed a substantial but variable increase in 5mC/5hmC at all genomic regions in Tet1/3 DKO embryos compared with controls (Fig. 5B), as shown for selected promoters and gene bodies in Fig. 5C and SI Appendix, Fig. S7C. We evaluated the variability of DNA modification (5mC/5hmC) at each CpG, eliminating CpGs covered by fewer than eight reads as well as CpGs with < 10% 5mC+5hmC (Fig. 5D). Tet1/3 DKO blastocysts displayed a much higher variability of DNA modification relative to controls, regardless of the actual level of modification present at each CpG.
Fig. 5.
The variable levels of DNA modification (5mC+5hmC) in Tet1/3 DKO E3.5 blastocysts parallel the transcriptional variability observed above. (A) Modification levels on each chromosome were plotted for three CTL and five Tet1/3 DKO E3.5 embryos based on RRBS analysis. Note that the DKOa blastocyst is the only one with a Y chromosome and that cytosines in control unmodified lambda DNA show more than 98% conversion to T in all embryos. (B) The levels of DNA modification at CpG islands (CGI), transcription factor binding sites (TFBS), enhancers, DNaseI accessible regions, promoter (±2 kb relative to the TSS), gene bodies (subdivided into 5′ UTR, exon, intron, 3′ UTR) and intergenic regions. Each circle represents one blastocyst, with the embryo ID shown within each circle: blue circles, control blastocysts; red circles, Tet1/3 DKO blastocysts. (C) Modification levels of part of the gene body of Sfi1 and the entire gene body of Prdm16 from individual control and Tet1/3 DKO blastocysts. Each bar represents one CpG, and the full length of each bar represents 100% modification (red, 5mC+5hmC; blue, C+5fC+5CaC). (D) The correlation plot depicts variances in the DNA modification level of individual CpGs from CTL and Tet1/3 DKO embryos. Each dot represents one CpG (more than eight reads and >10% 5mC+5hmC). The level of DNA modification at each CpG is graded from blue to red. Most dots fall above the diagonal regardless of modification level, indicating thatTet1/3 DKO blastocysts show greater variability of DNA modification compared with controls.
Together, our data indicate that loss of TET function results both in increased DNA modification as well as in increased variability of modification, consistent with the increased variability of gene expression observed in Fig. 4. Because RNA-seq and RRBS DNA methylation analysis cannot currently be performed on the same blastocysts, a direct correlation of gene expression with DNA modification status is technically out of reach.
Increased Levels and Variability of DNA Modifications at Repetitive Elements in Tet1/3 DKO Embryos Correlates with Altered and Variable Expression.
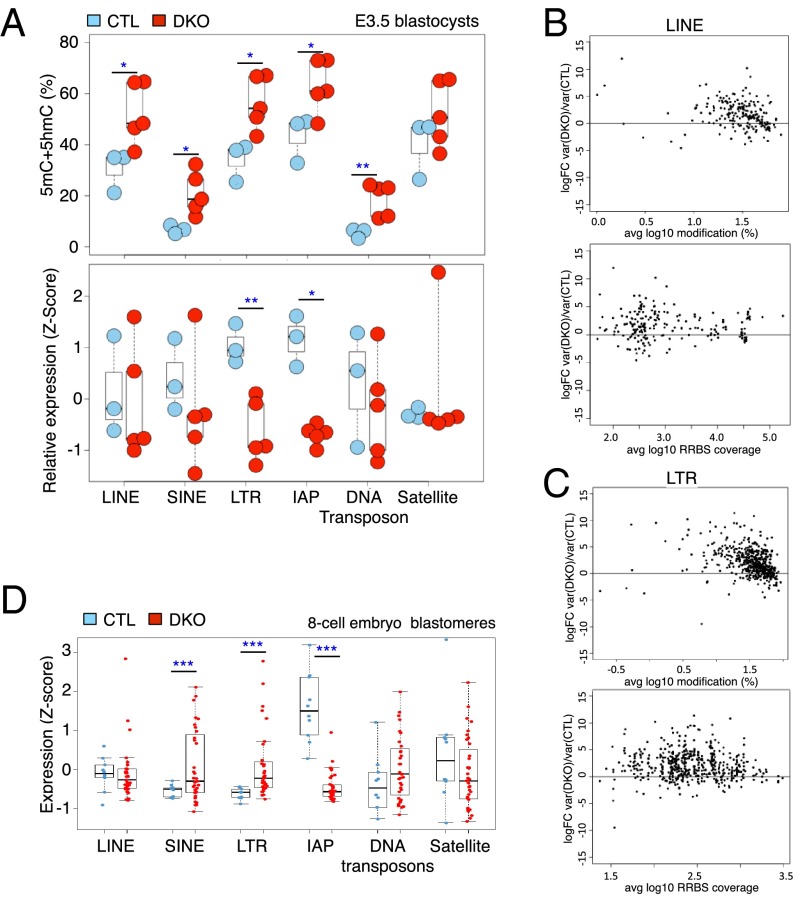
We examined 5mC/5hmC levels at repetitive elements in control and Tet1/3 DKO blastocysts. All elements—non-LTR (LINE, SINE) and LTR (LTR, IAP) retrotransposon groups, DNA transposons and satellite sequences—showed a variable increase in 5mC+5hmC in Tet1/3 DKO blastocysts compared with controls (Fig. 6A, Top). To illustrate this point in more detail, the greater variability of DNA modification in LINE and LTR elements of Tet1/3 DKO blastocysts is shown at the level of individual CpGs in Fig. 6 B and C. In parallel, we assessed the expression levels of these repetitive elements using our single-embryo RNA-seq data (obtained from a different set of blastocysts for the reason outlined above). The expression of LTR and IAP elements was significantly down-regulated in Tet1/3 DKO compared with control blastocysts (Fig. 6A, Bottom); expression of LINEs and SINEs showed a decrease in mean expression levels as well as an increase in the variability of expression.
Fig. 6.
Variability of DNA modification and expression of repetitive elements in Tet1/3 DKO early embryos. (A, Top) DNA modification (5mC+5hmC) levels associated with LINE, SINE, LTR, IAP, DNA transposon, and satellite sequences taken from RRBS data. Blue circles, control blastocysts; red circles, Tet1/3 DKO blastocysts. (Bottom) Expression levels of the corresponding elements taken from RNA-seq data using a separate set of blastocysts. Blue circles, control blastocysts; red circles, Tet1/3 DKO blastocysts (see Fig. 3). (B and C) The variance ratios of DNA modification for individual CpGs within LINEs (B) and LTRs (C) of CTL and Tet1/3 DKO embryos are plotted against average DNA modification level (Top) and RRBS coverage (Bottom). FC, fold change. (D) Expression of LINE, SINE, LTR, IAP, DNA transposon, and satellite sequences taken from RNA-seq data on 10 individual CTL and 35 individual Tet1/3 DKO blastomeres from eight-cell embryos. Note the strongly diminished expression of IAP elements, also observed for E3.5 blastocysts in A. Welch's t test was applied in A and D; *P, <0.05; **P, <0.01; ***P, <0.001.
Because single cells are technically not yet amenable to RRBS, we were unable to measure 5mC/5hmC in single blastomeres from control and Tet1/3 DKO eight-cell embryos. Instead, we assessed the expression levels of retrotransposons, DNA transposons, and satellite sequences at this stage (Fig. 6D). Confirming the results in blastocysts, expression of IAP elements was significantly down-regulated in Tet1/3 DKO compared with control eight-cell blastomeres, whereas expression of SINES and LTRs was increased (Fig. 6D).
Thus, Tet1/3 DKO embryos show variably increased 5mC+5hmC as well as an increase in the variability of expression of transposable elements compared with control embryos and may positively control IAP expression at the eight-cell and blastocyst stages of embryonic development.
Deficiency of Cholesterol Synthesis in Tet1/3 DKO Blastocysts Results in Failure of the Sonic-Hedgehog Pathway in Late Embryogenesis.
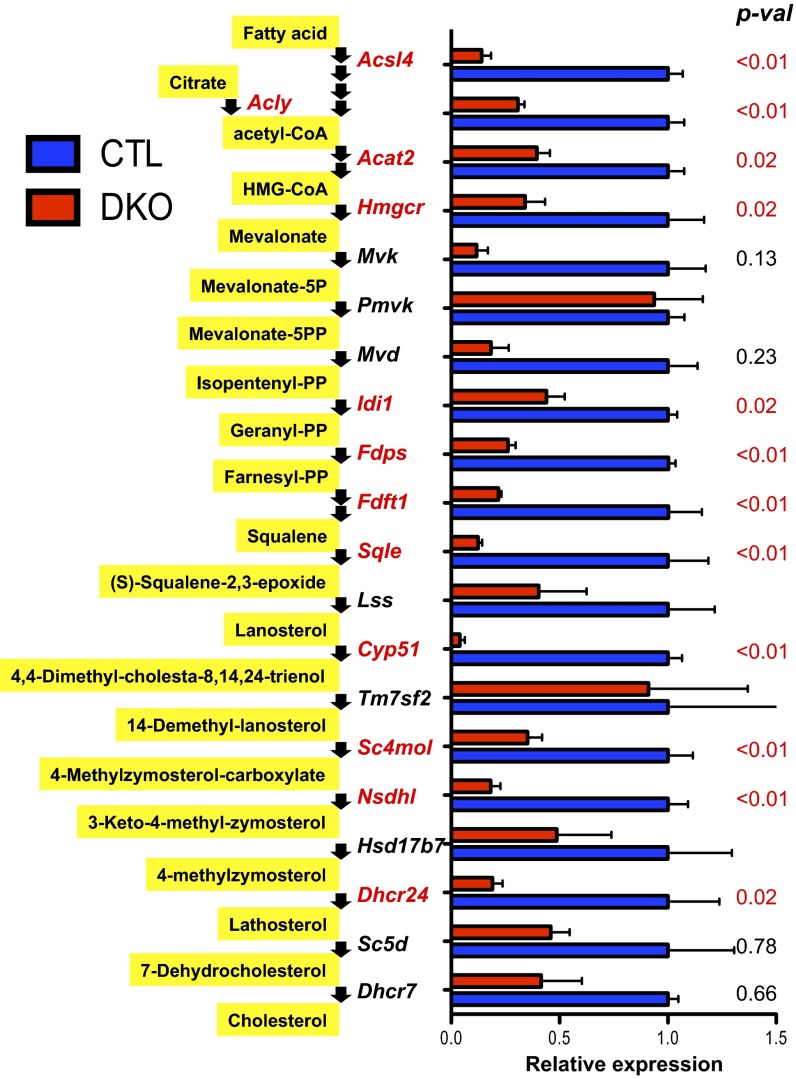
GO analysis for the 40 genes consistently differentially regulated in Tet1/3 DKO embryos (adjusted P value <0.01) identified a number of genes whose products regulate cellular lipid metabolism and cholesterol biosynthesis that were down-regulated in Tet1/3 DKO embryos relative to controls (Fig. 7 and SI Appendix, Fig. S8 A and B and Dataset S1). The transcription factor Srebp2, encoded by the Srebf2 gene, regulates the expression of a large number of these enzymes; it is normally resident in the endoplasmic reticulum but is escorted to the Golgi by a chaperone, Scap, and then cleaved to release its DNA-binding domain when cells become depleted of cholesterol (37). Four of the five Tet1/3 DKO embryos (DKO2–5) showed a strong reduction of Srebf2 mRNA expression; the fifth (DKO1) showed normal Srebf2 expression but strongly diminished expression of Scap mRNA (SI Appendix, Fig. S8C). Using antibodies to N-terminal and C-terminal regions of Srebp2, which detect total (ER and nuclei) and ER-localized forms of Srebp2, respectively (37), we confirmed that Srebp2 protein expression was also significantly down-regulated in Tet1/3 DKO embryos (SI Appendix, Fig. S8 D and E). However, there was no change in the unmethylated (C/5fC/5caC) status of the Srebp2 TSS and promoter at early embryonic stages in Tet1/3 DKO embryos compared with controls (SI Appendix, Fig. S8F).
Fig. 7.
Reproducible down-regulation of genes in the cholesterol biosynthetic pathway in Tet1/3 DKO E3.5 embryos compared with controls. Shown is the sequence of enzymes in the cholesterol biosynthesis pathway, indicating the degree of impaired expression based on RNA-seq data. The averaged normalized counts of three control and five Tet1/3 DKO embryos are plotted. Adjusted P values are shown at Right. Also see SI Appendix, Fig. S8.
Because sonic hedgehog (Shh) requires covalent modification by cholesterol for its activity (38), we speculated that defective cholesterol synthesis during the development of Tet1/3 DKO embryos might result in decreased functioning of the hedgehog-signaling pathway and contribute to developmental abnormalities. Indeed, the phenotypes of late stage (E10.5) Tet1/3 DKO embryos—poor forebrain formation and abnormal facial structures (SI Appendix, Fig. S8G)—resembled the phenotypically heterogeneous holoprosencephaly with consistent forebrain defects and facial abnormalities observed in Shh deficiency as well as in mice with defects in cholesterol synthesis (39, 40). Indeed mRNAs encoding signaling components [Patched1 (Ptch1), hedgehog interacting protein (Hhip), transcription factors (Gli1/2, Pax1/9), and other targets such as Cyclin D2 (Ccnd2)] of the Hedgehog signaling pathway were significantly decreased in E10.5 Tet1/3 DKO embryos relative to controls (SI Appendix, Fig. S8H).
Dysregulation of Imprinted Gene Expression at E10.5.
Even at this late stage, 5hmC levels were considerably lower in Tet1/3 DKO relative to control E10.5 embryos, suggesting incomplete compensation from Tet2 (SI Appendix, Fig. S9A, Left); however overall 5mC levels were not detectably affected in most embryos (SI Appendix, Fig. S9A, Right), suggesting that any changes in DNA modification occurred at a locus-specific level. We tested the DNA modification status of imprinted genes in genomic DNA of four control and four Tet1/3 DKO E10.5 embryos by BstUI digestion and qPCR quantification of methylated genomic DNA (41), after generating standard curves for amplification of short control and long differentially methylated regions (DMR)-containing amplicons (SI Appendix, Fig. S9B). Four different DMRs from two paternally and two maternally imprinted genes were analyzed for their level of DNA modification (5mC+5hmC) and for expression of the corresponding genes. Both maternally (Peg3 and Snrpn) and paternally (H19 and Igf2r) imprinted genes showed increased DNA modification at their DMRs and concomitantly decreased expression in E10.5 embryos (SI Appendix, Fig. S9C), suggesting that Tet1 and Tet3 regulate the expression and modification status of these imprinted genes during later embryonic development.
Discussion
Here we report the effects of TET loss of function in early embryogenesis. With one exception (20), most previous work in this area has focused on loss of function of individual TET proteins (17, 19, 25, 42). Because TET proteins clearly function redundantly (20, 25, 43), we preferred to examine mice with a very substantial loss of TET function during early embryonic development. We chose to delete both Tet1 and Tet3: Tet3 is present in zygotes and at the two-cell stage, whereas Tet1 is expressed from the two-cell stage to the blastocyst. We find (as expected) that the effects of double deletion are considerably more striking than the effects of individual deletion of either Tet1 or Tet3. More importantly, however, analysis of Tet1/3 DKO embryos revealed an unexpected phenotype of a global increase in transcriptional variability.
Even in a population of genetically identical cells, individual cells exhibit a remarkable degree of diversity, most easily visualized through flow cytometric profiles of protein or reporter expression, which often span one to two orders of magnitude. Variations in gene expression patterns within a single cell can be assessed by comparing the levels of two different reporter genes driven off the same promoter (intrinsic noise), whereas those that occur between cells (extrinsic noise) can be examined in many ways, including single-cell RNA sequencing, the technique we have used here (44–46). Cell-to-cell variability in gene expression reflects stochastic variation, differences in cell state or environmental input, and gene regulatory processes including the frequency and robustness of transcriptional initiation (46), the relative abundance of transcriptional regulators (47, 48), the size and frequency of transcriptional bursts (49), the spatial organization of the genome (50), and the relative efficiency of transcription vs. translation (51) (reviewed in refs. 45, 52).
Studies in yeast indicate that several epigenetic regulators can impose transcriptional consistency and suppress transcriptional noise. In many cases, this involves modulation of transcriptional elongation by RNA polymerase II (Pol II), a process linked to cotranscriptional histone modifications that regulate nucleosome disassembly and reassembly. Eviction of acetylated nucleosomes ahead of Pol II permits efficient elongation, whereas reassembly of deacetylated nucleosomes behind the elongating polymerase prevents initiation at spurious sites. Consistent with this scenario, mutations in histone chaperones that promote nucleosome assembly/disassembly, or in enzyme complexes that catalyze or erase histone modifications associated with gene bodies (e.g., H3K9/14 and H4 acetylation, H2B ubiquitylation, and H3K36 di- and trimethylation), affect transcriptional consistency in yeast (49, 53–55).
Our data suggest that in mammals, TET-mediated DNA modifications may play a similar role. In support of this hypothesis, 5hmC is present at the highest levels in gene bodies of the most highly expressed genes (43, 56–60), and TET proteins have been linked to many processes and histone modifications associated with transcriptional variability. (i) In yeast, the Set2 methyltransferase complex travels with RNA Pol II and methylates H3K36 in gene bodies; the H3K36me3 modification is recognized by the Sin3a/Rpd3 histone deacetylase (HDAC) complex, which permits nucleosome reassembly by deacetylating lysines on histones 3 and 4, thus preventing spurious initiation at intragenic sites (53). In mammalian cells, TET functions are linked to both types of histone modifications: high levels of 5hmC are present in the gene bodies of expressed genes; 5hmC distribution in gene bodies is strongly correlated with that of H3K36me3 (43, 56–60); and TET1 coimmunoprecipitates with SIN3A and HDAC1/2 in mouse ES cells (61). (ii) Also in yeast, H2B ubiquitylation has been implicated in nucleosome dynamics (54, 55), and a screen for chromatin regulators that altered the variability of reporter expression in yeast showed that H2B ubiquitylation recruited the Set3C HDAC complex, thereby limiting the high rate of Pol II elongation associated with transcriptional bursts (49). Similarly in mammalian cells, TET2 and TET3 have been linked to H2B ubiquitination through their association with the enzyme O-GlcNAc transferase (OGT) (62–64); OGT attaches O-linked N-acetylglucosamine to H2B (63), a modification required for subsequent H2B ubiquitylation (65). (iii) Finally, the linear extent (“breadth”) of H3K4me3 enrichment around gene promoters has been correlated with transcriptional elongation, Pol II pausing/processivity, increased DNase I accessibility, and increased transcriptional consistency in mammalian cells (66); the TET proteins TET2 and TET3 are implicated in this process because of their association with the SET/COMPASS complex, which contains the SETD1A lysine methyltransferase that deposits H3K4me3 (64).
In a recent elegant study, interference with one of two redundant pathways of gene regulation was shown to result in variable penetrance of an organismal phenotype, ultimately stemming from increased transcriptional noise (48). In Caenorhabditis elegans, end-1 and end-3 both control the expression of elt-2, a transcriptional regulator required for specification of the “E” cell, a precursor cell required for intestinal development. Mutations in upstream regulators (skn-1, med-1/2) that caused decreased expression of end-3 transcripts were also associated with an increased variability of end-1 expression, resulting in the emergence of a threshold effect that decreased elt2 expression and led to incomplete penetrance of the final phenotype, the total number of intestinal cells, and formation of a normal intestine. Although TET enzymes are likely to be ubiquitous global regulators that affect the expression of many genes in mammalian cells, the parallels are clear—double deficiency of Tet1 and Tet3, through strong interference with TET function and pronounced alterations in DNA cytosine modification, may not so much interfere with gene expression per se, as impose a global phenotype of increased transcriptional variability.
It seems self-evident that in a given cell type, transcriptional variability would affect certain gene networks more than others. Indeed, Tet1/3 DKO blastocysts showed reproducible down-regulation of a core set of genes involved in cholesterol biosynthesis and in Hedgehog signaling. Similarly, Tet1-deficient ES cells show clear skewing toward the trophectoderm lineage, both in cell culture and in teratomas (67), and Tet2-deficient hematopoietic stem/precursor cells show skewed differentiation toward the myeloid lineage (19, 68). If TET proteins maintain the consistency of gene transcription during cell lineage specification as suggested here, their loss might be expected to disturb preferentially the expression of gene products—including critical transcription factors and proteins in key signaling pathways—involved in determining cell fate.
Whether TET loss of function acts through increased DNA methylation, loss of oxi-mC production, or both remains unclear. The genome-wide DNA demethylation observed in early embryos appears to be a default pathway independent of TET function or oxi-mC modification: both the 5hmC-modified paternal genome and the unmodified maternal genome are passively demethylated at similar rates during the robust cell divisions that occur in the early embryo (69). Even in zygotes, demethylation of both maternal and paternal pronuclei requires DNA replication, a process only partially dependent on Tet3 (14, 15). For each observed outcome, the relative contributions of oxi-mC production and DNA demethylation may be distinguished by comparing cells deficient in DNA methyltransferases, which would be depleted for both 5mC and oxi-mC, with cells deficient in TET proteins, which lack only oxi-mC. Specific downstream mechanisms include altered recruitment of key transcription factors to their binding sites in DNA, either through direct modification of the binding sequences, modulation of chromatin accessibility, or both; retrotransposon modification and expression, because insertion of these elements is known to create alternative promoters for host genes during early embryogenesis (70, 71); and changes in local histone modifications, as suggested by an early study in which DNA methylation in the gene body was shown to regulate Pol II elongation efficiency through H3K4-di/tri methylation and H3K9/14 acetylation (72). Future studies will clarify these points.
Materials and Methods
Generation of Tet1- and Tet3-Deficient Mice.
The endogenous Tet1 locus was targeted for conditional Tet1, and Tet3 alleles were generated by excision of exons 8, 9, and 10 (SI Appendix, Fig. S1A, Top) and exon 2 respectively (SI Appendix, Fig. S1B).
Single-Embryo and Cell RNA Sequencing.
CTL and Tet1/3 DKO eight-cell embryos and blastocysts were collected at E2.5 and E3.5 respectively. Eight-cell embryos were dissociated, lysed, and converted into double stranded cDNA using SMARTer ultra low RNA kit for Illumina sequencing (Clontech). One nanogram of cDNA for each sample was used for preparing libraries using Nextera XT DNA sample preparation kit (IIlumina) and sequenced using an Illumina HISEQ 2500 instrument. RNA-seq data were mapped against mm9 using Tophat. Sequencing read counts per gene were calculated using htseq-count.
Supplementary Material
Acknowledgments
We thank Susan Togher and Ryan Hastie for animal husbandry and genotyping, Dr. Saguna Krishnaswami (J. Craig Venter Institute) and the Scripps sequencing core for help with single-embryo RNA sequencing, the La Jolla Institute (LJI) sequencing core for single-cell RNA sequencing and reduced-representation bisulfite sequencing, Dr. Klaus Rajewsky for support of S.B.K., Dr. Mohit Jain (University of California, San Diego) for valuable discussions, and Dr. Grzegorz Chodaczek (LJI microscope core) for technical advice. J.K. was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. W.A.P. was supported by the National Science Foundation predoctoral graduate research fellowship while this work was being performed, and subsequently by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. L.C. was the recipient of a Feodor-Lynen fellowship from the Alexander von Humboldt foundation. M.L. is supported by the Max Planck Society within its International Max Planck Research School for Computational Biology and Scientific Computing program (IMPRS-CBSC). A.T. was the recipient of an Irvington postdoctoral fellowship from the Cancer Research Institute. This work was supported by NIH R01 Grants AI044432 and HD065812 (to A.R.) and a Director’s New Innovator Award (DP2-OD-008646-01) (to S.K.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The study accession no. is: PRJEB9516. The study unique name is: ena-STUDY-MPIMG-02-06-2015-10:21:27:198-321.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510510112/-/DCSupplemental.
References
- 1.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pastor WA, Aravind L, Rao A. TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14(6):341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ooi SK, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122(Pt 16):2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286(41):35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, et al. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol. 2012;8(4):328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hackett JA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339(6118):448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackett JA, Surani MA. DNA methylation dynamics during the mammalian life cycle. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20110328. doi: 10.1098/rstb.2011.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 12.Wossidlo M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 13.Iqbal K, Jin SG, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA. 2011;108(9):3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen L, et al. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell. 2014;15(4):459–470. doi: 10.1016/j.stem.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo F, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15(4):447–458. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139(11):1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang RR, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13(2):237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudenko A, et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79(6):1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko M, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA. 2011;108(35):14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawlaty MM, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24(3):310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dawlaty MM, et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29(1):102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu X, et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 2014;14(4):512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Inoue A, Matoba S, Zhang Y. Transcriptional activation of transposable elements in mouse zygotes is independent of Tet3-mediated 5-methylcytosine oxidation. Cell Res. 2012;22(12):1640–1649. doi: 10.1038/cr.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macfarlan TS, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487(7405):57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9(2):166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaguchi S, et al. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492(7429):443–447. doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18(4):675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Biase F, Cao X, Zhong S. Cell fate inclination within 2-cell and 4-cell mouse embryos revealed by single-cell RNA sequencing. Genome Res. 2014;24(11):1787–1796. doi: 10.1101/gr.177725.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko M, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497(7447):122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichols J, et al. Complementary tissue-specific expression of LIF and LIF-receptor mRNAs in early mouse embryogenesis. Mech Dev. 1996;57(2):123–131. doi: 10.1016/0925-4773(96)00531-x. [DOI] [PubMed] [Google Scholar]
- 31.Ware CB, et al. Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development. 1995;121(5):1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- 32.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473(7347):389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5(1):e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Booth MJ, Raiber EA, Balasubramanian S. Chemical methods for decoding cytosine modifications in DNA. Chem Rev. 2015;115(6):2240–2254. doi: 10.1021/cr5002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484(7394):339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakanishi MO, et al. Trophoblast-specific DNA methylation occurs after the segregation of the trophectoderm and inner cell mass in the mouse periimplantation embryo. Epigenetics. 2012;7(2):173–182. doi: 10.4161/epi.7.2.18962. [DOI] [PubMed] [Google Scholar]
- 37.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109(9):1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Incardona JP, Eaton S. Cholesterol in signal transduction. Curr Opin Cell Biol. 2000;12(2):193–203. doi: 10.1016/s0955-0674(99)00076-9. [DOI] [PubMed] [Google Scholar]
- 39.Haas D, Muenke M. Abnormal sterol metabolism in holoprosencephaly. Am J Med Genet C Semin Med Genet. 2010;154C(1):102–108. doi: 10.1002/ajmg.c.30243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muenke M, Beachy PA. Genetics of ventral forebrain development and holoprosencephaly. Curr Opin Genet Dev. 2000;10(3):262–269. doi: 10.1016/s0959-437x(00)00084-8. [DOI] [PubMed] [Google Scholar]
- 41.Lorthongpanich C, et al. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science. 2013;341(6150):1110–1112. doi: 10.1126/science.1240617. [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature. 2013;504(7480):460–464. doi: 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2014;111(4):1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 45.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467(7312):167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304(5678):1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453(7194):544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463(7283):913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinberger L, et al. Expression noise and acetylation profiles distinguish HDAC functions. Mol Cell. 2012;47(2):193–202. doi: 10.1016/j.molcel.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCullagh E, Seshan A, El-Samad H, Madhani HD. Coordinate control of gene expression noise and interchromosomal interactions in a MAP kinase pathway. Nat Cell Biol. 2010;12(10):954–962. doi: 10.1038/ncb2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31(1):69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 52.Raser JM, O’Shea EK. Noise in gene expression: Origins, consequences, and control. Science. 2005;309(5743):2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123(4):581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 54.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31(1):57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 55.Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011;25(21):2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsagaratou A, et al. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc Natl Acad Sci USA. 2014;111(32):E3306–E3315. doi: 10.1073/pnas.1412327111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gan H, et al. Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat Commun. 2013;4:1995. doi: 10.1038/ncomms2995. [DOI] [PubMed] [Google Scholar]
- 59.Song CX, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29(1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vella P, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49(4):645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 63.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493(7433):561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deplus R, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32(5):645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fujiki R, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480(7378):557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benayoun BA, et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158(3):673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8(2):200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334(6053):194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peaston AE, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7(4):597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 71.Xie D, et al. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 2010;20(6):804–815. doi: 10.1101/gr.100594.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11(11):1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.