Significance
Proper function of RNA is critical to the health and maintenance of a cell, and its misregulation plays a critical role in the development of many disorders. To date, the ability to study RNA has been severely limited; many analytical techniques focus only on quantifying expression levels of transcripts and are not capable of reporting on intracellular localization, which has emerged as a critical component of RNA function. Similarly, techniques capable of probing RNA localization only offer snapshots in fixed cells. Herein we introduce a novel nanoconjugate, termed the Sticky-flare, capable of reporting on both of these critical components in live cells, thereby gaining a more complete picture or RNA function than any analytical technique to date.
Keywords: FISH, in situ hybridization, RNA detection, RNA localization, nanotechnology
Abstract
We report a novel spherical nucleic acid (SNA) gold nanoparticle conjugate, termed the Sticky-flare, which enables facile quantification of RNA expression in live cells and spatiotemporal analysis of RNA transport and localization. The Sticky-flare is capable of entering live cells without the need for transfection agents and recognizing target RNA transcripts in a sequence-specific manner. On recognition, the Sticky-flare transfers a fluorophore-conjugated reporter to the transcript, resulting in a turning on of fluorescence in a quantifiable manner and the fluorescent labeling of targeted transcripts. The latter allows the RNA to be tracked via fluorescence microscopy as it is transported throughout the cell. We use this novel nanoconjugate to analyze the expression level and spatial distribution of β-actin mRNA in HeLa cells and to observe the real-time transport of β-actin mRNA in mouse embryonic fibroblasts. Furthermore, we investigate the application of Sticky-flares for tracking transcripts that undergo more extensive compartmentalization by fluorophore-labeling U1 small nuclear RNA and observing its distribution in the nucleus of live cells.
The study of RNA is a critical component of biological research and in the diagnosis and treatment of disease. Recently, the localization of mRNA has been identified as an essential process for a number of cellular functions, including restricting the production of certain proteins to specific compartments within cells (1). For instance, synaptic potentiation, the basis of learning and memory, relies on the local translation of specific mRNAs in pre- and postsynaptic compartments (2). Likewise, the misregulation of RNA distribution is associated with many disorders, including mental retardation, autism, and cancer metastasis (3–5). However, despite the significant role of mRNA transport and localization in cellular function, the available methods to visualize these phenomena are severely limited. For example, FISH, the most commonly used technique to analyze spatial distribution of RNA, requires fixation and permeabilization of cells before analysis (6). As a result, analysis of dynamic RNA distribution is restricted to a single snapshot in time (7, 8). With such a limitation, understanding the translocation of RNA with respect to time, cell cycle, or external stimulus is difficult if not impossible. Furthermore, fixed cell analysis is a lengthy and highly specialized procedure due to the number of steps necessary to prepare a sample. Fixation, permeabilization, blocking, and staining processes each require optimization and vary based on cell type and treatment conditions, rendering FISH prohibitively complicated in many cases. Likewise, live cell analysis platforms such as molecular beacons require toxic transfection techniques, such as microinjection or lipid transfection, and are rapidly sequestered to the nucleus on cellular entry (9, 10). Recently more sophisticated live cell analyses have been developed that use genetic engineering to introduce exogenous hybrid genes via viral or plasmid transfection. These hybrid transcripts include added aptamer motifs that bind to subsequently introduced or coexpressed fluorescent reporters such as malachite green (11), GFP (12, 13), or GFP derivatives (14–16). Such techniques offer dynamic visualization of RNA; however, the reliance of genetic engineering and exogenous hybrid RNA sequences makes such techniques highly involved and does not allow for the quantification of endogenous gene expression. Thus, to accurately study the dynamics of endogenous RNA, a new type of analysis platform is required. Ideally such a platform should be capable of both quantifying transcript expression and tracking intracellular transcripts in real time, without the need for transfection agents or specialized techniques. Herein we present such a platform and use the newly developed construct to perform RNA quantification and real-time analysis of dynamic RNA translocation.
Previously our group introduced the concept of the Nanoflare, a spherical nucleic acid (SNA) conjugate capable of quantifying RNA concentration in live cells with single cell resolution (17–22). The Nanoflare consists of a 13-nm gold nanoparticle core functionalized with a densely packed, highly oriented shell of oligonucleotides designed to be antisense to a target RNA transcript. A fluorophore-conjugated reporter strand, termed the flare sequence, is subsequently hybridized to the antisense oligonucleotides via complementary base pairing. Hybridization of the flare sequence holds the fluorophore in close proximity to the gold core of the SNA, effectively quenching fluorescence. However, upon cellular entry the antisense capture sequences of the Nanoflare bind to targeted transcripts, forming longer, more stable duplexes. These binding events displace the flares from the gold surface, resulting in a discrete “turning on” of fluorescence, the extent of which is quantifiably related to the expression level of the target RNA. Importantly, the Nanoflare enters live cells via receptor-mediated endocytosis without the need for harmful transfection techniques (19, 23–28) and with negligible cytotoxicity (29) and immunogenicity (30). As a result, the Nanoflare has grown into a powerful and prolific tool in biology and medical diagnostics, with ∼1,600 unique forms commercially available today (sold under the SmartFlare trade name).
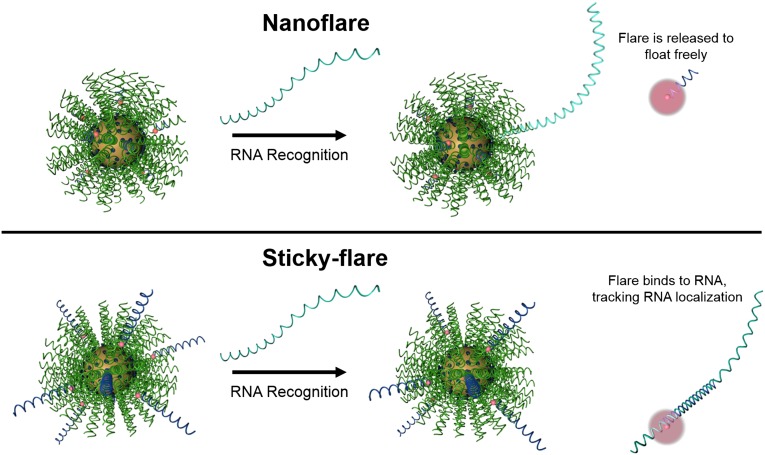
Conventional Nanoflares do not allow one to determine the spatial distribution of RNA within the cell. Indeed, Nanoflare-transcript binding results in a displacement of the fluorophore-labeled sequence from the nanoparticle construct, which spatially separates the fluorescent reporter and transcript (Fig. 1, Upper). However, with a simple design change where the flare sequence is made longer and complementary to the target RNA transcript, a new construct can be realized that can track and quantify RNA within a cell (Fig. 1, Lower). Herein, we report the synthesis and development of such a construct, termed the Sticky-flare, and investigate its use as a platform for RNA quantification and real-time tracking of transcripts as they are transported within live HeLa and mouse embryonic fibroblast (MEF) cells. These constructs and their activity in cells represent the only way of measuring both the quantity and location of mRNA in live cells.
Fig. 1.
Schematic of RNA recognition by the Nanoflare (Upper) and Sticky-flare (Lower). On recognition, the Nanoflare binds to RNA targets and releases a fluorophore-labeled oligonucleotide reporter (flare) to float freely in the cytoplasm. In contrast, flares from the Sticky-flare are longer and complementary to RNA targets, which allows them to bind transcripts and act as fluorescent labels for intracellular tracking.
Results and Discussion
Evaluation of Target Recognition by Sticky-Flares.
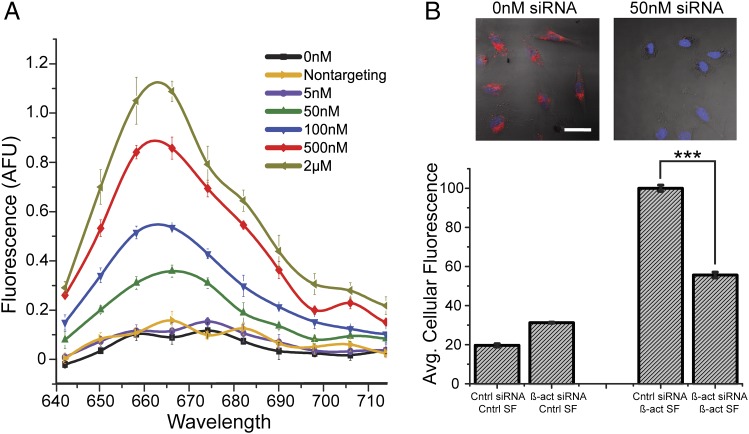
Sticky-flares were first evaluated in vitro for their utility in quantifying complementary nucleic acid targets in a sequence-specific manner. Solutions of β-actin targeting Sticky-flares (1 nM) were evaluated before and after the addition of fully-complementary targets, ranging from 5 nM to 2 μM in PBS. On addition of a complementary target, a resulting increase in fluorescence was observed in a manner proportional to the concentration of target added, showing the Sticky-flares can be used to quantify the concentration of complementary sequences in vitro. Importantly, the addition of noncomplementary target (2 μM) had no measurable effect (Fig. 2A).
Fig. 2.
Characterization of sequence-specific target recognition and quantification using the Sticky-flare. (A) In vitro assay quantifying the release of fluorescent flares resulting from sequence-specific recognition of a complementary target over a range of concentrations in PBS buffer and the lack of recognition when presented with a high concentration (2 µM) of a noncomplementary target (Sticky-flare concentration = 1 nM; sequence information given in Methods). (B) (Upper) Representative confocal images of HeLa cells treated with β-actin Sticky-flares; (Upper Left) cells treated with RNAiMAX (Life Technologies) but no siRNA and (Upper Right) cells treated with 50 nM siRNA designed to knockdown β-actin delivered via RNAiMAX. (Lower) Graph of β-actin knockdown in HeLa cells measured using Sticky-flares combined with flow cytometry. Cells treated with control siRNA (incorrect sequence for knocking down β-actin or any known gene) yields cells with the highest average fluorescence. When cells are treated with β-actin siRNA (50 nM), they exhibit an ∼45% reduction in average fluorescence (P < 0.001). The bars on the far left are control experiments probing fluorescence from cells treated with Sticky-flares that are not specific for β-actin mRNA or any known gene; they represent the background fluorescence from a Sticky-flare experiment.
Next, β-actin Sticky-flares were evaluated in a cell culture model by flow cytometry. HeLa human cervical cancer cells were treated with either control (nontargeting) siRNA (50 nM) delivered via Lipofectamine RNAiMAX transfection agent, anti–β-actin siRNA with RNAiMAX, or RNAiMAX alone for 24 h, after which the media was replaced with media containing Sticky-flares targeting either β-actin mRNA or U1 short nuclear RNA (negative control). After an additional 18-h incubation, the fluorescence of each cell was quantified. With β-actin Sticky-flares, we detected significant knockdown of RNA expression levels compared with cells treated with control siRNA or transfection agent alone, whereas the control Sticky-flares showed no significant difference with any treatment (Fig. 2B and Fig. S1).
Fig. S1.
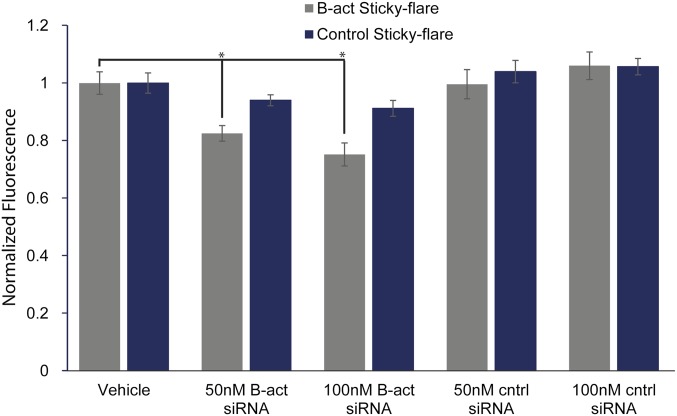
Detection of gene knockdown via Sticky-flares. β-Actin targeting Sticky-flares (gray) detects knockdown of the gene using varying quantities of β-actin siRNA, whereas control Sticky-flares (blue) remain largely unchanged. *P < 0.01.
Intracellular RNA Tracking by Sticky-Flares.
Our ability to track the spatial distribution of RNA with Sticky-flares was evaluated using confocal microscopy. Two genes with disparate intracellular function and localization patterns were chosen to analyze spatial distribution within cells: β-actin mRNA and U1 small nuclear RNA (snRNA). In previous reports, β-actin mRNA was found to localize at the growth cones of lamellae in embryonic fibroblasts (31). In contrast, U1 snRNA is imported into the nucleus, where it acts as a key component of the spliceosome (32).
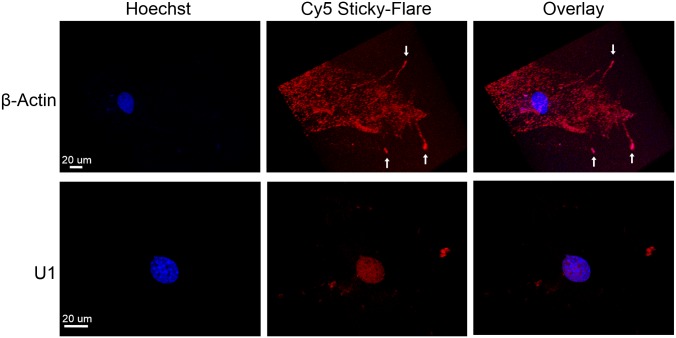
MEFs were cultured in glass-bottomed cell culture chambers with Sticky-flares for 12 h, after which the cells were treated with a nuclear stain (Hoechst) for 10 min and imaged in live form. Fluorescence from cells treated with β-actin Sticky-flares exhibited punctate fluorescence throughout the cell body and a demonstrable preference for the growth cone region of lamellae extensions (Fig. 3). Additional highly fluorescent regions can be seen within the lamellae extensions, marking β-actin RNA being actively transported to and from the growth cone. This active transport is further analyzed below. Importantly, Sticky-flares are not limited to use in live cells and can be used to verify RNA localization in fixed cells as a convenient control. Therefore, fixed and permeabilized MEFs were treated with Sticky-flares, confirming the growth cone specific localization observed in live cells (Fig. S2). Contrastingly, MEFs treated with Sticky-flares targeting U1 snRNA distinctly showed internuclear fluorescence. Importantly, SNA constructs are sequestered to the cytosol of live cells and cannot enter the nucleus on their own (33). Thus, the internuclear fluorescence of U1 Sticky-flares indicates specific release from the nanoparticle surface and subsequent transport of the fluorescent-labeled RNA target into the nucleus.
Fig. 3.
RNA localization in MEFs. β-Actin targeting Sticky-flares localize to the growth cone of growing lamellae, where β-actin RNA is found. Contrastingly, Sticky-flares targeting the U1 nuclear RNA localize to the nucleus. Red, Cy5 Sticky-flare; blue, nuclear stain (Hoechst).
Fig. S2.
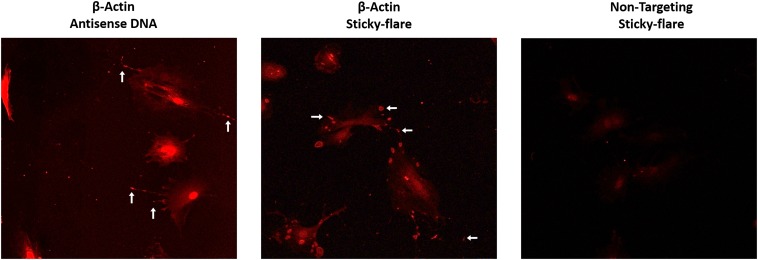
Sticky-flares in fixed cells. Mouse embryonic fibroblasts fixed, permeabilized, and treated with either antisense DNA (Left) or Sticky-flares (Center) detect β-actin mRNA at the leading edge of growing lamellae, similar to the localization of β-actin Sticky-flares in live cells.
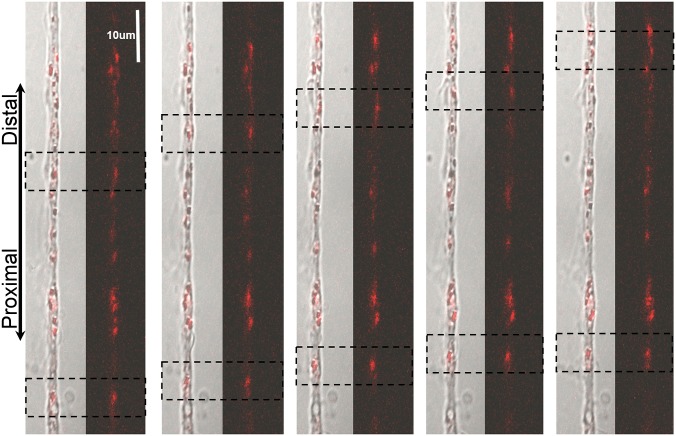
Beyond studying the final localization of mRNA strands, the facile, noninvasive nature of the Sticky-flare allows for real-time observation of dynamic RNA translocation in live cells. To demonstrate this, MEFs treated with β-actin Sticky-flares were imaged every 10 s with a confocal microscope for a total of 10 min. When the plane of imaging was focused on lamellae, transport of β-actin mRNA was observed primarily (but not exclusively) in the distal direction toward the growth cone (Fig. 4 and Movie S1). Furthermore, when focused directly at the body of the cell, RNA dynamics become even more evident, with hundreds of fluorophore-labeled β-actin sequences being transported throughout the cell (Movie S2).
Fig. 4.
Observation of dynamic β-actin mRNA transport in MEF cells. Endogenously expressed β-actin mRNA is transported distally toward the growth cone (red, Cy5-labeled Sticky-flare). Dashed boxes indicate the labeled RNA being tracked. Each panel indicates a 50-s advancement.
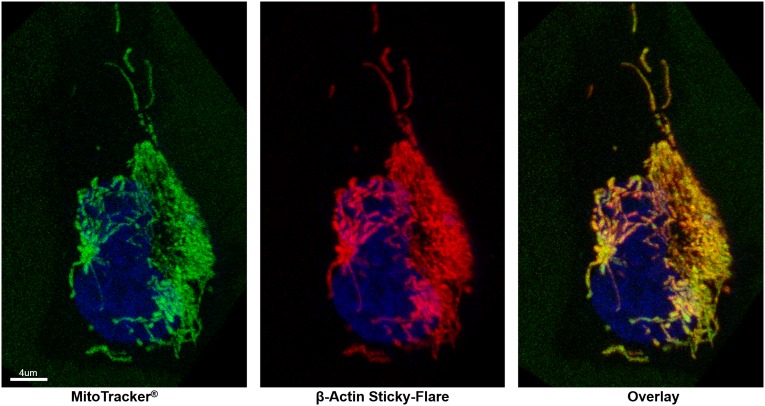
Similar analyses were performed in HeLa cells. In this case, β-actin Sticky-flares exhibited a starkly different intracellular distribution, showing a high degree of colocalization with mitochondria (Fig. 5). This colocalization of β-actin mRNA and mitochondria in HeLa cells has not been demonstrated before, and the reason behind such highly spatially restricted expression is not known. However, this distribution closely parallels the mitochondrial localization of K-RAS and GAPDH in HeLa cells, which has been reported in prior work (34).
Fig. 5.
β-Actin mRNA colocalizes with mitochondria in HeLa cells. Red, Cy5-labeled Sticky-flare; green, mitochondrial marker; blue, nuclear stain (Hoechst).
To observe dynamic RNA movement and to further probe the β-actin/mitochondrial colocalization, fluorescence was monitored in HeLa cells starved by culturing in Eagle’s balanced salt solution. On starvation, the mitochondrial network collapses and mitochondria and RNA both migrated toward the perinuclear region, forming a more punctate expression pattern and maintaining colocalization over the time observed (Movie S3).
Conclusion
We synthesized and introduced a novel nanoconjugate termed the Sticky-flare, which is a result of redesigning the conventional Nanoflare or SmartFlare to extend the capabilities of the unique SNA platform. The Sticky-flare uses the same targeting strategy of recognizing and quantifying RNA targets in live cells but has the additional functionality of being able to bind to target transcripts, enabling further analysis of RNA transport and localization. As such, this is the first demonstration, to our knowledge, of a single platform that enables a complete analysis of RNA function in live cells and overcomes many limitations of previous analytical techniques. Due to the similar nature of the Nanoflare and Sticky-flare constructs, future studies will explore the development of multiplexed Sticky-flare constructs that would be capable of targeting multiple transcripts at once, to allow for an internal control in RNA quantification and for simultaneous tracking of multiple transcripts. We anticipate the Sticky-flare will be a valuable tool for investigating proper RNA function and its misregulation in disease, and will make such studies accessible to a broader community, given the ease of its application in cell culture. Further, the Sticky-flare may improve analyses in other model systems where asymmetric RNA expression is an essential component, such as embryonic development, tissue and organ regeneration, and neurobiology.
Methods
Sticky-Flare Synthesis.
Oligonucleotides were synthesized using standard solid-phase phosphoramidite chemistry (Expedite 8909 Nucleotide Synthesis System; ABI). All reagents were purchased from Glen Research. Oligonucleotides were purified by reverse-phase HPLC. The oligonucleotide sequences used in this study are as follows: nontargeting alkyl-thiol, CGT CTA CCT TCG CGC AAA AAA A-alkane thiol; nontargeting flare, Cy5-GCG CGA AGG TAG ACG GAG TCG GTC GA; β-actin alkyl-thiol, CCG GCA TGT GCA A AAA AAA A-alkane thiol; β-actin flare, Cy5-TTG CAC ATG CCG GAG CCG TTG TCG ACG A; β-actin target, TCG TCG ACA ACG GCT CCG GCA TGT GCA A; U1 alkyl-thiol, TAT CCA TTG CAC TCC GG AAA AAA A-SH; U1 flare, Cy5-CCG GAG TGC AAT GGA TAA GCC TCG CC; U1 target, GGC GAG GCT TAT CCA TTG CAC TCC GG.
To prepare the DNA-functionalized SNA conjugates, alkylthiol-terminated oligonucleotides (3 μM) were combined with citrate-capped 13-nm gold particles (13 nM) and 0.01% Tween-20 and incubated for 1 h at room temperature. Next, phosphate buffer (pH = 7.4) and sodium chloride were added to a final concentration of 5 and 150 mM, respectively, and incubated overnight. Then, sodium chloride was added in 0.05-M increments over 3 h to achieve a final NaCl concentration of 300 mM, and the particles were shaken at room temperature for 4 h. Finally, the conjugates were purified by centrifugation and redispersed in PBS.
Flares were hybridized on the purified DNA-gold nanoparticless by adding a stoichiometric equivalent of 10 flares per nanoparticle. The solution was then heated to 70 °C and slowly cooled to room temperature overnight to facilitate hybridization. The resulting Sticky-flares were then sterilized using a 0.2-μm acetate syringe filter (GE Healthcare) to prevent cell contamination and stored at 4 °C.
Ex Vivo Quantification of Sequence-Specific Target Recognition.
Sticky-flares were incubated in PBS (Gibco) at a concentration of 1 nM. Subsequently, fully complementary oligonucleotide targets (DNA) were added to the solution at the concentrations listed in the text above. This mixture was allowed to incubate at room temperature for 5 min, after which the resulting fluorescence was quantified via a BioTek Synergy H4 fluorescence plate reader.
Cell Culture and Sticky-Flare Treatment.
HeLa cells were cultured in DMEM (Gibco) supplemented with 10% (vol/vol) FBS and 1% penicillin/streptomycin. Gene knockdown was effected by treating with 50 nM anti–β-actin (Santa Cruz Biotechnology) for 24 h with Lipofectamine RNAiMAX according to the recommended protocol. Control siRNA experiments (Santa Cruz Biotechnology) were conducted in a similar manner. Cells were then washed once with PBS and further incubated with 400 pM Sticky-flares in OptiMEM (Gibco) for an additional 24 h. Fluorescence of trypsinized cells was quantified by using a Guava Easycyte HT flow cytometer (Millipore). Confocal microscopy was performed with Zeiss 510 (Zeiss) and SP5 (Leica) confocal microscopes. Mitochondria were stained using CellLight Mitochondria-GFP (Life Technologies).
Fixing and permeabilization of MEFs was performed by treating cells with 4% (vol/vol) paraformaldehyde for 10 min, followed by 0.1% Triton-X for 5 min. Cells were then rinsed three times with PBS and treated with either 1 nM Sticky-flares or 100 nM antisense DNA strands for 1 h. Following this incubation, cells were rinsed a further three times with PBS and imaged in the same fashion as live cells.
Supplementary Material
Acknowledgments
Confocal analysis was performed at the Quantitative Bioelemental Imaging Center (Northwestern University). This project was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases Grants R01AR060810 and R21AR062898 and Center for Cancer Nanotechnology Excellence initiative of the National Institutes of Health under Award U54 CA151880. P.S.R. is grateful to the National Science Foundation for a graduate research fellowship under Grant DGE-1324585, and W.E.B. is grateful for the Ryan Fellowship from Northwestern University.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510581112/-/DCSupplemental.
References
- 1.Thomas MG, Pascual ML, Maschi D, Luchelli L, Boccaccio GL. Synaptic control of local translation: The plot thickens with new characters. Cell Mol Life Sci. 2014;71(12):2219–2239. doi: 10.1007/s00018-013-1506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiler IJ, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci USA. 1997;94(10):5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu-Yesucevitz L, et al. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31(45):16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassell GJ, Warren ST. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60(2):201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shestakova EA, Wyckoff J, Jones J, Singer RH, Condeelis J. Correlation of β-actin messenger RNA localization with metastatic potential in rat adenocarcinoma cell lines. Cancer Res. 1999;59(6):1202–1205. [PubMed] [Google Scholar]
- 6.Bauman JGJ, Wiegant J, Borst P, van Duijn P. A new method for fluorescence microscopical localization of specific DNA sequences by in situ hybridization of fluorochromelabelled RNA. Exp Cell Res. 1980;128(2):485–490. doi: 10.1016/0014-4827(80)90087-7. [DOI] [PubMed] [Google Scholar]
- 7.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348(6233):aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45(3):407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- 9.Leonetti JP, Mechti N, Degols G, Gagnor C, Lebleu B. Intracellular distribution of microinjected antisense oligonucleotides. Proc Natl Acad Sci USA. 1991;88(7):2702–2706. doi: 10.1073/pnas.88.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kedmi R, Ben-Arie N, Peer D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials. 2010;31(26):6867–6875. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 11.Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J Am Chem Soc. 2003;125(48):14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 12.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2(4):437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 13.Fusco D, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13(2):161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333(6042):642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat Methods. 2013;10(12):1219–1224. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filonov GS, Moon JD, Svensen N, Jaffrey SR. Broccoli: Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc. 2014;136(46):16299–16308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-flares: Probes for transfection and mRNA detection in living cells. J Am Chem Soc. 2007;129(50):15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prigodich AE, et al. Multiplexed nanoflares: mRNA detection in live cells. Anal Chem. 2012;84(4):2062–2066. doi: 10.1021/ac202648w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosi NL, et al. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312(5776):1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 20.Prigodich AE, et al. Nano-flares for mRNA regulation and detection. ACS Nano. 2009;3(8):2147–2152. doi: 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Will B, et al. 2012. Biochemistry and biomedical applications of spherical nucleic acids (SNAs). Nanomaterials for Biomedicine (American Chemical Society, Washington, DC), Vol 1119, pp 1-20.
- 22.Halo TL, et al. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc Natl Acad Sci USA. 2014;111(48):17104–17109. doi: 10.1073/pnas.1418637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi CHJ, Hao L, Narayan SP, Auyeung E, Mirkin CA. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc Natl Acad Sci USA. 2013;110(19):7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giljohann DA, et al. Gold nanoparticles for biology and medicine. Angew Chem Int Ed Engl. 2010;49(19):3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel PC, et al. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjug Chem. 2010;21(12):2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao L, Patel PC, Alhasan AH, Giljohann DA, Mirkin CA. Nucleic acid-gold nanoparticle conjugates as mimics of microRNA. Small. 2011;7(22):3158–3162. doi: 10.1002/smll.201101018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cutler JI, Auyeung E, Mirkin CA. Spherical nucleic acids. J Am Chem Soc. 2012;134(3):1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 28.Jensen SA, et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med. 2013;5(209):209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massich MD, Giljohann DA, Schmucker AL, Patel PC, Mirkin CA. Cellular response of polyvalent oligonucleotide-gold nanoparticle conjugates. ACS Nano. 2010;4(10):5641–5646. doi: 10.1021/nn102228s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massich MD, et al. Regulating immune response using polyvalent nucleic acid-gold nanoparticle conjugates. Mol Pharm. 2009;6(6):1934–1940. doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu G, et al. Interactions of elongation factor 1α with F-actin and β-actin mRNA: Implications for anchoring mRNA in cell protrusions. Mol Biol Cell. 2002;13(2):579–592. doi: 10.1091/mbc.01-03-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmo-Fonseca M, et al. In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J. 1991;10(7):1863–1873. doi: 10.1002/j.1460-2075.1991.tb07712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel PC, Giljohann DA, Seferos DS, Mirkin CA. Peptide antisense nanoparticles. Proc Natl Acad Sci USA. 2008;105(45):17222–17226. doi: 10.1073/pnas.0801609105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santangelo PJ, Nitin N, Bao G. 2005. Direct visualization of mRNA colocalization with mitochondria in living cells using molecular beacons. BIOMEDO 10(4):044025–044026.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.