Significance
We tend to think of our genome as an unchanging store of information; however, recent evidence suggests that genomes vary between different cells in the same organism. How these differences arise and what effects they have remain unknown, but clearly our genome can change. In a single-celled organism, genome changes occur at random, and advantageous changes slowly propagate by natural selection. However, it is known that the DNA encoding ribosomes can change simultaneously in a whole population. Here we show that signaling pathways that sense environmental nutrients control genome change at the ribosomal DNA. This demonstrates that not all genome changes occur at random and that cells possess specific mechanisms to optimize their genome in response to the environment.
Keywords: ribosomal DNA, homologous recombination, Sir2, copy number variation, TOR
Abstract
Repeated regions are widespread in eukaryotic genomes, and key functional elements such as the ribosomal DNA tend to be formed of high copy repeated sequences organized in tandem arrays. In general, high copy repeats are remarkably stable, but a number of organisms display rapid ribosomal DNA amplification at specific times or under specific conditions. Here we demonstrate that target of rapamycin (TOR) signaling stimulates ribosomal DNA amplification in budding yeast, linking external nutrient availability to ribosomal DNA copy number. We show that ribosomal DNA amplification is regulated by three histone deacetylases: Sir2, Hst3, and Hst4. These enzymes control homologous recombination-dependent and nonhomologous recombination-dependent amplification pathways that act in concert to mediate rapid, directional ribosomal DNA copy number change. Amplification is completely repressed by rapamycin, an inhibitor of the nutrient-responsive TOR pathway; this effect is separable from growth rate and is mediated directly through Sir2, Hst3, and Hst4. Caloric restriction is known to up-regulate expression of nicotinamidase Pnc1, an enzyme that enhances Sir2, Hst3, and Hst4 activity. In contrast, normal glucose concentrations stretch the ribosome synthesis capacity of cells with low ribosomal DNA copy number, and we find that these cells show a previously unrecognized transcriptional response to caloric excess by reducing PNC1 expression. PNC1 down-regulation forms a key element in the control of ribosomal DNA amplification as overexpression of PNC1 substantially reduces ribosomal DNA amplification rate. Our results reveal how a signaling pathway can orchestrate specific genome changes and demonstrate that the copy number of repetitive DNA can be altered to suit environmental conditions.
Eukaryotic genomes contain abundant multicopy sequences, ranging from low copy segmental duplications to the giant tandem arrays found at key functional regions such as centromeres, telomeres, and the ribosomal DNA (rDNA) (1). Copy number variation of protein coding genes has been linked with multiple diseases, suggesting copy number has significant effects on gene expression (2, 3). The budding yeast rDNA has been used extensively as a model system for dynamic copy number change in repetitive DNA. The rDNA consists of a tandem array of ∼180 tandem copies, each containing genes for the 35S and 5S preribosomal RNAs. rDNA copy number is stable in a population, but recombination between rDNA copies is frequent because of the presence of a recombination-stimulating HOT1 element in each copy (4–7). The HOT1 element includes a unidirectional replication fork barrier dependent on the Fob1 protein that halts replication forks moving in the opposite direction to RNA Pol I (8); Fob1 is required both for ectopic HOT1 activity and for rDNA recombination (9).
The primary model for Fob1-stimulated recombination involves breakage of a replication fork stalled at the replication fork barrier, leaving a single-ended double-strand break that can initiate break-induced replication (BIR) with the sister chromatid (10). The rate of recombination between copies is regulated by the histone deacetylase (HDAC) Sir2 (11–13), and recombination through this pathway is strictly dependent on the homologous recombination (HR) machinery (14). Frequent recombination events are required to maintain rDNA homogeneity (15, 16) and result in the loss of markers integrated in the rDNA (7). However, this HR-dependent pathway regulated by Sir2 is nondirectional; repeat gain and loss occurs at equivalent rates, so no change in average copy number is observed over time (13).
Nonetheless, concerted increases in rDNA copy number occur in Saccharomyces cerevisiae populations with low or limiting rDNA copy number (10, 17), as well as in a variety of other organisms (18, 19), showing that an rDNA amplification pathway must also exist. This may overlap with the Sir2-regulated pathway but must be distinct, as complete de-regulation of rDNA BIR in sir2Δ mutants does not cause constitutive rDNA amplification (13). In contrast, constitutive rDNA amplification has been reported in cells lacking histone H3 K56 acetyltransferase activity, suggesting H3 K56 acetylation may control entry to an rDNA amplification pathway (20, 21). Surprisingly, rDNA amplification can occur in the absence of critical HR proteins, including strand exchange factor Rad52 (20), suggesting two mechanistically separable pathways exist: the HR-dependent BIR pathway and a non-HR-dependent amplification pathway.
Advantageous copy number changes are generally assumed to occur at random and then spread through a population by natural selection. However, increasing rDNA copy number in yeast does not provide a detectable growth advantage under laboratory conditions, so rapid rDNA amplifications cannot be explained by such a mechanism (10, 22). This implies the existence of a mechanism that can monitor rDNA copy number and instigate rDNA amplification when required. The target of rapamycin (TOR) pathway stimulates marker loss from the rDNA via the nondirectional BIR pathway (23, 24) and is also known to modulate H3 K56 acetylation in the rDNA (25). Because the TOR pathway responds to environmental nutrient availability (26) and represses rDNA recombination during caloric restriction (24, 27), we asked whether TOR signaling controls rDNA amplification. Here we show that rDNA amplification in budding yeast occurs through two pathways that are coordinately regulated by TOR signaling, providing a clear demonstration that the copy number of certain loci can be tailored to suit the current environment.
Results
rDNA Amplification Is Controlled by the TOR Pathway.
rDNA copy number is stably maintained at 150–200 repeats in wild-type yeast, and cells with low rDNA copy numbers (fewer than ∼80 copies) undergo rapid amplification toward the wild-type level (10, 17). However, low copy number rDNA arrays cannot amplify in the absence of Fob1, and amplification in fob1Δ cells is initiated by the introduction of a Fob1 expression plasmid (17, 22, 28). We exploited this assay to test whether TOR signaling is required for rDNA amplification in cells with ∼35 rDNA repeats (rDNA35), which, in accord with previous data, have only a minimal growth defect compared with isogenic cells with 180 rDNA copies (SI Appendix, Fig. S1 A and B).
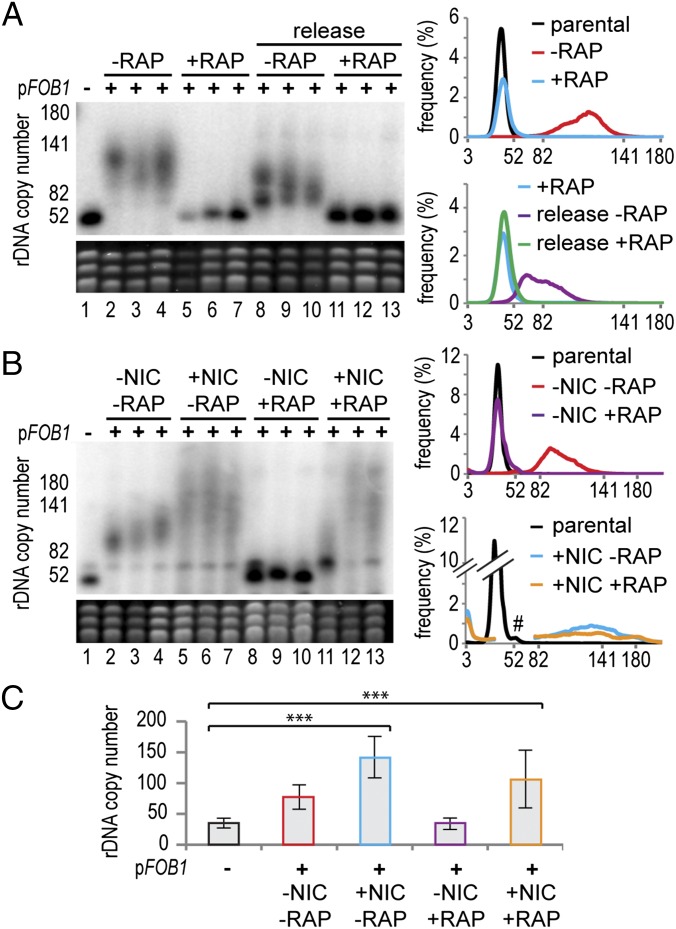
The rDNA array occupies ∼40% of chromosome XII in wild-type yeast, and the migration of chromosome XII by pulsed-field gel electrophoresis (PFGE) is routinely used to assay rDNA copy number. Heterogeneous chromosome XII signals indicate rDNA copy number heterogeneity in the population (e.g., Fig. 1A, compare lanes 1 and 2). Other chromosomes are shown by ethidium staining to control for loading and genome stability. Multiple clones are routinely tested, and the PFGE data can be combined into average rDNA copy number distribution plots (e.g., Fig. 1A, Upper right, derived from Fig. 1A, Left, lanes 1–7).
Fig. 1.
The TOR pathway controls rDNA amplification. (A) rDNA35 cells in which FOB1 is deleted (lane 1) were transformed with a pFOB1 plasmid that expresses FOB1 from the endogenous promoter. Half of the transformation mix was plated without rapamycin (lanes 2–4), and half with rapamycin (lanes 5–7), with three colonies from each transformation analyzed after three restreakings (∼60 generations). Cells from lanes 5–7 were restreaked four times without rapamycin (lanes 8–10) or with rapamycin (lanes 11–13). Cells were grown to stationary phase in liquid culture with or without rapamycin, they were lysed, and chromosomes were separated by PFGE. (Upper) Chromosome XII, of which rDNA constitutes ∼40% in a wild-type cell. (Lower) Ethidium stain of other chromosomes. Graphs show the rDNA copy number distribution averaged across clones of the same genotype, calculated from the PFGE data. (B) rDNA35 cells were transformed with pFOB1, plated on rapamycin (RAP) and/or nicotinamide (NIC) and analyzed as in A. #Region removed because of cross hybridization to chromosome IV. (C) Histogram showing qPCR quantification of rDNA amplification in the presence of nicotinamide (NIC) and/or rapamycin (RAP). Error bars represent 95% confidence interval (CI). ***P < 0.01 by one-way ANOVA. n = 5.
rDNA35 cells, which lack the FOB1 gene, were transformed with a plasmid expressing FOB1 from the endogenous promoter (pFOB1), and multiple transformants were grown in the presence or absence of the TOR inhibitor rapamycin. rDNA35 cells underwent rapid rDNA amplification on introduction of the FOB1 plasmid; however, this amplification process was completely repressed by rapamycin (Fig. 1A, lanes 1–7 and upper distribution plot). The rapamycin-treated cells were then restreaked on plates with or without rapamycin for a further ∼60 generations, and after drug removal, the rDNA amplified rapidly (Fig. 1A, lanes 8–13 and lower distribution plot). Rapamycin is therefore a potent but reversible inhibitor of rDNA amplification.
TOR Modulates rDNA Amplification Independent of Growth Rate.
Rapamycin treatment causes slow growth, and although the cells in Fig. 1A were grown for equivalent generations, it is possible that rDNA amplification simply reflects growth rate. Alternatively, rapamycin may block rDNA amplification through the activity of Sir2 or other enzymes (23–25). To distinguish these possibilities, we tested whether HDAC inhibition could separate the effects of rapamycin on growth and rDNA copy number.
Treatment with the Sir2 inhibitor nicotinamide did not increase growth rate in the presence or absence of rapamycin (SI Appendix, Fig. S2 A and B). However, growth of rDNA35 cells in the presence of nicotinamide caused faster rDNA amplification with more population heterogeneity (Fig. 1B, compare lanes 2–4 and 5–7), showing that nicotinamide enhances the rDNA amplification pathway, as has previously been demonstrated for the BIR pathway (29, 30). Importantly, rapamycin was unable to block rDNA amplification in the presence of nicotinamide, showing that rapamycin inhibits rDNA amplification through a nicotinamide-sensitive pathway that is separable from growth rate (Fig. 1B, lanes 8–13, and compare distribution plots without and with nicotinamide).
The comparison of heterogeneous rDNA distributions by PFGE can be subjective, and we therefore developed a quantitative PCR (qPCR) assay to quantitate average rDNA copy number in genomic DNA (SI Appendix, Fig. S3). This assay allows analysis of multiple independent samples derived as far as possible from independent clones, facilitating statistical analysis of changes in copy number. In accordance with the PFGE data, this assay demonstrated that nicotinamide treatment allowed significant rDNA amplification in the presence or absence of rapamycin (Fig. 1C).
Rapamycin treatment also affects RNA Pol I transcription, which is required for rDNA amplification (5, 6, 10). We therefore analyzed the level of the RNA Pol I primary transcript 35S and intermediates in the 18S rRNA synthesis pathway. As expected, based on growth rates, rapamycin treatment decreased ribosome synthesis, but nicotinamide had no effect on 35S levels in the presence or absence of rapamycin, nor did it lead to any change in the pattern of ribosome synthesis intermediates that would indicate increased transcription (SI Appendix, Fig. S2C). This shows that the repression of rDNA amplification by rapamycin does not stem from reduced RNA pol I transcription.
These experiments demonstrate that the repression of rDNA amplification by rapamycin cannot be attributed to defects in growth or RNA pol I transcription and is instead mediated through a nicotinamide-sensitive pathway.
Rapamycin Acts Through Multiple HDACs.
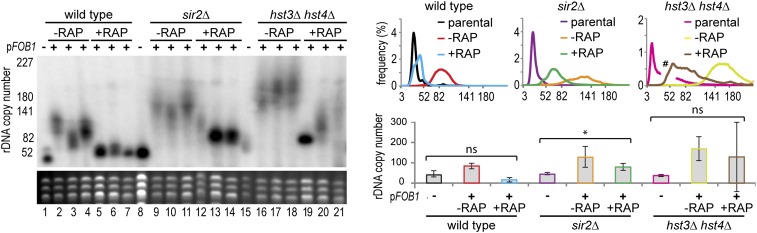
Nicotinamide inhibits Sir2 (30) as well as the Sir2 homologs Hst3 and Hst4 (31), all of which affect rDNA recombination rate (11, 21). To determine which of these HDACs controls rDNA amplification, we tested whether rapamycin represses rDNA amplification in rDNA35 cells lacking Hst3 and Hst4, which have a largely degenerate activity, or lacking Sir2.
As before, rDNA amplification was strongly repressed in rDNA35 cells treated with rapamycin (Fig. 2, lanes 1–7 and upper distribution plot). However, significant rDNA amplification occurred in rDNA35 sir2Δ cells in the presence of rapamycin, although this was very limited compared with nontreated cells (Fig. 2, lanes 8–14, middle distribution plot and histogram). Therefore, rapamycin acts partially but not completely through Sir2 to repress rDNA amplification. In rDNA35 hst3Δ hst4Δ cells, the ability of rapamycin to block amplification was also compromised, and more heterogeneous rDNA expansions were observed, indicating a higher recombination rate (Fig. 2, lanes 15–21 and lower distribution plot), although still less than in nonrapamycin-treated cells and with very high clone-to-clone variability (see analysis of more clones in SI Appendix, Fig. S4A). Single hst3Δ and hst4Δ mutants in the rDNA35 background showed only small amplifications in rapamycin-treated cells, consistent with the degenerate activity of these enzymes (SI Appendix, Fig. S4B). We also tested the triple-mutant rDNA35 sir2Δ hst3Δ hst4Δ; however, this mutant showed massive clonal rDNA variation, even in the absence of rapamycin (SI Appendix, Fig. S4C).
Fig. 2.
TOR controls rDNA amplification through HDAC modulation. SIR2 or HST3 and HST4 were deleted in rDNA35, and cells were transformed with pFOB1 and plated on rapamycin as in Fig. 1. rDNA distribution plots were calculated from data in SI Appendix, Fig. S4A, which contains the same samples, but with more rapamycin-treated clones. #Region removed because of cross-reaction to chromosome IV. Histogram shows qPCR quantification, error bars represent 95% CI. *P < 0.05 by Student's t test, comparing parental to pFOB1-transformed rapamycin-treated cells. n = 4. ns, not significant. The large confidence interval for rapamycin-treated hst3Δ hst4Δ cells is indicative of massive phenotypic variation between clones (SI Appendix, Fig. S4A).
These results demonstrate that neither Sir2 nor Hst3/4 are fully responsible for the repression of rDNA amplification by rapamycin, showing that multiple pathways contribute to rDNA amplification.
H3 K56 Acetylation Represses the Non-HR-Dependent Pathway.
Hst3 and Hst4 are the HDACs for H3 K56, which has been implicated in non-HR-independent rDNA amplification (20). However, only loss of histone chaperone Asf1 has been shown to cause non-HR-dependent amplification, and Asf1 affects many processes in addition to H3 K56 acetylation (32), leaving it unclear whether H3 K56 acetylation actually regulates the non-HR-dependent pathway.
To confirm this, we used an existing plasmid shuffle assay (33) to introduce H3 K56 mutations in wild-type and rad52Δ backgrounds. Both H3 K56R and H3 K56Q mutants, which mimic permanently deacetylated or acetylated lysine, respectively, underwent significant rDNA amplification in the absence of the critical HR protein Rad52, showing that defects in H3 K56 acetylation instigate rDNA amplification by the non-HR-dependent pathway (SI Appendix, Fig. S5A). H3 K56R mutants displayed a stronger phenotype than H3 K56Q mutants; however, this difference is not specific to rDNA recombination; promotion of HR-dependent sister chromatid recombination depends on an H3 K56 acetylation-deacetylation cycle and is more seriously impaired by H3 K56R than H3 K56Q mutations (34). Recent genetic evidence also shows that although hypo- and hyperacetylation of H3 K56 both inhibit HR, they do so through different mechanisms (35).
Different H3 K56 mutants therefore impair HR and reciprocally enhance non-HR-dependent rDNA amplification, showing that the H3 K56 acetylation cycle acts to repress non-HR-dependent rDNA amplification.
rDNA Amplification Depends on Two Mechanisms.
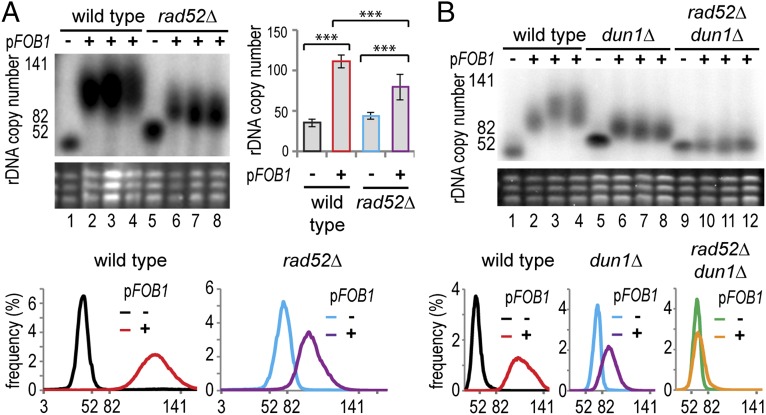
Because the H3 K56 acetylation cycle represses rDNA amplification through the non-HR-dependent pathway and rapamycin acts through the H3 K56 HDACs Hst3 and Hst4 to repress normal rDNA amplification (Fig. 2 and SI Appendix, Fig. S5A), we suspected the non-HR-dependent pathway may contribute to normal rDNA amplification. To test this, we assayed rDNA amplification in an HR-deficient rad52Δ background.
Surprisingly, rDNA amplification in rDNA35 cells transformed with pFOB1 occurred in the presence or absence of Rad52 (Fig. 3A). qPCR confirmed this result but also revealed that rDNA amplification was significantly faster in RAD52 than in rad52Δ cells (Fig. 3A, histogram). The experiment was then repeated over ∼300 generations; the difference between RAD52 and rad52Δ cells was not maintained after the initial ∼60 generations, and at later times, both cell types underwent amplification at similar rates (0.015–0.02 new repeats per repeat per generation; SI Appendix, Fig. S5B). This demonstrates that rDNA amplification in normal cells can occur in the absence of Rad52.
Fig. 3.
rDNA amplification occurs by multiple mechanisms. (A) Wild-type and rad52Δ rDNA35 cells lacking FOB1 were transformed with the plasmid pFOB1 and analyzed as in Fig. 1. Histogram shows qPCR quantification, and error bars represent 95% CI. ***P < 0.01 by one-way ANOVA. n = 3. (B) rDNA amplification in rDNA35 cells carrying dun1Δ and dun1Δ rad52Δ mutations, transformed with pFOB1 and analyzed as in Fig. 1.
This does not, however, demonstrate that the non-HR-dependent pathway is active when Rad52 is present; to assess the contribution of the non-HR-dependent pathway in cells expressing Rad52, we deleted the replicative kinase Dun1. Loss of Dun1 has little effect on Rad52-dependent HR but completely inhibits the non-HR-dependent amplification pathway (20, 36, 37). rDNA35 dun1Δ cells underwent rDNA amplification after the introduction of pFOB1, but again, to a lesser extent than wild-type (Fig. 3B, lanes 1–8 and distribution plots). The partial suppression of amplification seen in rad52Δ and in dun1Δ cells suggested that both HR-dependent and non-HR-dependent pathways are active in normal cells. To confirm this, we created rDNA35 dun1Δ rad52Δ double mutants and indeed found that amplification was completely repressed, showing that rDNA amplification occurs simultaneously through the HR-dependent and non-HR-dependent pathways (Fig. 3B, lanes 9–12 and distribution plot).
Combined with the data on rapamycin action in HDAC mutants, this demonstrates that both the HR-dependent BIR pathway and the non-HR-dependent amplification pathway mediate rDNA amplification in normal cells with low rDNA copy number, and that both pathways are coordinately regulated by TOR signaling.
Modulation of rDNA Amplification in Response to the Environment.
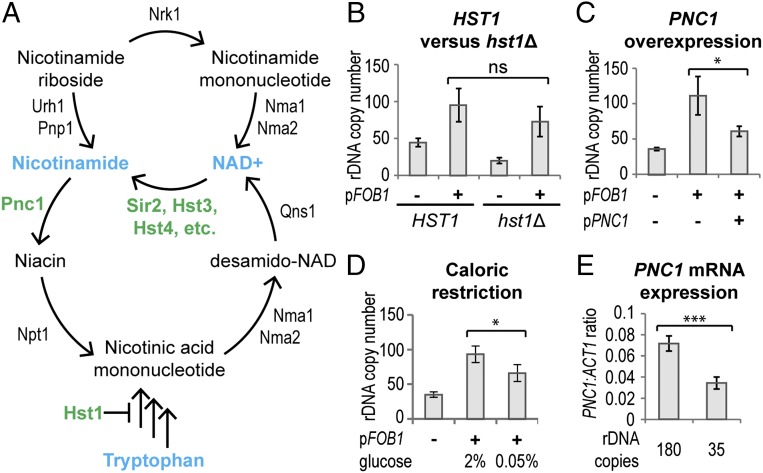
Sir2, Hst3, and Hst4 share a common reaction mechanism in which the substrate acetyl is transferred to a NAD+ cofactor, yielding nicotinamide and O-acetyl-ADP ribose. Nicotinamide is recycled to NAD+ via a salvage pathway that interfaces with the salvage pathway for nicotinamide riboside and with the de novo NAD+ synthesis pathway (Fig. 4A). Nicotinamide inhibits Sir2, Hst3, and Hst4 (29, 31), so the activity of these enzymes could be coordinately regulated through either NAD+ or nicotinamide levels; both options have been proposed to explain how Sir2 extends lifespan under caloric restriction (27, 29). To probe the role of these metabolites in rDNA amplification, we manipulated the NAD+ biosynthesis pathways and measured rDNA amplification using qPCR.
Fig. 4.
rDNA amplification is regulated by the NAD+ salvage pathway. (A) Schematic of NAD+ salvage pathways; key proteins and metabolites for this work are shown in green and blue, respectively. (B) rDNA amplification in rDNA35 HST1 and rDNA35 hst1Δ cells transformed with pFOB1, grown for 60 generations, and then assayed by qPCR. The difference between HST1 pFOB1 and hst1Δ pFOB1 stems from the difference in copy number before plasmid transformation and is not significant. Error bars represent 95% CI. n = 5 without pFOB1, n = 6 with pFOB1. (C) Amplification of rDNA35 cells cotransformed with pFOB1 and a high copy PNC1 plasmid or an empty vector. Cells were grown as in Fig. 1 and analyzed as in B. n = 6. (D) rDNA35 cells were transformed with pFOB1 and grown as in Fig. 1 on 2% or 0.05% glucose media and then analyzed as in B. n = 4 (parental and 0.05%), n = 9 (2%). (E) Expression of PNC1 mRNA relative to ACT1 in rDNA180 and rDNA35 cells measured by qPCR. Error bars represent 95% CI. ***P < 0.01 by Student's t test. n = 4.
We first tested whether increasing intracellular NAD+ could repress rDNA amplification. Hst1 autoregulates de novo NAD+ biosynthesis, and hst1Δ cells have increased NAD+ levels (38); however, deletion of HST1 in rDNA35 had no clear effect after excluding a difference in starting rDNA copy number between the HST1 and hst1Δ strains (Fig. 4B), showing that changes in NAD+ concentration have no significant effect on rDNA amplification rate. We then tested whether decreasing nicotinamide would have a stronger effect, as has been observed for marker loss through the Sir2-regulated BIR pathway (23, 29). Nicotinamide is metabolized by the nicotinamidase Pnc1, so we introduced the PNC1 gene on a high copy plasmid at the same time as the FOB1 plasmid. Comparing rDNA amplification in cells overexpressing PNC1 to empty vector controls showed that decreasing nicotinamide levels strongly repressed rDNA amplification (Fig. 4C). This demonstrates that the rate of rDNA amplification can be modulated by changes in PNC1 expression.
Caloric restriction induces PNC1 overexpression, which represses the BIR pathway (24), suggesting that rDNA amplification rate may be modulated by caloric restriction. To confirm this, we transformed pFOB1 into rDNA35 cells and grew them in normal glucose [2% (wt/vol)] or under caloric restriction (0.05% glucose), leading to PNC1 overexpression as expected (SI Appendix, Fig. S6A). After 60 generations of growth, we observed that caloric restriction significantly repressed rDNA amplification (Fig. 4D), showing that the rate of rDNA copy number change is directly linked to environmental nutrient availability. As with rapamycin treatment, caloric restriction impairs growth and reduces RNA pol I transcription; however, the repression of rDNA amplification is clearly separable from growth, as overexpression of PNC1 reduces rDNA amplification, but not growth rate (SI Appendix, Fig. S6 B and C and Fig. S2C).
The effect of PNC1 expression on rDNA amplification led us to question whether PNC1 levels are altered in cells with low rDNA copy number, which would be an important indicator of an active mechanism responding to low rDNA copy number. Indeed, PNC1 mRNA is significantly reduced in rDNA35 cells compared with isogenic rDNA180 controls (Fig. 4E). This does not fully explain the rDNA amplification phenotype of rDNA35 cells, as amplification is not completely suppressed by caloric restriction, whereas the PNC1 mRNA level is fully restored (compare Figs. 4D and SI Appendix, Fig. S6A), but clearly shows that the activity of Sir2 and Hst3/4 is selectively reduced in these cells through an increase in nicotinamide concentration.
Taken together, our results show that rDNA amplification is a tightly controlled process that is modulated in response to nutrient availability. rDNA amplification requires TOR signaling, which simultaneously controls the activity of multiple HDACs. These HDACs in turn regulate HR-dependent and non-HR-dependent rDNA recombination pathways that are both required for efficient rDNA amplification (Fig. 5).
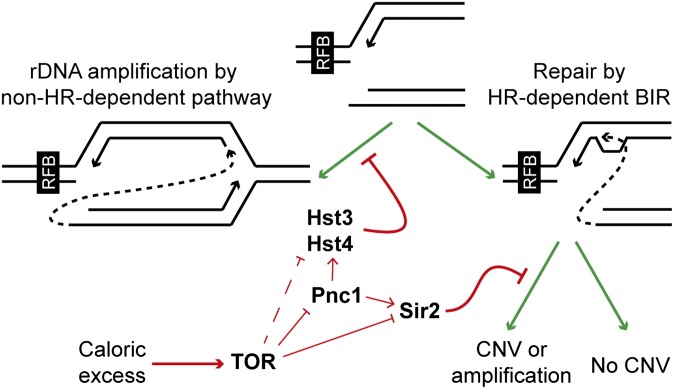
Fig. 5.
Regulation of rDNA amplification in response to caloric excess. Replication forks stalled at the replication fork barrier that have undergone cleavage (Center top) can enter the HR-dependent BIR pathway or the non-HR-dependent amplification pathway, which is repressed by Hst3 and Hst4. The BIR pathway can result in nondirectional copy number variation or amplification, but copy number variation through this pathway is repressed by Sir2. In response to excess nutrients, TOR signaling represses Sir2, Hst3, and Hst4 through suppression of PNC1 expression, but also represses Sir2 and potentially Hst3/Hst4 through a Pnc1-independent mechanism, leading to copy number amplification.
Discussion
Control of rDNA Amplification in Response to the Environment.
It has long been known that some organisms can amplify rDNA copy number, indicating the existence of controlled mechanisms for copy number change (18, 19). Here we have demonstrated that rDNA amplification in budding yeast is regulated by the TOR pathway and is performed by at least two recombination pathways under the control of multiple HDACs.
The rapamycin-sensitive Target of Rapamycin Complex 1 (TORC1) orchestrates budding yeast cell growth in response to nutrient levels (reviewed in ref. 39), and therefore the repression of rDNA amplification by rapamycin or caloric restriction firmly links rDNA copy number to nutrient availability. TOR inhibition can alter the rate of marker loss from the rDNA (23, 24); however, this occurs through the BIR pathway, which acts primarily to homogenize rDNA sequences, and it is not clear why rDNA homogenization should respond to the environment. In contrast, cells with suboptimal rDNA copy number are forced to up-regulate RNA pol I transcription to maintain ribosome synthesis, and rDNA amplification is a logical response in this situation; although ribosome synthesis can be enhanced temporarily by increasing RNA pol I transcription, this strategy is harmful in the long term (22, 40). Controlled rDNA amplification is therefore a response to available nutrients being in excess compared with ribosome synthesis capacity. Caloric restriction has been extensively investigated in yeast (41), but conversely, the effects of nutrient or caloric excess are largely unexplored because of complications from the osmolarity of high-glucose solutions (42). We observe that cells with low rDNA copy number in normal glucose media show reduced expression of PNC1, a gene that is overexpressed on caloric restriction and is required for lifespan extension (24, 29). Interestingly, SIR2 down-regulation has previously been noted in cells with low rDNA copy number, which would also reduce lifespan (43). These data suggest that exposure of yeast to caloric excess produces a specific transcriptional response, which may be very significant, given the conserved relationship among calorie availability, TOR signaling, and longevity in eukaryotes (reviewed in ref. 44).
PNC1 repression is not entirely responsible for rDNA amplification in rDNA35 cells, as overexpression of PNC1 only partially reverses the phenotype. In contrast, rapamycin totally inhibits rDNA amplification through Sir2, Hst3, and Hst4, showing that TOR also modulates the activity of one or more of these enzymes in a Pnc1-independent manner. The association of Sir2 with the rDNA increases on rapamycin treatment, suggesting that TOR displaces Sir2 from the rDNA through an unknown mechanism (23), and the same may be true for Hst3 and Hst4. TOR signaling therefore affects rDNA copy number through multiple mechanisms.
rDNA copy number amplification departs from the standard model of adaptation through random mutation followed by selection, as there is no growth difference between low and normal rDNA copy number cells under our experimental conditions (SI Appendix, Fig. S1 and refs. 10 and 22). Instead, we show that rDNA copy number is regulated by signaling events that are clearly separable from growth, providing the first example to our knowledge of a signaling pathway that can specifically regulate copy number. This raises the fascinating possibility that copy number of other regions of the genome may also be controllable in response to environmental conditions.
rDNA Amplification Through a Noncanonical Recombination Pathway.
Copy number change through Rad52-independent mechanisms has been reported in a number of systems, but has been considered an undesirable consequence of defective genome stability, occurring at a frequency of less than one in a million cells (45–47). Here we have shown that this process can be effectively controlled, occurring in a concerted manner across a population of cells.
Although Sir2, Hst3, and Hst4 are structurally related, they have very different effects on recombination. Sir2 regulates expression of ncRNAs in the rDNA spacer, removing cohesin and allowing a broken replication fork to undergo BIR with unmatched repeats (10). In contrast, Hst3 and Hst4 control recombination pathway choice at stalled replication forks; disturbance of the H3 K56 acetylation cycle prevents HR with a sister chromatid (34), and at the rDNA instigates non-HR-dependent recombination, leading to amplification. Because the non-HR-dependent pathway causes constitutive gain of rDNA copies, cells could regulate rDNA amplification by modulating H3 K56 acetylation (see model Fig. 5). This method of regulation may seem unlikely, as loss of Hst3 and Hst4 leads to general genome instability (31, 34); however, the loss of HDAC activity need not be complete. HR proteins are excluded from the nucleolus (48), and the highly repetitive rDNA is an excellent substrate for non-HR-dependent recombination (20). Therefore, a reduction in H3 K56 HDAC activity that has little effect on the rest of the genome could well drive non-HR-dependent rDNA amplification.
Materials and Methods
Detailed methods are given in SI Appendix, Materials and Methods.
Cells were grown on synthetic media, rapamycin (SCBT) was used at 25 nM, and nicotinamide (Sigma) at 5 mM. PFGE used standard methods, and qPCR for BUD23 and 25S rDNA were performed on EcoRI digested genomic DNA.
Supplementary Material
Acknowledgments
We thank Ann Kirchmaier for strains and Peter Rugg-Gunn, Sarah Elderkin, and Alex Murray for critical reading of the manuscript. This work was funded by the Wellcome Trust (Grants 088335 and 093735). C.V.J. is funded through an MRC studentship, and M.A.K. is supported by an Erwin Schroedinger fellowship (J 3341) from Austrian Science Fund (FWF) (Austria).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505015112/-/DCSupplemental.
References
- 1.Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72(4):686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craddock N, et al. Wellcome Trust Case Control Consortium Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464(7289):713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 4.Keil RL, Roeder GS. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39(2 Pt 1):377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 5.Stewart SE, Roeder GS. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9(8):3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voelkel-Meiman K, Keil RL, Roeder GS. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48(6):1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- 7.Szostak JW, Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980;284(5755):426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- 8.Brewer BJ, Fangman WL. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55(4):637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1(5):465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12(24):3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56(5):771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi T, Ganley AR. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309(5740):1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117(4):441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- 14.Johzuka K, Horiuchi T. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells. 2002;7(2):99–113. doi: 10.1046/j.1356-9597.2001.00508.x. [DOI] [PubMed] [Google Scholar]
- 15.Ganley AR, Kobayashi T. Monitoring the rate and dynamics of concerted evolution in the ribosomal DNA repeats of Saccharomyces cerevisiae using experimental evolution. Mol Biol Evol. 2011;28(10):2883–2891. doi: 10.1093/molbev/msr117. [DOI] [PubMed] [Google Scholar]
- 16.Eickbush TH, Eickbush DG. Finely orchestrated movements: Evolution of the ribosomal RNA genes. Genetics. 2007;175(2):477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi T, Nomura M, Horiuchi T. Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21(1):136–147. doi: 10.1128/MCB.21.1.136-147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown DD, Dawid IB. Specific gene amplification in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science. 1968;160(3825):272–280. doi: 10.1126/science.160.3825.272. [DOI] [PubMed] [Google Scholar]
- 19.Gall JG. Free ribosomal RNA genes in the macronucleus of Tetrahymena. Proc Natl Acad Sci USA. 1974;71(8):3078–3081. doi: 10.1073/pnas.71.8.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houseley J, Tollervey D. Repeat expansion in the budding yeast ribosomal DNA can occur independently of the canonical homologous recombination machinery. Nucleic Acids Res. 2011;39(20):8778–8791. doi: 10.1093/nar/gkr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ide S, Saka K, Kobayashi T. Rtt109 prevents hyper-amplification of ribosomal RNA genes through histone modification in budding yeast. PLoS Genet. 2013;9(4):e1003410. doi: 10.1371/journal.pgen.1003410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol. 2003;23(5):1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ha CW, Huh WK. Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2011;39(4):1336–1350. doi: 10.1093/nar/gkq895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5(10):e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Fan M, Pfeffer LM, Laribee RN. The histone H3 lysine 56 acetylation pathway is regulated by target of rapamycin (TOR) signaling and functions directly in ribosomal RNA biogenesis. Nucleic Acids Res. 2012;40(14):6534–6546. doi: 10.1093/nar/gks345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lempiäinen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21(6):855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289(5487):2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 28.Cioci F, et al. Silencing in yeast rDNA chromatin: Reciprocal relationship in gene expression between RNA polymerase I and II. Mol Cell. 2003;12(1):135–145. doi: 10.1016/s1097-2765(03)00262-4. [DOI] [PubMed] [Google Scholar]
- 29.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423(6936):181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277(47):45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 31.Celic I, et al. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol. 2006;16(13):1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 32.Mousson F, Ochsenbein F, Mann C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma. 2007;116(2):79–93. doi: 10.1007/s00412-006-0087-z. [DOI] [PubMed] [Google Scholar]
- 33.Miller A, Yang B, Foster T, Kirchmaier AL. Proliferating cell nuclear antigen and ASF1 modulate silent chromatin in Saccharomyces cerevisiae via lysine 56 on histone H3. Genetics. 2008;179(2):793–809. doi: 10.1534/genetics.107.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muñoz-Galván S, Jimeno S, Rothstein R, Aguilera A. Histone H3K56 acetylation, Rad52, and non-DNA repair factors control double-strand break repair choice with the sister chromatid. PLoS Genet. 2013;9(1):e1003237. doi: 10.1371/journal.pgen.1003237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadyrova LY, et al. A reversible histone H3 acetylation cooperates with mismatch repair and replicative polymerases in maintaining genome stability. PLoS Genet. 2013;9(10):e1003899. doi: 10.1371/journal.pgen.1003899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haghnazari E, Heyer WD. The DNA damage checkpoint pathways exert multiple controls on the efficiency and outcome of the repair of a double-stranded DNA gap. Nucleic Acids Res. 2004;32(14):4257–4268. doi: 10.1093/nar/gkh717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fasullo M, Koudelik J, AhChing P, Giallanza P, Cera C. Radiosensitive and mitotic recombination phenotypes of the Saccharomyces cerevisiae dun1 mutant defective in DNA damage-inducible gene expression. Genetics. 1999;152(3):909–919. doi: 10.1093/genetics/152.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bedalov A, Hirao M, Posakony J, Nelson M, Simon JA. NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23(19):7044–7054. doi: 10.1128/MCB.23.19.7044-7054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189(4):1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327(5966):693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- 41.Schleit J, Wasko BM, Kaeberlein M. Yeast as a model to understand the interaction between genotype and the response to calorie restriction. FEBS Lett. 2012;586(18):2868–2873. doi: 10.1016/j.febslet.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaeberlein M, Andalis AA, Fink GR, Guarente L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol Cell Biol. 2002;22(22):8056–8066. doi: 10.1128/MCB.22.22.8056-8066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michel AH, Kornmann B, Dubrana K, Shore D. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 2005;19(10):1199–1210. doi: 10.1101/gad.340205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan JE, Kolodner RD. Rapid analysis of Saccharomyces cerevisiae genome rearrangements by multiplex ligation-dependent probe amplification. PLoS Genet. 2012;8(3):e1002539. doi: 10.1371/journal.pgen.1002539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4(9):e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schacherer J, de Montigny J, Welcker A, Souciet JL, Potier S. Duplication processes in Saccharomyces cerevisiae haploid strains. Nucleic Acids Res. 2005;33(19):6319–6326. doi: 10.1093/nar/gki941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres-Rosell J, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9(8):923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.