Significance
Tumor necrosis factor (TNF) is a cytokine involved in a large array of inflammatory diseases such as arthritis and psoriasis. We have found that the insertion of a retrotransposon in the 3′ untranslated region of Tnf impairs its normal regulation and causes polyarthritis and heart valve disease. We have identified several CCCH-containing zinc-finger proteins as new regulators of Tnf expression, as well as a new regulatory element within the TNF 3′UTR. Although anti-TNF therapies are successful in controlling some inflammatory diseases, their cost and the development of resistance in some patients show that other treatment options are required. Our findings provide an important foundation toward understanding how Tnf expression is regulated and identify potential targets for therapeutic intervention.
Keywords: rheumatoid arthritis, heart valve disease, mRNA stability, CCCH ZFP, gene regulation
Abstract
Rheumatoid arthritis (RA) and ankylosing spondylitis (AS) are chronic inflammatory diseases that together affect 2–3% of the population. RA and AS predominantly involve joints, but heart disease is also a common feature in RA and AS patients. Here we have studied a new spontaneous mutation that causes severe polyarthritis in bone phenotype spontaneous mutation 1 (BPSM1) mice. In addition to joint destruction, mutant mice also develop aortic root aneurism and aorto-mitral valve disease that can be fatal depending on the genetic background. The cause of the disease is the spontaneous insertion of a retrotransposon into the 3′ untranslated region (3′UTR) of the tumor necrosis factor (TNF), which triggers its strong overexpression in myeloid cells. We found that several members of a family of RNA-binding, CCCH-containing zinc-finger proteins control TNF expression through its 3′UTR, and we identified a previously unidentified regulatory element in the UTR. The disease in BPSM1 mice is independent of the adaptive immune system and does not appear to involve inflammatory cytokines other than TNF. To our knowledge, this is the first animal model showing both polyarthritis and heart disease as a direct result of TNF deregulation. These results emphasize the therapeutic potential of anti-TNF drugs for the treatment of heart valve disease and identify potential therapeutic targets to control TNF expression and inflammation.
Rheumatoid arthritis (RA) and ankylosing spondylitis (AS) are related and overlapping chronic inflammatory diseases that affect joints and severely impair mobility of the patients (1, 2). RA patients generally present with a symmetric polyarthritis of the peripheral joints. RA affects more women than men, typically leads to bone and cartilage erosion, and is often associated with the presence of rheumatoid factor in the serum. By contrast, AS is a disease of the sacroiliac joint and the spine with variable involvement of peripheral joints. AS is more prevalent in men than women, generally rheumatoid factor-negative, and characterized by new cartilage and bone formation that leads to ankylosis of the spine. The causes of RA and AS are ill-defined, but genetic influences have a major role in both diseases. It is generally accepted that RA and AS are auto-inflammatory diseases that involve many inflammatory cytokines. Interestingly, although disease-modifying antirheumatic drugs have been useful in the treatment of RA, they have shown limited effect in AS. However, anti-TNF therapies have shown a high efficacy in both diseases. Both RA and AS are systemic diseases that can also affect several other organs. Extra-articular manifestations of RA include vasculitis, lung inflammation, formation of s.c. nodules, and heart disease (3), whereas psoriasis, inflammatory bowel disease (IBD), and heart valve disease are commonly associated with AS (4–6). We describe here a new mouse model with characteristics of both RA and AS.
Results
A New Model of Spontaneous Polyarthritis.
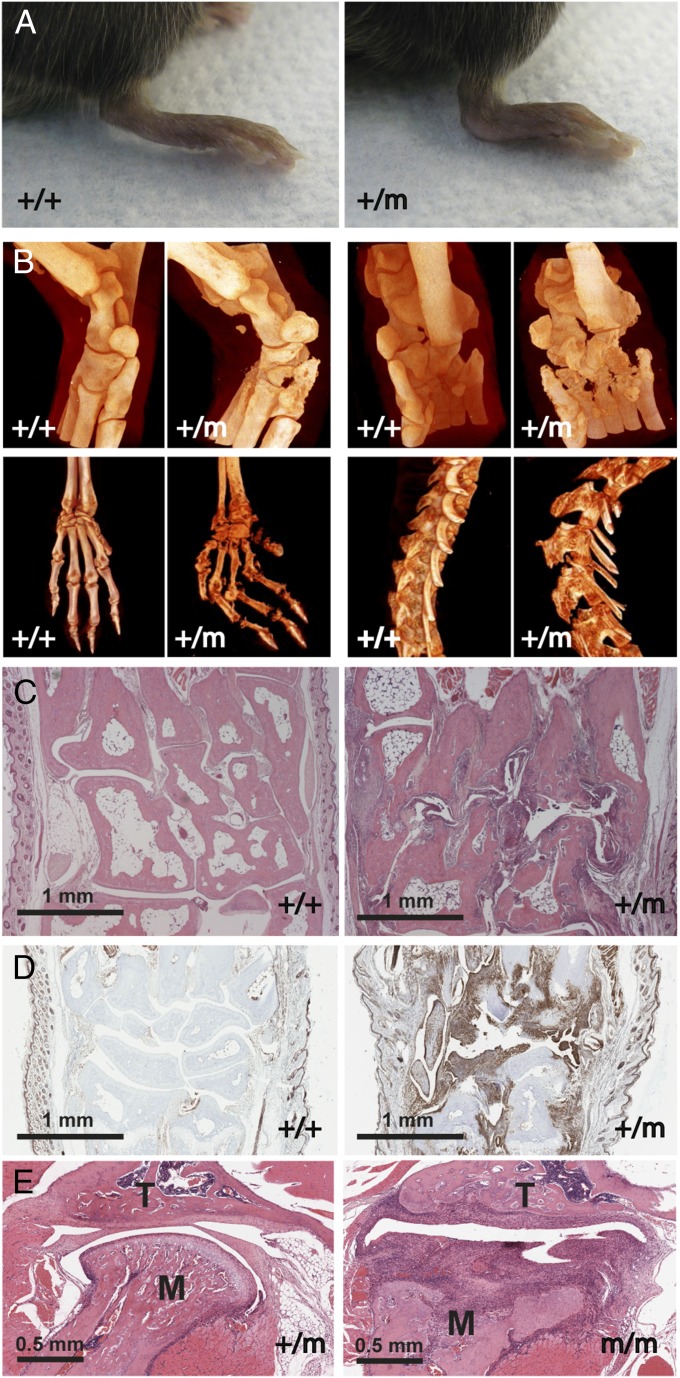
A dominant mutation that leads to deformed limbs and progressive paralysis of the hind legs coupled to an abnormal curvature of the spine at the level of the rib cage occurred spontaneously in our mouse colony. Ankle deformation was the most obvious early sign of the disease and occurred without acute swelling or redness (Fig. 1A). As the disease progressed, affected animals gradually lost their grasping strength and their forelimbs became angled at wrist level whereas the hind-limb digits became immobile. Around 7 mo, a pronounced hunchback was associated with the progressive paralysis of the hind limbs. Histological examination revealed a severe symmetrical, erosive chronic polyarthritis, reminiscent of human RA and AS. It primarily affected the peripheral joints (ankles, metatarsal, and interphalangeal joints) (Movie S1A and Movie S1B) with less severe damage in central joints (knees and shoulders). The diseased joints showed extensive pannus invasion, bone destruction, and cartilage erosion (Fig. 1C). Pannus tissue was dominated by F4/80+ cells (Fig. 1D), and most affected joints were devoid of lymphocytes and neutrophils. No sex bias was observed, and bone phenotype spontaneous mutation 1 (BPSM1) mice did not contain IgM or IgG rheumatoid factors (data not shown). The hunchback was due to the severe erosion of the thoracic vertebrae T10–T13 (Fig. 1B), the rearmost anchor points of the trapezius muscles. Tomography of the spine of an 180-d-old affected mouse showed the 13th (floating) ribs entering the neural canal instead of being attached to the centrum (Movie S1C and Movie S1D); this is probably the cause of hind-limb paralysis. Bone erosion appeared maximal in joints with the highest load factor, suggesting that mechanical stress may be an important determinant of the severity of the disease. No signs of inflammation were found in the skin, liver, blood vessels, eyes, or gut of these mice, and no evidence for cartilage or bone repair was found.
Fig. 1.
BPSM1 mutant mice spontaneously develop chronic polyarthritis. (A) Hindpaws of 100- d-old BPM1+/m mice are luxated at the level of the ankle. (B) Micro X-ray computed tomography of ankles, paws, and spine of 170-d-old WT (+/+) and affected (+/m) BPSM1 mice showing extensive bone erosion and limb deformation in the mutants. (C) Representative sections through metatarsal bones stained with hematoxylin and eosin showing bone erosion and penetration of pannus tissue in ankle joints. (D) Representative sections through the wrists of 90-d-old mice show strong F4/80 staining of the pannus tissue in BPSM1+/m animals. (E) Sections through the temporomandibular joints of 43-d-old BPSM1+/m and BPSM1m/m mice exemplify the dramatic acceleration of disease development in homozygote BPSM1 mutant mice. M, mandible condyle; T, temporal bone.
Development of the disease was dramatically accelerated in BPSM1 homozygote animals, in which ankles were luxated and wrists were clubbed already at 3 wk of age. Temporomandibular joints were also severely affected in these mice (Fig. 1E), which became paralyzed around 6 wk of age.
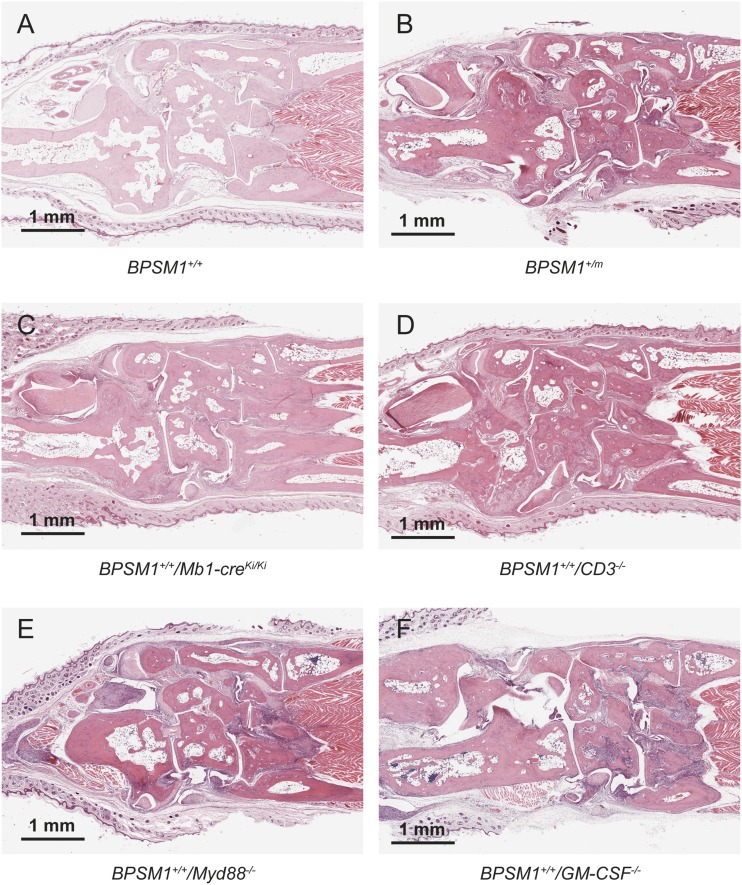
Loss of B or T cells [through crosses with Mb1-cre (7) or CD3ε KO mice (8), respectively] did not change the kinetics or the severity of the disease (Fig. S1). This finding is reminiscent of murine models of arthritis caused by human TNF overexpression or DNaseII deficiency (9, 10). Loss of Myd-88 (11) also had no impact, excluding an important role for IL1R and Toll-like receptors TLR2 and TLR4 in the BPSM1 disease (Fig. S1). Granulocyte/macrophage colony-stimulating factor (GM-CSF) antibodies are presently evaluated in phase 2 clinical trials for the treatment of RA (12). However, loss of GM-CSF (13) did not significantly alter the kinetics or severity of disease in BPSM1 mice (Fig. S1).
Fig. S1.
Disease in BPSM1 mice is independent of the adaptive immune system, Myd88, or GM-CSF. Sections through the ankle of 100-d-old WT (A), BPSM1+/m (B), BPSM1+/+/Mb1-creKi/Ki (C), BPSM1+/+/CD3−/− (D), BPSM1+/+/Myd88−/− (E), and BPSM1+/+/GM-CSF−/− (F) mice showing similar bone erosion in all of the mice carrying the BPSM1 mutation.
Arthritis in BPSM1 Mice Is Due to a New Mutation in the TNF Gene.
TNF is a central cytokine in RA and AS, and anti-TNF therapies have been successful in the control of these diseases, although about 30% of RA patients do not respond to such treatments (14), and recently developed mouse models have demonstrated that RA can also develop without TNF (15).
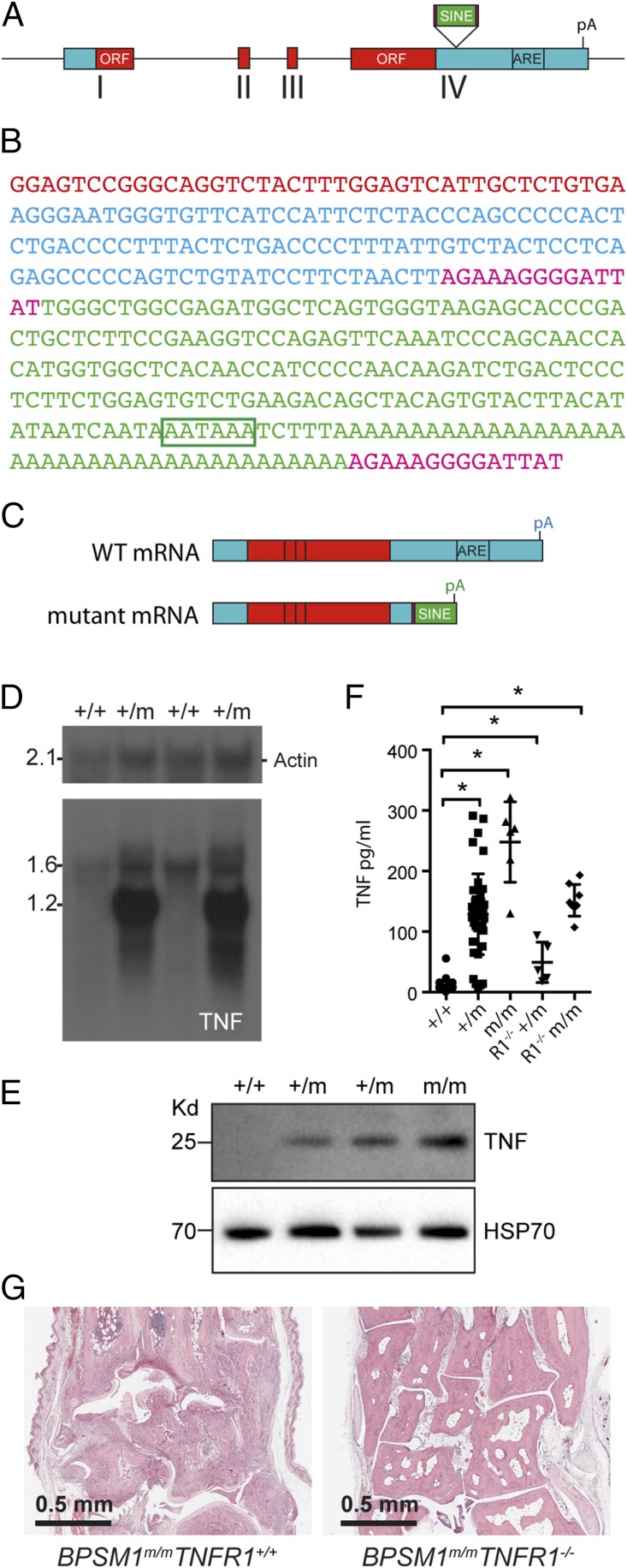
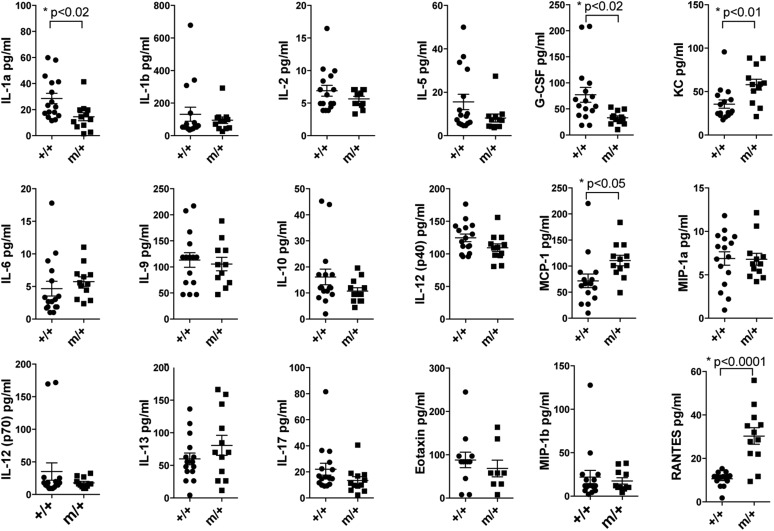
Intercrosses of BPSM1 with TNF-deficient mice (16) showed a strong genetic linkage between the BPSM1 and the TNF loci, indicating that the BPSM1 mutation must reside within the TNF locus or in its close proximity. Exome and whole-genome sequencing helped to identify the insertion of a small interspersed element (SINE) in the 3′UTR of TNF (Fig. 2 A and B) in every affected BPSM1 mouse (n > 1,200). No other mutation was identified in the 10 Mbp either side of TNF. Insertion of the SINE provides a new polyadenylation signal that terminates TNF mRNA prematurely, effectively truncating 650 nt from WT TNF 3′UTR (Fig. 2C). Importantly, the ORF is not modified. The 3′UTR of TNF contains several conserved AU-rich elements (ARE) that are partly responsible for the rapid turnover of its mRNA (17). Insertion of the SINE thus removes the normal negative regulation imposed by the 3′UTR. Accordingly, although TNF mRNA was barely detectable in unstimulated bone marrow-derived macrophages (BMDM) from wild-type (+/+) mice, an abnormally short TNF mRNA was highly expressed in unstimulated BMDM from affected BPSM1+/m mice (Fig. 2D). Similarly, membrane TNF was undetectable in unstimulated wild-type BMDM and was readily detected in unstimulated heterozygous and homozygous BPSM1 BMDM (Fig. 2E). Accordingly, high levels of circulating TNF were detected by ELISA in the serum of affected BPSM1+/m and BPSM1m/m mice (Fig. 2F). Elevated TNF levels are often accompanied by high levels of other proinflammatory cytokines such as IL1β, IL6, IL18, or IL23 (18), but none of these cytokines were particularly elevated in affected BPSM1+/m sera (Fig. S2). Loss of TNFR1 completely prevented the development of RA in both BPSM1+/m and BPSM1m/m mice (Fig. 2G), even though high levels of TNF were still present in the sera of TNFR1−/−/BPSM1+/m and TNFR1−/−/BPSM1m/m mice (Fig. 2F). This shows that TNFR1 but not TNFR2 is critical for the development of arthritis in BPSM1 mice.
Fig. 2.
A SINE retrotransposon causes TNF overexpression in BPSM1 mutant mice. (A) Schematic representation of WT TNF gene, exons I–IV, ORF appearing as red boxes, and site of insertion of the SINE retrotransposon upstream of the ARE. (B) Sequence of the inserted SINE retrotransposon: red shows the end of the TNF ORF; blue shows the TNF 3′UTR up to the insertion point; 14-nt direct repeats are shown in purple; SINE sequence is shown in green; and poly(A) signal is boxed. (C) Schematic representation of WT and mutant TNF mRNAs, showing the truncation of the 3′UTR and loss of regulatory sequences in the mutant. (D) Northern blot analysis of mRNAs from BMDM of the indicated genotypes using Actin (loading control) and TNF-specific probes. (E) Western blot analysis of lysates from BMDM of the indicated genotypes using HSP70 (loading control) and TNF-specific antibodies. (F) Quantitation of serum TNF levels in mice of the indicated genotype by ELISA. (G) Ankle joint sections showing that loss of TNFR1 prevents RA in BPSM1+/m and BPSM1m/m mice.
Fig. S2.
Serum concentration of cytokines and chemokines in BPSM1 mice. Cytokines and chemokines were measured in mouse sera by multiplex bead array using the Bio-Plex mouse cytokine panel (Bio-Rad Laboratories).
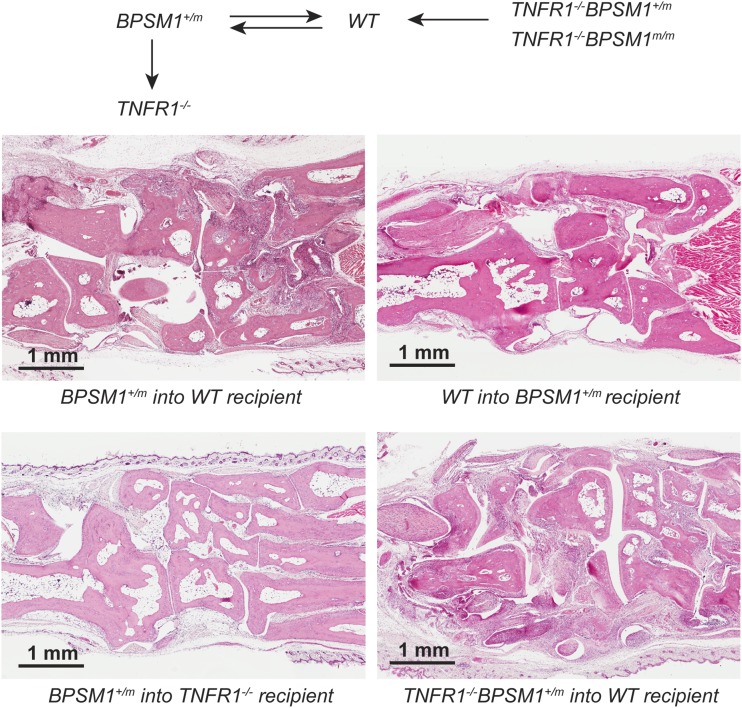
Bone marrow transplantation experiments (results summarized in Table 1 and Fig. S3) showed that lethally irradiated WT mice transplanted with BPSM1+/m bone marrow developed arthritis within 5 mo, showing that hematopoietic cells are the source of excess TNF. Conversely, 5-wk-old BPSM1+/m mice transplanted with WT bone marrow did not develop the disease over the following 5 mo. Thus, the presence of the mutation in radiation-resistant synoviocytes is not sufficient for disease development. We then transplanted BPSM1+/m bone marrow into lethally irradiated WT or TNFR1−/− recipients and examined them after transplantation. Although severe disease developed within 5 mo in all WT recipients of BPSM1+/m bone marrow, TNFR1-deficient recipients remained disease-free during that period. The presence of TNFR1 on synoviocytes is thus necessary for the development of the disease. Remarkably, all of the WT recipients of bone marrow from TNFR1−/−BPSM1m/m healthy donors developed the disease within 2 mo. This suggests that the F4/80+ cells that make the bulk of the pannus tissue are most likely macrophage-like type A synoviocytes and not macrophages originating from the bone marrow. Similar bone marrow transplantation experiments performed with TNFΔARE mice also indicated that the primary and sufficient targets of TNF in this pathology are synovial cells (19). Moreover, a similar synovium hyperplasia was described in mice transgenically expressing human IL1α (20).
Table 1.
Summary of bone marrow transplantation experiments
| Donor | BPSM1+/m (sick) | WT | BPSM1+/m (sick) | TNFR1−/− BPSM1m/m (healthy) |
| Recipient | WT | BPSM1+/m | TNFR1−/− | WT |
| Disease | Yes | No | No | Yes |
Fig. S3.
Bone marrow transplantation experiments. Schematic representation of the bone marrow transplantation experiments and histological examples in the ankle joints. Donors and recipients are indicated below each picture.
In summary, it appears that the polyarthritis observed in BPSM1 mice is due to the abnormal proliferation of synoviocytes in response to high levels of TNF secreted by myeloid cells in conjonction with a high mechanical stress.
Stability of TNF mRNA Is Affected by Multiple Zinc-Finger Proteins.
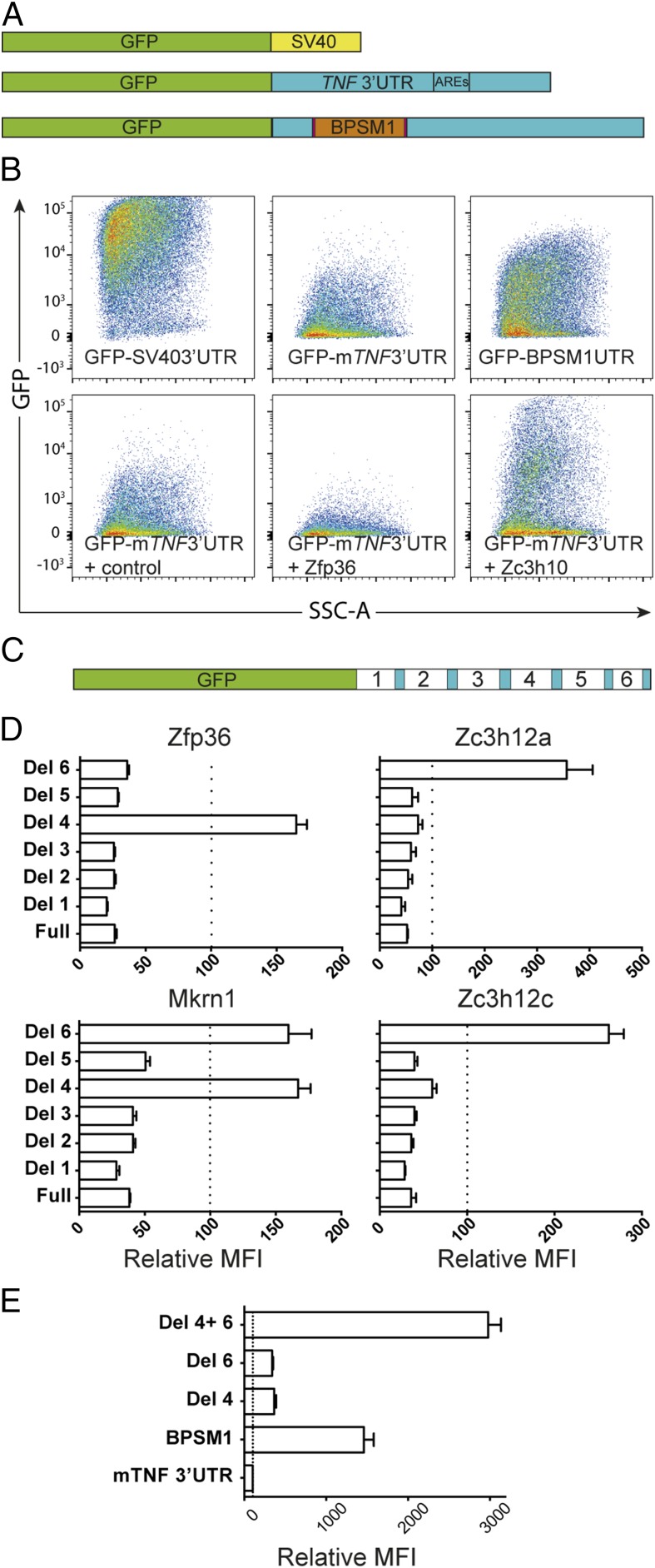
Deletion of six ARE from the 3′UTR of TNF leads to chronic inflammatory arthritis in knock-in mice (17). Mice deficient in tristetraprolin (TTP1, also known as Zfp36), a CCCH-containing zinc-finger protein (ZFP), develop cachexia, arthritis, and autoimmunity, and these pathologies are all prevented by treatment with TNF antibodies (21). Intriguingly, however, myeloid-specific deletion of Zfp36 failed to produce a similar phenotype (22). Zfp36 plays a role in the regulation of steady-state TNF levels by controlling mRNA stability and translation (17). Many CCCH-containing ZFP are expressed in macrophages and have been proposed to play a role in regulating their effector functions (23). To investigate the potential interaction of all CCCH ZFP on TNF 3′UTR, we engineered GFP reporter constructs derived from the pGL3-promoter reporter (Promega) (Fig. 3A). Substitution of the SV40 late poly(A) signal by TNF 3′UTR (either mouse or human WT sequence) drastically reduced GFP expression (Fig. 3B). Coexpression of Zfp36 further decreased GFP levels in accordance with its previously reported negative regulation of TNF. The construct with the BPSM1-derived TNF 3′UTR expressed higher GFP levels than the vector with the WT TNF 3′UTR, confirming that the retrotransposon insertion in the BPSM1 mice disrupts the normal attenuation of TNF expression (Fig. 3B).
Fig. 3.
Several ZFP regulate the expression of TNF through its 3′UTR. (A) Schematic representation of the GFP reporter constructs used to assay the effect of ZFP. (B) Representative FACS data showing the effect of the TNF 3′UTR on the expression of a GFP reporter, its down-regulation by Zfp36, and its up-regulation by Zc3h10. (C) Schematic representation of the deletions in the reporters used to define the regulatory regions in the TNF 3′UTR. (D) Regions 4 and 6 contain elements through which Zfp36, Zc3h12a, Zc3h12c, and Mkrn1 exert their regulatory function. (E) Strong synergy between regulatory regions 4 and 6. Data in D and E represent mean ± SEM of three independent experiments.
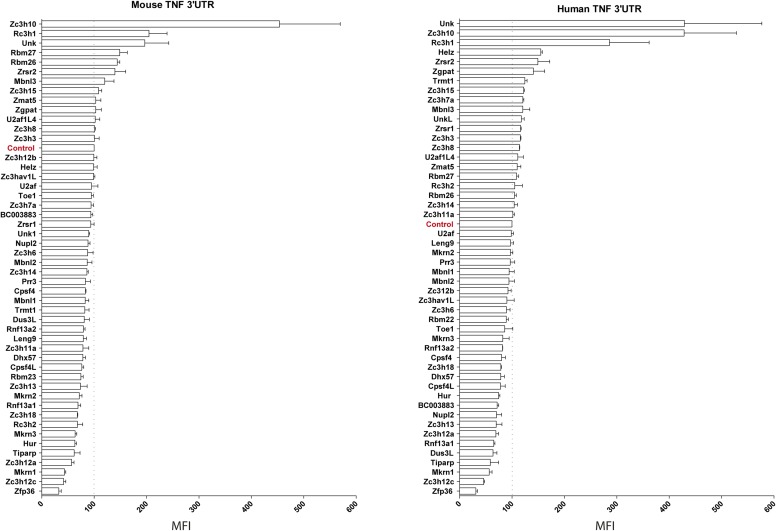
We next used these reporters to investigate the regulatory potential of 50 different CCCH-containing ZFP on the expression of GFP in 293T cells (Fig. S4). Three additional ZFP (Mkrn1, Zc3h12a, Zc3h12c) were found to repress the expression of GFP-mTNF3′UTR or GFP-hTNF3′UTR, whereas Rc3h1 (Roquin), Zc3h10, and Unkempt consistently increased the expression of GFP from these reporters (Fig. 3B and Fig. S4).
Fig. S4.
ZFP regulate the expression of TNF through its 3′UTR. The regulatory effect of 50 CCCH-containing ZFP on the expression of GFP-mTNF3′UTR- and GFP-hTNF3′UTR reporters was evaluated by cotransfection in HEK293T cells. A mCherry expression vector was used as a transfection control and an empty vector (Control) was used to measure the basal expression of the reporters. GFP mean fluorescence intensity (MFI) was measured in mCherry-positive cells by FACS, and results are expressed as relative MFI compared with control. Results represent the mean ± SEM of three independent experiments.
To further explore the nature of this regulation of TNF expression by ZFP, we engineered six deletions in the TNF 3′UTR of the reporter (Fig. 3C). As expected, Zfp36 lost its ability to down-regulate the expression of the reporter upon deletion of the ARE (Del 4) and was not affected by any of the other deletions (Fig. 3D). By contrast, Zc3h12a and Zc3h12c retained their effect upon deletion of the ARE, but lost it upon deletion of 76 nt located at the 3′ end of the 3′UTR, upstream of the polyadenylation signal (Del 6). The impact of Mkrn1 was affected by the loss of region 4 or region 6. Deletion of both regions 4 and 6 from the reporter with mTNF 3′UTR (Del4 Del6) led to maximal GFP expression in 293T cells, suggesting a synergistic effect of these two elements on the silencing of TNF expression (Fig. 3E). These data reveal that multiple ZFP can affect the steady-state expression of the TNF gene through its 3′UTR and that this regulation involves a previously unidentified region of the 3′UTR that we call the New Regulatory Element (NRE). Thus, potential genetic causes of TNF deregulation extend far beyond the −308A promoter single-nucleotide variant commonly examined in genome-wide association study studies, and searches for a disease-causing mutation in the TNF gene should include its 3′UTR, as well as variations in all of the genes (e.g., ZFP) that regulate TNF expression.
BPSM1 Mice Develop Aortic Root Aneurism and Aortic and Mitral Valve Inflammation and Fibrosis.
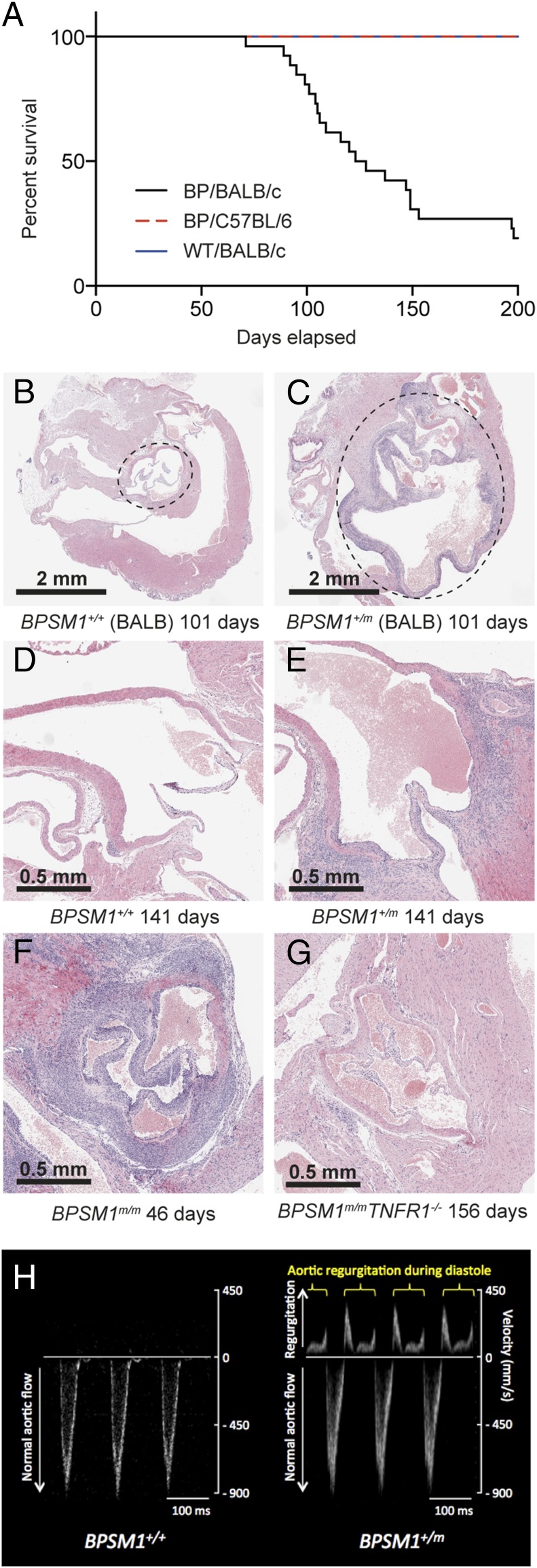
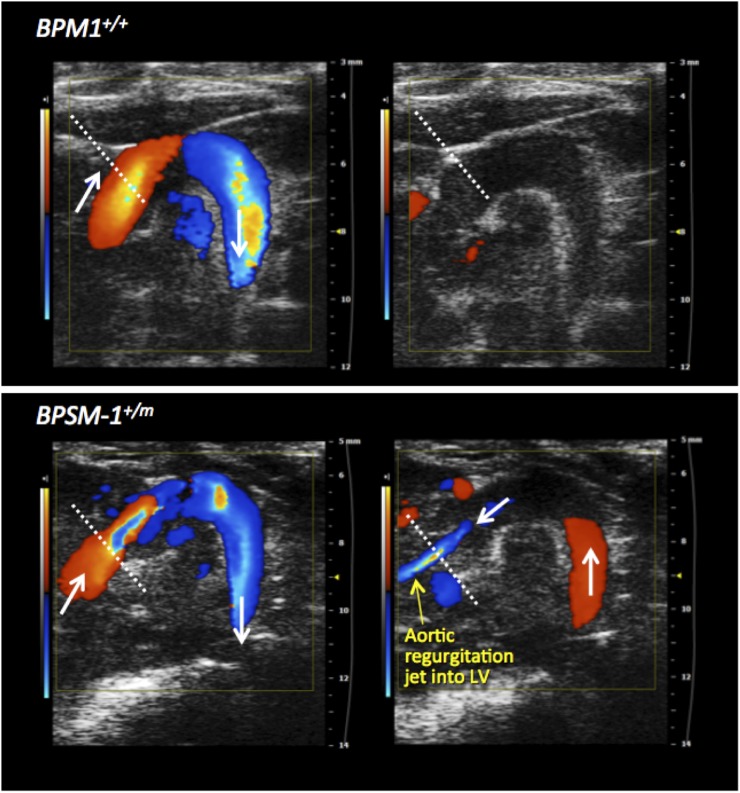
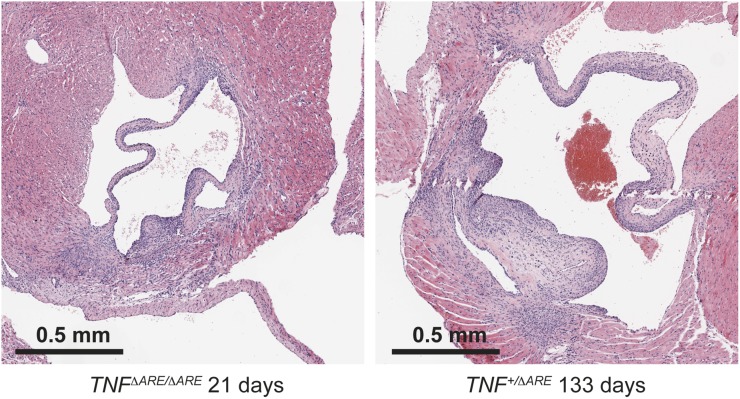
Cardiovascular diseases are commonly associated with inflammatory conditions, particularly RA and AS (6, 24). BPSM1 mice were backcrossed onto the BALB/c genetic background for the purpose of chromosomal mapping. After six rounds of backcrossing, many of the mutant mice died suddenly between 90 and 160 d of age, a phenomenon that was never observed on the C57BL/6 background (Fig. 4A). Aortic root aneurisms were found in all BALB/c mice with the BPSM1 mutation, as well as dramatic inflammation, thickening, and fibrosis involving the aortic and mitral valves (Fig. 4B). Aortic root diameter in BPSM1 mice on the BALB/c background could reach 4 mm (1–1.5 mm in WT BALB/c or WT C57BL/6 mice; Fig. 4 B and C). A similar valve inflammation was found in the BPSM1 mice (both heterozygotes and homozygotes) of the C57BL/6 background, but the aortic aneurism was much less pronounced (Fig. 4 D and E), and none of these animals died of aortic dissection up to 200 d of age (Fig. 4A). We found no sign of inflammation in any other part of the aorta or elsewhere in the heart. The tricuspid and pulmonary valves were unaffected, further suggesting that mechanical stress is an important factor in the development of the disease because pressure is much greater in the left ventricle than in the right ventricle. We did not find any cardiac abnormality in a collagen-induced model of RA. The heart disease of BPSM1 mice (even homozygotes) was entirely prevented by loss of TNFR1 (Fig. 4G). Echocardiography revealed that valve disease in BPSM1 mice manifests mainly as regurgitation, indicated by the presence of a backward aortic blood flow (Fig. 4H and Fig. S5). A similar inflammation of the heart valves was found in TNFΔARE mice, although this had not been reported before (Fig. S6).
Fig. 4.
BPSM1 mutant mice develop aortic root aneurism and aorto-mitral valve inflammation. (A) Kaplan–Meier curve showing the early death of BPSM1 mutant mice on the BALB/c background (black) compared with WT BALB/c mice (blue) or BPSM1 mutant mice on the C57BL/6 background (red). (B and C) Transverse sections through the heart show inflammation of the valves and the dramatic enlargement of the aortic root (dotted circle) in an affected (+/m) BPSM1 mutant mouse on the BALB/c background compared with a WT control (+/+). (D and E) Longitudinal sections through the heart of 141-d-old unaffected (+/+) and affected (+/m) BPSM1 mice of the C57BL/6 background showing inflammation of the valves and limited enlargement of the aortic root. (F and G) The spectacular valve inflammation present in a 46-d-old homozygous (m/m) affected BPSM1 mouse is completely prevented by the loss of TNFR1, even in a 156-d-old homozygote mutant. (H) Transaortic blood velocity measured by echocardiography shows regurgitation in BPSM1+/m mice.
Fig. S5.
Aortic regurgitation in BPSM1+/m mice. The presence or absence of aortic valve regurgitation was evaluated by color Doppler from a modified right parasternal view of the aortic arch. (Left) Blood flow upon valve opening. (Right) The flow upon valve closure 45 ms later. Aortic regurgitation appears as a retrograde jet from the aorta into the left ventricle.
Fig. S6.
Heart valve disease in TNFΔARE mice. Transverse sections through the heart show inflammation of the mitral valves in a 21-d-old TNFΔARE/ΔARE homozygote and a 133-d-old TNF+/ΔARE heterozygote on the C57BL/6 genetic background.
Discussion
We describe here the consequences of the insertion of a SINE in the 3′UTR of TNF. More than 200 copies of this particular retrotransposon or highly related sequences (>95% identity) are found throughout the mouse genome, either as stand-alone entities (i.e., Tmem176a) or integrated in other genes such as Als2, Chpt1, or Mfsd11. Although previously thought of as “junk DNA,” SINEs are now considered major drivers of evolution as their integration into genes can lead to new functionalities for those genes. In the BPSM1 mice, the insertion of the SINE causes a strong overexpression of TNF due to the replacement of the regulatory sequence contained in TNF 3′UTR by the UTR and the polyadenylation signal contained in the SINE. Not surprisingly, BPSM1 mice show a phenotype similar to that of the previously described TNFΔARE mice (17), with a few interesting differences. BPSM1 mice in our mouse facility failed to develop gut inflammation, whereas it was observed in the original report about TNFΔARE mice. As our studies definitively show that genetic background can have a major influence on the phenotype, the fact that BPSM1 mice are on a pure C57BL/6 background whereas TNFΔARE mice were maintained on a mixed 129SvxC57BL/6 background could be an explanation. However, Kontoyiannis et al. also observed that arthritis in TNFΔARE mice develops even in the absence of significant gut pathology, i.e., in the RAG−/− background (17). Because RAG−/− mice are kept under specific pathogen-free conditions, it is also possible that the absence of IBD in this case may have been due to a difference of microbiota, which plays a critical role in the aetio-pathology of IBD (25). We have placed BPSM1 mice in a different, “dirtier,” mouse facility to test this hypothesis, but have so far failed at inducing IBD in these mice.
We have found that TNF overexpression causes a dramatic inflammation of the aortic and mitral valves, accompanied by a fatal aortic aneurism when mice are kept on the BALB/c genetic background. Even though it was not reported before, we found that TNFΔARE mice also present with inflammation of the same heart valves. This phenotype of TNFΔARE mice was previously overlooked probably because it is not fatal on the mixed 129SvxC57BL/6 background. Although such an endeavor is beyond the scope of the present study, it would be of great interest to identify the modifier locus that causes the aortic root aneurism in BPSM1+/m mice on the BALB/c background, as this could provide a further clue about the mechanism that causes this condition.
TNF 3′UTR is known to contain two distinct elements bound by at least two different CCCH-containing ZFP. Zfp36 binds the ARE in region 4, whereas Roquin binds the constitutive decay element (CDE) in region 5 (26). Many other CCCH-containing ZFP are expressed in myeloid cells and participate in their activation (23). Our reporter assay led to the identification of new regulators of TNF and, more importantly, of an NRE located in region 6 at the 3′ end of TNF 3′UTR, just before the polyadenylation site. We showed that the NRE acts in synergy with the ARE. Insertion of the SINE shortly after the ORF effectively truncates most of the 3′UTR from the gene and thereby eliminates the control by ARE, CDE, and NRE in our BPSM1 mice. It might be argued that loss of the ARE is responsible for most of the phenotype because BPSM1 mice are very similar to TNFΔARE mice. However, a closer examination of the cloning strategy used to generate TNFΔARE mice shows that the NRE is also destroyed in these mice. Indeed, the loxP-neo-loxP cassette used for selection of embryonic stem cells was inserted within the BstEII restriction site located within the NRE (17). Although this probably explains why BPSM1 mice are so similar to the TNFΔARE mice, it also casts a doubt about the conclusions on the importance of Zfp36 and the ARE in TNF regulation and leaves open the possibility that other factors such as Zc3h12a, Zc3h12c, Mkrn1, Unk, and Zc3h10 might have a similar importance in it. It might also explain why myeloid-specific Zfp36 deficiency did not phenocopy the complete Zfp36 deficiency and in particular did not cause arthritis (22). The identification of Zc3h10 and Unk as positive regulators of TNF expression is particularly interesting as it offers two new potential therapeutic targets to control TNF expression.
RA is a complex disease that does not always involve TNF (15), and it will be of particular interest to elucidate whether mice with disruptions of Nlrp3 inflammasome/IL-1 signaling also develop heart disease. Recent research revealed striking similarities in the mechanisms that control heart valve cell differentiation and those involved in cartilage, tendon, and bone development (27). It is therefore logical that a phenomenon that causes histopathology and dysfunction of one system would also affect the other; this provides insight into the reason why heart disease is more common in arthritis patients.
Collectively, our results show that 30 y after TNF was first cloned, we are still discovering key pathways governing its regulation. Once fully elucidated, we shall be able to screen for alterations in these regulatory mechanisms and hence aid in the identification of patients most likely to respond to anti-TNF therapies. We anticipate that our BPSM1 mice will be very valuable for the preclinical evaluation of therapies relevant to heart valve disease and rheumatoid arthritis.
Materials and Methods
Mice.
All animal experiments were conducted with the approval of the Animal Ethics Committee of the Walter and Eliza Hall Institute. BPSM1 mice were the result of a spontaneous mutation. The generation of mice devoid of CD3ε, Myd88, GM-CSF, TNF, TNF-R1, as well as that of Mb1-cre has been described (7, 8, 11, 13, 16, 28). Conclusions were reached after examining at least 10 mice of each genotype. All mice were on the C57BL/6 genetic background with the exception of BP/BALB/c mice.
Bone Marrow-Derived Macrophages.
Bone marrow cell suspensions were cultured for 7 d in DMEM + 10% FBS (vol/vol) + 20% (vol/vol) L929 mouse fibroblast conditioned medium. RNA from the resulting BMDM were prepared using a Dynabeads mRNA DIRECT Purification Kit (Life Technologies). Northern blot was performed with the NorthernMax-Gly Kit (Life Technologies). RT-PCR was carried out with forward 5′-TGCCTATGTCTCAGCCTCTT-3′ and reverse 5′-GCCTTGTCCCTTGAAGAGAA-3′ primers for TNF and with forward 5′-TGTATGCCTCTGGTCGTACC-3′ and reverse 5′-CAACGTCACACTTCATGATGG-3′ primers for actin.
Bone Marrow Transplantation Experiments.
Recipients were lethally irradiated (2 × 550 rad) and injected with 1–2 × 106 bone marrow cells.
TNF Western Blot and ELISA.
Secreted TNF and IL-6 were measured from BMDM culture medium or mouse serum using the murine TNF and IL-6 ELISA Ready-Set-Go reagents (eBioscience) and read using the Chameleon plate reader (Hidex). For the Western blot, TNF antibodies were from the same kit as above, and the HSP70 antibody was mouse monoclonal mAb N6 (gift from Robin Anderson, Peter MacCallum Cancer Research Institute, Melbourne).
Analysis of Serum Cytokines and Chemokines.
Cytokines and chemokines were measured in mouse sera by multiplex bead array using the Bio-Plex mouse cytokine 23-plex panel (Bio-Rad Laboratories), according to the manufacturer’s instructions. The assay was read on a Bio-Plex 200 instrument and analyzed using Bio-Plex Manager version 5.0 software.
TNF 3′UTR Reporter Assays.
GFP reporter constructs were engineered by replacing the Luciferase-coding region of the pGL3-promoter vector (Promega) with EGFP and then introducing either the murine (WT or BPSM1-derived) or the human TNF 3′UTR between the Xba1 and BamH1 sites (complete sequences upon request). HEK293T cells were transiently transfected using Fugene 6 (Promega) with GFP-TNF3′UTR reporter constructs, a pGL3-mCherry control construct and one of the 50 CCCH ZFP pCMV6 plasmids (Origene), and analyzed 3 d later using flow cytometry on a LSR IIW (BD Biosciences). GFP mean fluorescence intensity was calculated on live mCherry-positive cells and compared with empty vector control.
Micro X-Ray Computed Tomography.
The micro X-ray computed tomography measurements were carried out using an Xradia micro XCT200 system (Carl Zeiss X-ray Microscopy, Inc.). TXMReconstructor software was used to reconstruct a 3D image from the 2D projection dataset, and a TXM3DViewer (Carl Zeiss X-ray Microscopy, Inc.) was used for the 3D visualization and creating the movies.
Echocardiography.
In vivo aortic ultrasound images were obtained using a Vevo 2100 Imaging System (VisualSonics) with a 40-MHz probe. Mice were anesthetized using 1.4–1.8% isoflurane in room air during image acquisition and were in a supine position on the heating pad of the Vevo system, with continuous monitoring of the rectal temperature and ECG. The presence or absence of aortic valve regurgitation was evaluated by color Doppler from a modified right parasternal view of the aortic arch, and Doppler velocity was sampled from the transverse aorta.
Supplementary Material
Acknowledgments
We thank J. Stanley, S. O’Connor, T. Bertenis, and G. Siciliano for animal care and expertise; S. Wilcox, M. Robati, and A. Lin for technical assistance; J. Corbin for automated blood analysis; B. Malissen (Marseille Luminy) for CD3ε KO mice; M. Reth (Max-Planck Institute of Immunobiology) for Mb1-cre KI mice; S. Akira (Osaka University Immunology Frontier Research Center) for Myd-88 KO mice; T. Mak (The Campbell Family Institute for Breast Cancer Research) for TNFR1 KO mice; I. Wicks for GM-CSF KO mice; J. Silke for TNF KO mice; G. Thomas for TNFΔARE mice; and A. Strasser for critically reviewing the manuscript. This work was supported by the Australian National Health and Medical Research Council (NHMRC; Program Grant 461221, Research Fellowship 1042629); the Leukemia and Lymphoma Society (Specialized Center of Research Grant 7015); the Arthritis Australia Zimmer fellowship; and infrastructure support from the NHMRC (Independent Research Institutes Infrastructure Support Scheme) and the Victorian State Government (Operational Infrastructure Support).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508399112/-/DCSupplemental.
References
- 1.Lee DM, Weinblatt ME. Rheumatoid arthritis. Lancet. 2001;358(9285):903–911. doi: 10.1016/S0140-6736(01)06075-5. [DOI] [PubMed] [Google Scholar]
- 2.van der Horst-Bruinsma IE, Lems WF, Dijkmans BA. A systematic comparison of rheumatoid arthritis and ankylosing spondylitis. Clin Exp Rheumatol. 2009;27(4) Suppl 55:S43–S49. [PubMed] [Google Scholar]
- 3.Roldan CA, DeLong C, Qualls CR, Crawford MH. Characterization of valvular heart disease in rheumatoid arthritis by transesophageal echocardiography and clinical correlates. Am J Cardiol. 2007;100(3):496–502. doi: 10.1016/j.amjcard.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 4.Voskuyl AE. The heart and cardiovascular manifestations in rheumatoid arthritis. Rheumatology (Oxford) 2006;45(Suppl 4):iv4–iv7. doi: 10.1093/rheumatology/kel313. [DOI] [PubMed] [Google Scholar]
- 5.Palazzi C, Salvarani C, D’Angelo S, Olivieri I. Aortitis and periaortitis in ankylosing spondylitis. Joint Bone Spine. 2011;78(5):451–455. doi: 10.1016/j.jbspin.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Peters MJ, van der Horst-Bruinsma IE, Dijkmans BA, Nurmohamed MT. Cardiovascular risk profile of patients with spondylarthropathies, particularly ankylosing spondylitis and psoriatic arthritis. Semin Arthritis Rheum. 2004;34(3):585–592. doi: 10.1016/j.semarthrit.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA. 2006;103(37):13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malissen M, et al. Altered T cell development in mice with a targeted mutation of the CD3-ε gene. EMBO J. 1995;14(19):4641–4653. doi: 10.1002/j.1460-2075.1995.tb00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keffer J, et al. Transgenic mice expressing human tumour necrosis factor: A predictive genetic model of arthritis. EMBO J. 1991;10(13):4025–4031. doi: 10.1002/j.1460-2075.1991.tb04978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawane K, et al. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443(7114):998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- 11.Adachi O, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9(1):143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 12.Nair JR, Edwards SW, Moots RJ. Mavrilimumab, a human monoclonal GM-CSF receptor-α antibody for the management of rheumatoid arthritis: A novel approach to therapy. Expert Opin Biol Ther. 2012;12(12):1661–1668. doi: 10.1517/14712598.2012.732062. [DOI] [PubMed] [Google Scholar]
- 13.Stanley E, et al. Granulocyte/macrophage colony-stimulating factor-deficient mice show no major perturbation of hematopoiesis but develop a characteristic pulmonary pathology. Proc Natl Acad Sci USA. 1994;91(12):5592–5596. doi: 10.1073/pnas.91.12.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubbert-Roth A, Finckh A. Treatment options in patients with rheumatoid arthritis failing initial TNF inhibitor therapy: A critical review. Arthritis Res Ther. 2009;11(Suppl 1):S1. doi: 10.1186/ar2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vande Walle L, et al. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature. 2014;512(7512):69–73. doi: 10.1038/nature13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Körner H, et al. Distinct roles for lymphotoxin-alpha and tumor necrosis factor in organogenesis and spatial organization of lymphoid tissue. Eur J Immunol. 1997;27(10):2600–2609. doi: 10.1002/eji.1830271020. [DOI] [PubMed] [Google Scholar]
- 17.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: Implications for joint and gut-associated immunopathologies. Immunity. 1999;10(3):387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 18.Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol. 2008;9(4):378–387. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 19.Armaka M, et al. Mesenchymal cell targeting by TNF as a common pathogenic principle in chronic inflammatory joint and intestinal diseases. J Exp Med. 2008;205(2):331–337. doi: 10.1084/jem.20070906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niki Y, et al. Macrophage- and neutrophil-dominant arthritis in human IL-1 alpha transgenic mice. J Clin Invest. 2001;107(9):1127–1135. doi: 10.1172/JCI11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor GA, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4(5):445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 22.Qiu LQ, Stumpo DJ, Blackshear PJ. Myeloid-specific tristetraprolin deficiency in mice results in extreme lipopolysaccharide sensitivity in an otherwise minimal phenotype. J Immunol. 2012;188(10):5150–5159. doi: 10.4049/jimmunol.1103700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang J, Song W, Tromp G, Kolattukudy PE, Fu M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One. 2008;3(8):e2880. doi: 10.1371/journal.pone.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarzi-Puttini P, Atzeni F, Shoenfeld Y, Ferraccioli G. TNF-alpha, rheumatoid arthritis, and heart failure: A rheumatological dilemma. Autoimmun Rev. 2005;4(3):153–161. doi: 10.1016/j.autrev.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Hold GL, et al. Role of the gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World J Gastroenterol. 2014;20(5):1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leppek K, et al. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013;153(4):869–881. doi: 10.1016/j.cell.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 27.Lincoln J, Lange AW, Yutzey KE. Hearts and bones: Shared regulatory mechanisms in heart valve, cartilage, tendon, and bone development. Dev Biol. 2006;294(2):292–302. doi: 10.1016/j.ydbio.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 28.Pfeffer K, et al. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell. 1993;73(3):457–467. doi: 10.1016/0092-8674(93)90134-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.