Significance
Given the importance of autophagy in a number of human diseases, we have identified small-molecule modulators of autophagy that affect disease-associated phenotypes in relevant cell types. BRD5631 and related compounds can serve as tools for studying how autophagy regulates immune pathways, and for evaluating the therapeutic potential of modulating autophagy in a variety of disease contexts. Deeper investigation into their mechanisms of action may reveal proteins and pathways that could serve as relevant targets for future therapeutic discovery.
Keywords: high-throughput screening, autophagy, small-molecule probes, Crohn’s disease
Abstract
Studies of human genetics and pathophysiology have implicated the regulation of autophagy in inflammation, neurodegeneration, infection, and autoimmunity. These findings have motivated the use of small-molecule probes to study how modulation of autophagy affects disease-associated phenotypes. Here, we describe the discovery of the small-molecule probe BRD5631 that is derived from diversity-oriented synthesis and enhances autophagy through an mTOR-independent pathway. We demonstrate that BRD5631 affects several cellular disease phenotypes previously linked to autophagy, including protein aggregation, cell survival, bacterial replication, and inflammatory cytokine production. BRD5631 can serve as a valuable tool for studying the role of autophagy in the context of cellular homeostasis and disease.
Macroautophagy (hereafter autophagy), an evolutionarily conserved catabolic process in which cytosolic content is engulfed and degraded in lysosomes, has been implicated in several human diseases, including inflammatory disorders such as Crohn’s disease (CD), immune responses to Mycobacterium tuberculosis (Mtb), α1-antitrypsin deficiency, nonalcoholic fatty liver disease, and neurodegenerative disorders (1). Autophagy is frequently initiated by ULK1-mediated phosphorylation of Beclin1, leading to activation of the lipid kinase VPS34, which in turn leads to engagement of two ubiquitin (Ub)-like conjugation systems that modify ATG12 and ATG8 (LC3) to trigger expansion of autophagosomal membranes (2). Once initiated, degradation by autophagy can be either nonselective for bulk intracellular components (canonical) or selective for cargo such as damaged organelles (mitophagy), invasive pathogens (xenophagy), or protein aggregates (aggrephagy) (3).
Although many types of cells use autophagy, disease phenotypes linked to autophagy-related genes are often restricted primarily to specific organ systems. CD-associated genetic variants that regulate autophagy have been linked to phenotypes involving the intestinal epithelium and cytokine-producing innate immune cells that may reflect high microbial load. More specifically, disease-associated alleles in genes that regulate bacterial recognition (NOD2, IRGM) and autophagy (ATG16L1) are associated with reduced clearance of intracellular bacteria (4). Furthermore, expression of a coding variant in ATG16L1 (T300A) correlates with compromised bacterial defense, increased release of inflammatory cytokines (IL-1β), and reduced formation of antimicrobial granules in gut Paneth cells (5, 6). Similar organ-associated phenotypes have been observed for variants linked to other diseases. Mutations in the autophagy gene WDR45 have been associated with static encephalopathy of childhood with generation in adulthood (SENDA), and SENDA disease phenotypes (iron accumulation and cerebral atrophy) appear to be restricted to the brain despite WDR45 expression in skeletal muscle (7). Brain-related disease phenotypes have also been observed in patients with a form of hereditary spastic paraparesis bearing a recessive mutation in the putative autophagy regulator TECPR2, and in patients suffering from cerebellar atrophy associated with mutations in the autophagy modulator SNX14 (8, 9).
Small-molecule enhancers of autophagy are increasingly being tested for their beneficial effects on disease phenotypes in relevant organ systems and cell types (10). For example, different enhancers exhibit distinct profiles of activity in innate and adaptive immune pathways, including host defense against pathogens (11). Phenothiazine-derived antipsychotics (prochlorperazine edisylate) and first-line antibiotics (isoniazid and pyrazinamide) promote autophagy-dependent activity against Mtb, the latter involving modulation of proinflammatory responses in Mtb-infected macrophages (12, 13). Likewise, the autophagy-inducing peptide Tat-Beclin1 enhances autophagy-dependent antiviral activity in cell and animal models (14). In the context of protein aggregation disorders, carbamazepine (CBZ)-induced autophagy reduces accumulation of α1-antitrypsin in liver cells and rescues hepatic fibrosis in mice (15), whereas rapamycin- or trehalose-induced autophagy clears α-synuclein and mutant huntingtin aggregates in neurodegenerative disease models, among others (16). Autophagy defects linked to the lysosomal storage disorder Niemann–Pick type C1 disease (NPC1; caused by mutation in the NPC1 protein) can be restored by stimulating autophagy with rapamycin or CBZ, which is cytoprotective in NPC1 disease-relevant cells (17, 18). In the context of CD, small molecules have been shown to promote favorable immune phenotypes. Treatment with the PDK1 inhibitor AR-12 in macrophages (19) or rapamycin in epithelial cells (20) promotes bacterial clearance, and treatment with rapamycin leads to reduced IL-1β levels in innate immune cells and animals, possibly via degradation of pro-IL-1β or its processing machinery (21).
Though modulating the mTOR pathway appears to affect disease phenotypes in several contexts, mTOR signaling is also critical for nutrient sensing, cell growth, and other fundamental processes. For several reasons, it may be beneficial to avoid perturbation of this critical cellular pathway in treating disease; for example, its inhibition has also been linked to infection (22). Additional small molecules have been shown to induce autophagy independently of mTOR (e.g., trifluoperazine, niguldipine, ABT-737). These compounds span a range of annotated protein targets and signaling pathways, but the relevance of the annotated targets to promotion of autophagy often remains undefined and may in some cases constitute impediments for further development (1, 10). Identification of small-molecule modulators of canonical and selective autophagy in disease-relevant cell types with novel and distinct mechanisms of action will contribute to studying the relevance of autophagy in disease and the potential of autophagy enhancement as a therapeutic strategy.
Here, we describe the identification of novel enhancers of autophagy using a high-throughput screen (HTS) of 59,541 small molecules prepared by stereoselective diversity-oriented chemical synthesis (23). We demonstrate that representative hits promote autophagy without perturbing mTOR signaling or lysosomal function, and display autophagy-dependent activity in a number of disease-relevant models, including suppression of NPC1-triggered apoptosis in human induced pluripotent stem cell (hiPSC)-derived neurons, enhancement of bacterial colocalization with autophagic markers and clearance in epithelial cells, and suppression of microbial product-mediated IL-1β secretion by macrophages. One probe, BRD5631, is capable of rescuing defects in bacterial colocalization with LC3 and suppressing IL-1β production in cells harboring the CD-associated allele of ATG16L1 (T300A). BRD5631 and related compounds constitute a novel set of mTOR-independent autophagy probes for studying the regulation of autophagy and its impact on disease pathways.
Results
High-Throughput Screening Identifies Novel Small-Molecule Modulators of Autophagy.
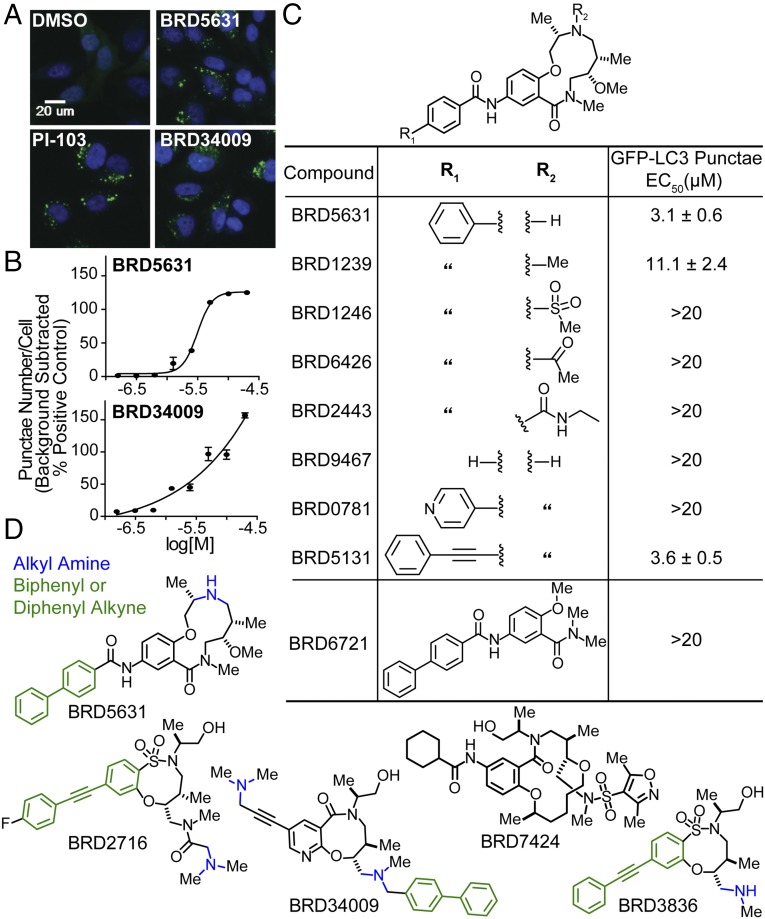
To discover novel small-molecule modulators of canonical autophagy, we performed a HTS of 59,541 stereochemically and skeletally diverse compounds derived from diversity-oriented synthesis (DOS). These molecules are enriched for sp3-hybridized atoms relative to conventional commercial libraries (24–26), resulting in topographically rich 3D structures. The primary HTS measured autophagosome number in HeLa cells stably expressing GFP-LC3 (Fig. 1A). LC3 normally displays a diffuse cytosolic pattern (LC3-I), but upon induction of autophagy, LC3 is posttranslationally modified (LC3-II) and accumulates on autophagosome membranes. This change can be visualized and quantified by counting the number of GFP-LC3 punctae per cell by microscopy (27). Significance of changes was assessed by comparing the distribution of punctae per cell for each compound (test distribution) to that of DMSO (null distribution), and computing the area under the curve of the test distribution beyond the critical value corresponding to 95% confidence of the null distribution.
Fig. 1.
HTS identifies DOS-derived small-molecule autophagy modulators. (A and B) HeLa cells stably expressing GFP-LC3 were treated with compounds in 8-point dose (4 h), and the number of GFP punctae per cell was quantified by fluorescence microscopy and automated image analysis. Representative images of cells treated with DMSO, PI-103 (2.5 µM), BRD5631 (10 µM), or BRD34009 (10 µM) are shown. Data points of dose curves are presented as the mean ± SD, n = 3 from a representative experiment. (C) Results of GFP-LC3 punctae formation assay show that BRD5631 analogs corroborate structure-activity relationship trends observed in the primary HTS. Data are the mean ± SD of three independent experiments. (D) Chemical structures of five prioritized compounds.
To assess data quality, we confirmed that the distributions of punctae per cell for the negative control (DMSO) and the positive control (PI-103, a dual mTOR/PI3K inhibitor) showed strong and statistically significant separation. Typically, ∼70% of PI-103–treated cells exhibited numbers of punctae per cell greater than the 95% critical threshold for the DMSO distribution. We identified ∼2,000 hits that increased the average number of GFP-LC3 punctae per cell to a level ≥30% of the positive control (background-subtracted). A total of 998 of these hits were selected based on activity and structure and tested for effects on viability of HeLa cells (72-h treatment) using CellTiter-Glo; 85% of hits showed little to modest toxicity (<25% effect) at 10 μM (data not shown).
The number of GFP-LC3 punctae in the cell can be increased by either activating autophagy or by inhibiting autophagy at late steps in the process, such as by disrupting lysosome function. We prioritized 400 representative hits based on potency and chemical structure for testing in a secondary assay for autophagosome formation and maturation (flux) that relies on visualizing LC3 protein tagged with both mCherry and GFP. The GFP signal is attenuated in autolysosomes, while the mCherry signal remains stable (28); as a result, autophagosomes (GFP+/mCherry+) and autolysosomes (GFP−/mCherry+) can be distinguished and counted by high-throughput imaging (SI Appendix, Fig. S1). The number of GFP−/mCherry+ punctae (presumed autolysosomes) was increased by 250 hits (putative activators) and decreased by 80 hits (putative late-stage inhibitors) from the primary screen.
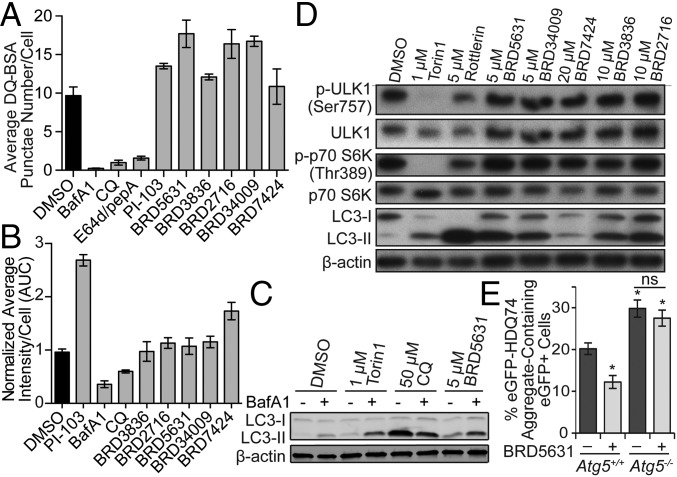
In analyzing the screening data, we noted that the hit rate for compounds having an alkyl amine (7%) was higher than that for all compounds (3%), an effect enhanced by the additional presence of a single lipophilic group, such as diphenyl alkyne (61%), biphenyl (26%), cyclohexane (21%), or naphthalene (37%) (SI Appendix, Fig. S2). By preparing and testing analogs of a potent hit, BRD5631 (Fig. 1 B and C), we confirmed that both structural features were important for its activity. Only analogs containing an amine retained activity, and both removal of the terminal phenyl group and its replacement with a pyridyl group eliminated activity of BRD5631. Compounds containing a lipophilic group and a basic amine can accumulate within lysosomes (lysosomotropism) and potentially interfere with lysosomal function, cell viability, and autophagy (29, 30). To determine whether our hits disrupt lysosomal function, we first tested their effect on lysosomal proteases by measuring processing of a fluorogenic substrate (DQ-BSA) (31). For these experiments, we selected four structurally representative hits, including BRD5631, as well as one hit lacking an amine (Fig. 1D), that display a range of activities in the GFP-LC3 assay (Fig. 1 A and B and SI Appendix, Fig. S3A) and mCherry-GFP-LC3 assay (SI Appendix, Fig. S1). As expected, control compounds that inhibit lysosomal proteases directly (E64d/pepstatin A) or indirectly by increasing lysosomal pH [bafilomycin A1 (BafA1) and chloroquine (CQ)] significantly decreased both DQ-BSA punctae number and intensity. In contrast, none of the selected hits reduced either the number or intensity of DQ-BSA punctae at doses that induce GFP-LC3 punctae (Fig. 2A and SI Appendix, Fig. S3B). Furthermore, the compounds did not disrupt accumulation of a fluorescent basic amine probe used to stain acidic compartments (LysoTracker Red; Fig. 2B and SI Appendix, Fig. S3C) or significantly affect viability at autophagy-modulating doses (<10% effect; SI Appendix, Fig. S3D) (32). These data suggest that BRD5631 and other hits do not increase GFP-LC3 punctae formation by broadly disrupting lysosomal function.
Fig. 2.
Functional characterization of autophagy modulators. (A) Compounds were evaluated in the DQ-BSA assay. BafA1 (100 nM), CQ (50 μM), E64d/pepA (10 μg/mL each), PI-103 (10 μM), and DOS compounds (10 μM each). Data are presented as mean ± SD, n = 3. (B) Compounds were evaluated in the LysoTracker assay in 8-point dose by twofold serial dilution from the following starting concentrations: PI-103 and DOS compounds (20 μM each), BafA1 (200 nM), CQ (100 μM). Data are presented as average area under the curve ± SD, n = 2. (C) HeLa cells were treated with indicated compounds (7 h). For the last 4 h, either DMSO or BafA1 (100 nM) was added, then protein samples were harvested for immunoblot analysis. (D) HeLa cells were treated with compounds (4 h), then harvested for immunoblot analysis. (E) Atg5+/+ and Atg5−/− MEFs were transfected with eGFP-HDQ74 construct followed by treatment with ±BRD5631 (10 µM; 48 h). Percentage of eGFP+ cells with eGFP-HDQ74 aggregates was quantified by fluorescence microscopy. Data are presented as mean ± SEM, n = 3. *P < 0.05, ns, not significant, compared to untreated Atg5+/+ MEFs, unless indicated otherwise, unpaired Student t test. All data are representative of multiple independent experiments.
We then characterized BRD5631 and the other selected compounds more deeply in mechanistic and functional assays for autophagy. Consistent with the results from microscopy, we found that BRD5631 led to increased levels of LC3-II by Western blot (Fig. 2C). When cotreated with the vacuolar H+-ATPase inhibitor BafA1 (a known late-stage inhibitor of autophagy), BRD5631 was able to increase LC3-II levels above those observed with BafA1 alone, suggesting that BRD5631 stimulates formation of new autophagosomes (Fig. 2C). These data are consistent with its ability to increase numbers of autolysosomes (mCherry+/GFP−) in the mCherry-GFP-LC3 assay. Similar results were observed with other screening hits to a lesser degree (SI Appendix, Fig. S3E), with the exception of BRD34009, which induced LC3-II formation but showed modest inhibitory activity in the mCherry-GFP-LC3 assay.
Because inhibition of mTOR signaling is known to promote autophagy, we determined whether the compounds affected phosphorylation of mTOR-regulated proteins, including p70 S6 kinase 1 (S6K1) (Thr389) (33) and the autophagy-initiating kinase ULK1 (Ser757) (34). Consistent with previous reports (33, 35), we confirmed that treatment with the mTOR kinase inhibitor torin1 and mTOR pathway modulator rottlerin led to decreased phosphorylation of S6K1 and ULK1 (Fig. 2D). In contrast, BRD5631 and other screening hits did not significantly affect S6K1 or ULK1 mTOR-dependent phosphorylation levels at 10 μM, suggesting that they do not enhance autophagy by directly inhibiting mTOR or the mTOR signaling pathway.
BRD5631 Decreases the Presence of Aggregates in Cells Expressing Poly-Q Repeats (eGFP-HDQ74).
We assessed the ability of BRD5631 to promote clearance of an autophagic cargo using the well-established substrate, mutant huntingtin (eGFP-HDQ74) (36). Transient overexpression of eGFP-HDQ74 in autophagy-deficient cells [Atg5−/− mouse embryonic fibroblasts (MEFs)] causes increased aggregate formation compared with their wild-type counterparts (Atg5+/+ MEFs) due to its inability to be degraded by autophagy (37). Treatment with BRD5631 significantly reduced the number of eGFP-HDQ74-positive cells in Atg5+/+ MEFs; this reduction was not observed in Atg5−/− MEFs, suggesting BRD5631 promotes clearance of mutant huntingtin in an autophagy-dependent manner (Fig. 2E). We also measured changes in the level of another specific autophagy substrate, p62 (38), in response to BRD5631 in these cells. Under steady-state conditions, Atg5−/− MEFs displayed increased p62 levels compared with Atg5+/+ MEFs. BRD5631 substantially increased LC3-II levels in Atg5+/+ MEFs as expected; however, it increased p62 levels in Atg5+/+ MEFs without any detectable effect in Atg5−/− MEFs (SI Appendix, Fig. S4A). Transcriptional up-regulation of p62 in response to autophagy activation has been previously reported (39), complicating interpretation of its levels as a readout for autophagic flux. We also observed that BRD5631 increased the transcript level of p62 in Atg5+/+ MEFs (SI Appendix, Fig. S4B), suggesting a complex effect of BRD5631 on p62 levels in this cellular context.
BRD5631 Suppresses NPC1-Induced Cell Death in a hiPSC-Derived Neuronal Model of Niemann–Pick Type C1 Disease.
We have recently shown that small-molecule enhancers of autophagy, such as CBZ, can rescue impairment of autophagic flux observed in NPC1, a lipid/lysosomal storage disease exhibiting accumulation of cholesterol in the late endosomal/lysosomal compartments and associated with degeneration of the cerebellum and liver in patients (17, 18). These enhancers also suppress cell death in NPC1 patient-specific hiPSC-derived neuronal and hepatic cells, which normally display cholesterol accumulation, impaired autophagic flux, and increased cell death in the absence of any external stressors relative to control cells (18). Using this system, we assessed the ability of the prioritized compounds to rescue the NPC1-associated loss of cell viability relative to the positive control (100 μM CBZ). We found that BRD5631, BRD2716, and BRD34009 significantly reduced cell death in NPC1 hiPSC-derived neurons (Fig. 3A). Interestingly, these compounds did not rescue increased cell death in NPC1 hiPSC-derived hepatic cultures (SI Appendix, Fig. S5). We further examined the impact of these compounds on autophagic flux by measuring p62 levels in NPC1 hiPSC-derived neurons, which display accumulation of this autophagy substrate relative to control cells (18) (Fig. 3B). In contrast to the results obtained in MEFs, treatment with BRD5631, BRD2716, and BRD34009 reduced levels of endogenous p62 in NPC1 neuronal cells (Fig. 3B). The correlation between the neuroprotective effects of BRD5631, BRD2716, and BRD34009 and clearance of autophagy substrates in NPC1 neuronal cells suggests a possible role for increased autophagic flux in rescuing cells from the disease phenotype.
Fig. 3.
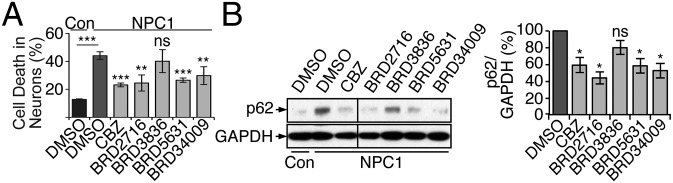
A subset of autophagy modulators rescue impairments in NPC1 disease hiPSC-derived neurons. (A) Control (WIBR3-derived) neurons were treated with DMSO, and NPC1 hiPSC (WIBR-IPS-NPC1–derived) neurons were treated with DMSO, DOS compounds (10 μM each), or CBZ (100 μM) for 3 days. Cell death was quantified using TUNEL assay and apoptotic nuclear morphology. Data are presented as the mean ± SD and are representative of two independent experiments, each run in triplicate. (B) Cell populations treated as described in A were subjected to immunoblot analysis. All samples were run in the same gel and blotted at the same time. The line denotes where one intervening lane was excised. Densitometric analysis shows p62 levels relative to GAPDH in NPC1 hiPSC-derived neurons. The control condition was set to 100%, and data represent the mean ± SEM of samples from the same experiment in A. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant, compared to DMSO-treated NPC1 cells, unless indicated otherwise, unpaired Student t test.
BRD5631 Enhances Bacterial Clearance in an Autophagy-Dependent Manner.
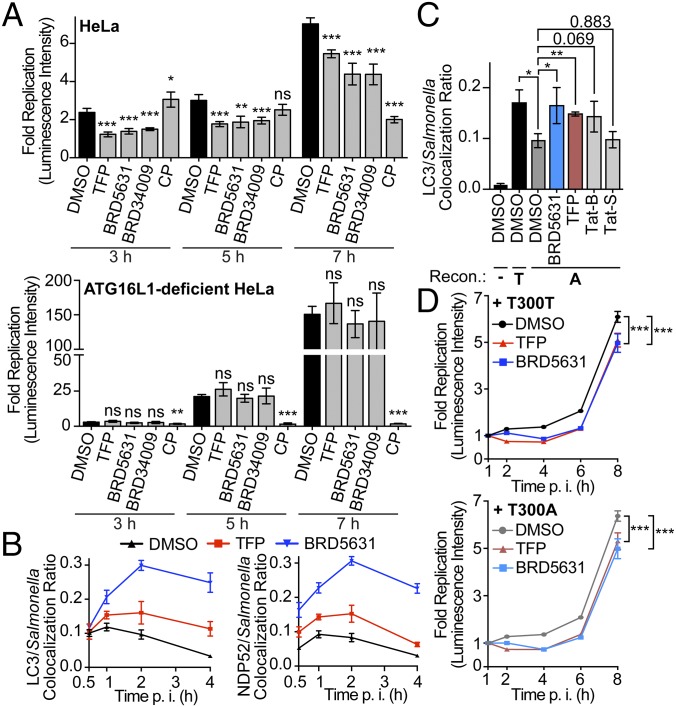
Several bacterial pathogens are selectively targeted by autophagy for degradation (bacterial xenophagy), including Salmonella, Shigella, Mtb, and group A Streptococcus (40). Furthermore, compromised antibacterial autophagy has been implicated by studies of disease-associated genes (ATG16L1, IRGM), potentially contributing to inflammation via an increased microbial burden within the intestinal epithelium (4). To determine if BRD5631 and other hits can promote antibacterial autophagy, we first measured their ability to inhibit bacterial replication using a bioluminescent strain of Salmonella enterica serovar Typhimurium infecting mammalian epithelial cells (HeLa). Three of five prioritized autophagy modulators (BRD5631, BRD34009, BRD2716) inhibited Salmonella replication over time, beginning as early as 3 h postinfection (p.i.), to similar or greater levels than an autophagy modulator reported to have anti-Salmonella activity, trifluoperazine (TFP) (41) (Fig. 4A and SI Appendix, Fig. S6 A and B). Traditional CFU assays confirmed the results of the luminescence assay (SI Appendix, Fig. S6C). None of the prioritized small molecules displayed general bactericidal activity in the absence of host cells or cytotoxicity to host cells at the concentration used for the clearance assay (SI Appendix, Fig. S6 D and E).
Fig. 4.
Autophagy modulators clear invasive bacteria. (A) Salmonella survival in cells was assessed in the bioluminescent bacterial replication assay. Luminescence intensity in live cells was measured at each indicated time point, and fold replication values were calculated for each well as raw luminescence intensity divided by the intensity at 1 h p.i. Data are presented as mean ± SD, n = 4 from a representative experiment. (B) HeLa cells were pretreated with compound (3 h) and infected with Salmonella. Cells were then fixed and subjected to immunostaining at indicated time points. Colocalization ratios were calculated to compare Salmonella colocalized with LC3 or NDP52 to total Salmonella. Data are presented as mean ± SD, n = 4 from a representative experiment. (C) ATG16L1-deficient HeLa cells were reconstituted with ATG16L1 T300T (T) or T300A (A), seeded on coverslips, pretreated, infected, and fixed and stained (1 h p.i.). Data are presented as mean ± SD of three independent experiments. At least 100 bacteria per coverslip and triplicated coverslips in each experiment were scored. (D) ATG16L1-deficient HeLa cells were reconstituted as described in C and infected, treated, and imaged as described in A. In A–D, all compounds were treated at 10 μM. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant, unpaired Student t test.
To assess the dependence of replication inhibition on autophagy, we tested the antibacterial activity of hits on a clonal ATG16L1-deficient HeLa cell line generated using the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system (42) (SI Appendix, Fig. S6F). ATG16L1 forms a complex with ATG5-ATG12 conjugates to promote conjugation of LC3 to phosphatidylethanolamine and elongation of autophagosomal membranes (43). As expected, ATG16L1-deficient cells displayed significantly greater levels of bacterial replication than the parental line (SI Appendix, Fig. S6G), and an antibiotic that directly targets bacteria [chloramphenicol (CP)] was able to suppress bacterial replication in both cell lines. When we treated the ATG16L1-deficient cells with BRD5631 and other hits, no reduction in bacterial replication was observed (Fig. 4A and SI Appendix, Fig. S6A). Although the interpretation may be complicated by the increased rate of replication in the knockout cells, these data suggest that autophagy is required for the effect of BRD5631 on bacterial clearance.
To determine whether compounds that inhibit replication of intracellular Salmonella affect its association with the antibacterial autophagy machinery, we measured the colocalization of dsRed-expressing Salmonella with both LC3 and NDP52, an adaptor protein that targets galectin-8- or Ub-coated Salmonella to autophagy (40). As reported previously, treatment with TFP enhanced the maximal colocalization of Salmonella and LC3 and NDP52 observed at 1 h p.i., which was maintained at later time points (4 h p.i.) when the colocalization events subsided in vehicle control-treated cells (Fig. 4B). BRD5631 was able to moderately enhance colocalization events at 1 h p.i. and later time points to a higher level than TFP (Fig. 4B).
The CD risk allele ATG16L1 T300A has been linked to impaired colocalization of Salmonella and LC3 in genetically modified HeLa cells (44). To determine if BRD5631 is able to overcome defects in colocalization of Salmonella and LC3 in the presence of this risk allele, we performed the colocalization assay in ATG16L1-deficient HeLa cells reconstituted with wild-type ATG16L1 or ATG16L1 T300A (SI Appendix, Fig. S6H). As reported, LC3 recruitment was diminished in the absence of ATG16L1. Reintroducing ATG16L1 T300A partially rescued the colocalization ratio, but significantly less than the ratio in cells reconstituted with wild-type ATG16L1 (Fig. 4C). TFP and BRD5631, as well as the autophagy-inducing peptide Tat-Beclin1, moderately enhanced the colocalization of LC3 and Salmonella in ATG16L1 T300A cells (Fig. 4C). Similarly, BRD5631 and TFP inhibited bacterial replication in the presence of ATG16L1 T300A (Fig. 4D).
BRD5631 Suppresses IL-1β Secretion in an Autophagy-Dependent Manner.
Previous studies have demonstrated a role for autophagy in the regulation of IL-1β levels. Although activation of autophagy by rapamycin has been shown to induce pro–IL-1β degradation and decreased IL-1β secretion in macrophages (21), the effect of mTOR-independent modulation of autophagy on cytokine production has not been systematically explored. To determine whether the mTOR-independent autophagy modulators discovered in our screen could regulate IL-1β release in response to microbial TLR4 and NOD2 ligands, we tested BRD5631 and other hits in IFN-γ–primed macrophages stimulated with lipopolysaccharide (LPS) and muramyl dipeptide (MDP). LPS-MDP stimulated robust IL-1β secretion from immortalized bone marrow-derived macrophages (Fig. 5A). The antiinflammatory glucocorticoid dexamethasone (Dex) and the caspase1 inhibitor VX-765, compounds that target transcriptional and posttranslational steps in IL-1β production, respectively, significantly reduced IL-1β release (Fig. 5A). All tested DOS-derived autophagy modulators reduced IL-1β secretion at 10 μM (Fig. 5A). Furthermore, the effect of these compounds on IL-1β secretion was dose-dependent and not associated with cytotoxicity (SI Appendix, Fig. S7 A and B).
Fig. 5.
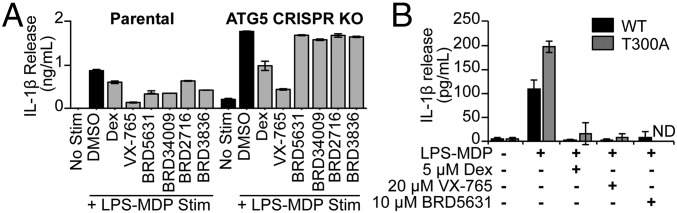
Autophagy modulators suppress IL-1β secretion. (A) IL-1β secretion was measured in immortalized murine bone marrow-derived macrophages primed with IFN-γ (100 ng/mL) (16 h) and stimulated with LPS (10 ng/mL)-MDP (10 μg/mL) (24 h), both in the presence of Dex (5 μM), VX-765 (20 μM), or DOS compounds (each 10 μM). Secreted IL-1β was detected by ELISA. Data are presented as mean ± SD, n = 3 from a representative experiment of three independent experiments. (B) IL-1β secretion was measured in murine splenic CD11b+ macrophages (ATG16L1 WT or T300A) primed with IFN-γ (100 ng/mL) (6 h) and then treated with IFN-γ (100 ng/mL)-LPS (2 ng/mL)-MDP (10 μg/mL) (24 h), both in the presence of Dex, VX-765, or BRD5631. Secreted IL-1β was detected by ELISA. Data are presented as mean ± SD, n = 4 mice. ND, not detected.
To determine whether the observed reduction in IL-1β release was autophagy-mediated, we assayed their IL-1β–repressive activity in immortalized bone marrow-derived macrophages in which Atg5 had been deleted by CRISPR. Consistent with Atg5 deletion, these cells show reduced levels of ATG5-ATG12 conjugates and LC3-II (SI Appendix, Fig. S7C). IL-1β secretion following LPS-MDP stimulation was significantly higher in the Atg5 CRISPR line compared with parental controls, consistent with previous reports for other autophagy-deficient cell models (5) (SI Appendix, Fig. S7D). All four autophagy modulators suppressed IL-1β release in the parental cell line with little to no effect in the Atg5 CRISPR cell line, suggesting that activities for these compounds are autophagy-mediated (Fig. 5A).
The CD risk allele ATG16L1 T300A has also been linked to increased levels of IL-1β in patients (45). To determine whether DOS-derived compounds are able to reduce IL-1β levels in the presence of this disease allele, we evaluated the activity of the compounds in ATG16L1 T300A cells derived from knock-in mice. Consistent with previous observations (5), splenic CD11b+ macrophages from ATG16L1 T300A mice demonstrated increased IL-1β release compared with wild-type controls (Fig. 5B). Interestingly, BRD5631 significantly reduced the elevated IL-1β levels observed in ATG16L1 T300A cells (Fig. 5B).
Discussion
Human genetic studies have linked autophagy to several diseases, and depending on the context, impairment of autophagy can lead to distinct phenotypes in specific cell types and organ systems. For example, CD-associated genetic variants in ATG16L1 have implicated autophagy defects as contributing to compromised antibacterial activity in epithelial cells, elevated IL-1β secretion from monocytes/macrophages, reduced Paneth cell function, and impaired antigen presentation in dendritic cells (4). These observations, as well as examples from other diseases, such as Niemann–Pick disease, TB infection, α1-antitrypsin deficiency, and neurodegenerative disorders, suggest that small-molecule modulation of canonical and/or selective autophagy in disease-relevant cell types could be therapeutically beneficial (1, 17).
Though several small-molecule enhancers of autophagy have been described, the mechanisms of action necessary to impact these disease-related immune pathways are not well understood. Autophagy induction via inhibition of mTOR signaling has been shown to promote the autophagic clearance of bacteria (20) and suppress IL-1β secretion by targeting pro–IL-1β for degradation (21). Though mTOR modulators are used therapeutically (10), they have been linked to several dose-limiting adverse effects, suggesting the importance of discovering novel, mTOR-independent modulators of autophagy. To this end, we have identified a set of novel DOS-derived small molecules that promote autophagy independently of mTOR, and that modulate a number of cell type-specific disease phenotypes.
Several compounds discovered in our screen for autophagy enhancers contain a biphenyl or diphenyl alkyne group and a basic amine. Although basic amines are thought to bias compounds for accumulation in acidic cellular compartments such as lysosomes (46), our data suggest that these molecules do not reduce cell viability, disrupt lysosomal protease function, or perturb accumulation of LysoTracker Red, making them distinct from known lysosomotropic molecules. It is certainly possible that the compounds accumulate at higher concentrations within acidic compartments and exert a specific function on a lysosomal protein or signal to regulate autophagy. Indeed, an analogous model has been proposed to underlie the activity of functional inhibitors of acid sphingomyelinase (ASM), which cause extrusion and consequent inactivation of ASM from the inner lysosomal membrane (47). Whether small molecules that operate through such a mechanism could be exploited in a therapeutic context remains unclear, but it is worth noting that similar structural moieties are present within previously reported FDA-approved enhancers of autophagy [e.g., calcium channel modulators (niguldipine) and D2 receptor modulators (TFP)] (48), arguing that such agents can be tolerated by humans. Further studies are needed to investigate how BRD5631 promotes autophagy in cells, and which regulatory pathways are involved in this process.
Importantly, BRD5631 appears to modulate several disease-associated phenotypes, including decreasing numbers of aggregate-expressing cells, reducing apoptosis of neurons in NPC1 models (at 10-fold lower concentration than CBZ), enhancing bacterial clearance, and decreasing IL-1β secretion. As such, studying BRD5631 and its mechanism of action may reveal therapeutically beneficial mechanisms for modulating autophagy in the context of human disease. For example, in CD patients, the risk allele ATG16L1 T300A has been linked to elevated IL-1β levels (45). Furthermore, the ATG16L1 T300A knock-in mouse model recapitulates many of the autophagy-relevant phenotypes observed in CD patients, including the overproduction of IL-1β in response to microbial ligands, providing a valuable system to test the impact of our compounds in the presence of a CD risk allele (5). We demonstrated that BRD5631 robustly suppressed the elevated IL-1β observed in ATG16L1 T300A macrophages from knock-in mice, revealing this molecule can modulate relevant phenotypes in the context of a human disease allele. Given BRD5631 itself is not well-tolerated in vivo (data not shown), the successful development of analogs that separate toxicity from autophagy enhancement would be a critical step to determine whether BRD5631 modulates autophagy in a therapeutically relevant manner.
The evaluation of autophagy enhancement as a therapeutic strategy will ultimately require careful investigation of its complex relationship to cell death and cell survival in different physiological contexts. For example, autophagy has been proposed to promote cell survival in the context of cancer and cell death in the context of hypoxia–ischemia. Small-molecule probes that modulate autophagy will be critical for studying the pathway in different disease-relevant contexts and for understanding how it can be targeted to yield new medicines that are both safe and effective.
Materials and Methods
For the primary HTS, HeLa cells stably expressing GFP-LC3 were seeded in 384-well plates (16 h) and treated with compounds (4 h). Cells were fixed in 3.7% (wt/vol) paraformaldehyde for 15 min, washed, and DNA stained with Hoechst 33342 (Sigma). High-throughput imaging (20×) was performed on an ImageXpress Micro automated microscope (Molecular Devices) and the number of GFP punctae per cell was quantified using MetaXpress high-content image analysis software. For the DQ-BSA assay, HeLa cells were pulsed with DQ-BSA (Life Technologies) (1 h), DQ-BSA was washed away, and cells were treated with compounds (6 h). Nuclei were stained with Hoechst 33342 (30 min) and live cells imaged as above. For the LysoTracker assay, compound-treated (4 h), live HeLa cells were stained with LysoTracker Red dye (Life Technologies) and Hoechst 33342 (1 h) and imaged as above. The percentage of eGFP+ cells with eGFP-HDQ74 aggregates was assessed as described (37). Neurons were generated from neural precursors, which were derived from hESCs and NPC1 hiPSCs using an embryoid body-based protocol, and analysis of neuronal cell death was performed as described (18). Bioluminescent bacterial replication and CFU assays were performed as described (41). For colocalization assays, HeLa cells were seeded in antibiotic-free media (16 h). After compound pretreatment (3 h), cells were infected with Salmonella expressing DsRed (Clontech), retreated with compounds, and at indicated p.i. time points, cells were fixed, stained, and imaged to quantify the number of LC3+ and NDP52+ bacteria. iBMDM line was generated from BMDMs infected with a J2 retrovirus carrying the v-myc and v-raf oncogenes and cultured for 14 d in M-CSF-conditioned media followed by 14 d without M-CSF. CD11b+ cells were isolated by positive selection using CD11b microbeads (Miltenyi) and spleens derived from C57 BL/6 Atg16l1wt/wt and Atg16l1T300A/T300A mice. IL-1b levels were measured by ELISA (BD Biosciences). T300A knock-in mice were generated and maintained as previously described (5). Animals were housed in a pathogen-free facility, and all procedures were performed in accordance with the institutional animal care and use committee at Massachusetts General Hospital. Additional details on reagents, chemical synthesis, and experimental protocols are available in SI Appendix.
Supplementary Material
Acknowledgments
We are grateful to David Rubinsztein for providing the eGFP-HDQ74 construct, and to Noboru Mizushima for providing the Atg5+/+ and Atg5−/− MEFs. We thank the Compound Management Team for providing compound plates and synthetic intermediates, and the members of the R.J.X. and S.L.S. laboratories for insightful discussions. Research reported in this publication was supported by the Leona M. and Harry B. Helmsley Charitable Trust Grant 500203 (to S.L.S. and R.J.X.); the National Institute of Allergy and Infectious Diseases’ Center for Excellence in Translational Research Grant U19AI109725 (to S.L.S., A.F.S., and R.J.X.); Birmingham Fellowship (S.S.); the National Niemann-Pick Disease Foundation Foundation (D.M.), Skoltech Center (R.J.); and NIH Grants DK097485 (to R.J.X.), U01CA176152 (to S.L.S.), R01-NS088538 (to R.J.), and MH104610 (to R.J.). S.L.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: Rudolf Jaenisch is an advisor to Stemgent and Fate Therapeutic.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512289112/-/DCSupplemental.
References
- 1.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11(9):709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16(6):495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 4.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- 5.Lassen KG, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci USA. 2014;111(21):7741–7746. doi: 10.1073/pnas.1407001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456(7219):259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saitsu H, et al. 2013. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet 45(4):445–449, 449e1.
- 8.Oz-Levi D, et al. Mutation in TECPR2 reveals a role for autophagy in hereditary spastic paraparesis. Am J Hum Genet. 2012;91(6):1065–1072. doi: 10.1016/j.ajhg.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akizu N, et al. Biallelic mutations in SNX14 cause a syndromic form of cerebellar atrophy and lysosome-autophagosome dysfunction. Nat Genet. 2015;47(5):528–534. doi: 10.1038/ng.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Y, Ren X, Hait WN, Yang JM. Therapeutic targeting of autophagy in disease: Biology and pharmacology. Pharmacol Rev. 2013;65(4):1162–1197. doi: 10.1124/pr.112.007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw SY, et al. Selective modulation of autophagy, innate immunity, and adaptive immunity by small molecules. ACS Chem Biol. 2013;8(12):2724–2733. doi: 10.1021/cb400352d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim JJ, et al. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe. 2012;11(5):457–468. doi: 10.1016/j.chom.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Sundaramurthy V, et al. Integration of chemical and RNAi multiparametric profiles identifies triggers of intracellular mycobacterial killing. Cell Host Microbe. 2013;13(2):129–142. doi: 10.1016/j.chom.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Shoji-Kawata S, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494(7436):201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hidvegi T, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329(5988):229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 16.Sarkar S. Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: Autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem Soc Trans. 2013;41(5):1103–1130. doi: 10.1042/BST20130134. [DOI] [PubMed] [Google Scholar]
- 17.Sarkar S, et al. Impaired autophagy in the lipid-storage disorder Niemann–Pick type C1 disease. Cell Reports. 2013;5(5):1302–1315. doi: 10.1016/j.celrep.2013.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maetzel D, et al. Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann–Pick Type C patient-specific iPS cells. Stem Cell Rep. 2014;2(6):866–880. doi: 10.1016/j.stemcr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu HC, et al. Eradication of intracellular Salmonella enterica serovar Typhimurium with a small-molecule, host cell-directed agent. Antimicrob Agents Chemother. 2009;53(12):5236–5244. doi: 10.1128/AAC.00555-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tattoli I, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11(6):563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Harris J, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286(11):9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motzer RJ, et al. RECORD-1 Study Group Efficacy of everolimus in advanced renal cell carcinoma: A double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372(9637):449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen TE, Schreiber SL. Towards the optimal screening collection: a synthesis strategy. Angew Chem Int Ed Engl. 2008;47(1):48–56. doi: 10.1002/anie.200703073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcaurelle LA, et al. An aldol-based build/couple/pair strategy for the synthesis of medium- and large-sized rings: Discovery of macrocyclic histone deacetylase inhibitors. J Am Chem Soc. 2010;132(47):16962–16976. doi: 10.1021/ja105119r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerard B, et al. Synthesis of a stereochemically diverse library of medium-sized lactams and sultams via S(N)Ar cycloetherification. ACS Comb Sci. 2011;13(4):365–374. doi: 10.1021/co2000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitzgerald ME, et al. Build/couple/pair strategy for the synthesis of stereochemically diverse macrolactams via head-to-tail cyclization. ACS Comb Sci. 2012;14(2):89–96. doi: 10.1021/co200161z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 29.Nadanaciva S, et al. A high content screening assay for identifying lysosomotropic compounds. Toxicol In Vitro. 2011;25(3):715–723. doi: 10.1016/j.tiv.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Ashoor R, Yafawi R, Jessen B, Lu S. The contribution of lysosomotropism to autophagy perturbation. PLoS One. 2013;8(11):e82481. doi: 10.1371/journal.pone.0082481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vázquez CL, Colombo MI. Assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (MDC) and DQ-BSA. Methods Enzymol. 2009;452:85–95. doi: 10.1016/S0076-6879(08)03606-9. [DOI] [PubMed] [Google Scholar]
- 32.Lemieux B, Percival MD, Falgueyret JP. Quantitation of the lysosomotropic character of cationic amphiphilic drugs using the fluorescent basic amine Red DND-99. Anal Biochem. 2004;327(2):247–251. doi: 10.1016/j.ab.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 33.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284(12):8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balgi AD, et al. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One. 2009;4(9):e7124. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 37.Sarkar S, Ravikumar B, Rubinsztein DC. Autophagic clearance of aggregate-prone proteins associated with neurodegeneration. Methods Enzymol. 2009;453:83–110. doi: 10.1016/S0076-6879(08)04005-6. [DOI] [PubMed] [Google Scholar]
- 38.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahani MH, Itakura E, Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy. 2014;10(3):431–441. doi: 10.4161/auto.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J, Brumell JH. Bacteria-autophagy interplay: A battle for survival. Nat Rev Microbiol. 2014;12(2):101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conway KL, et al. Atg16l1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology. 2013;145(6):1347–1357. doi: 10.1053/j.gastro.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujita N, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19(5):2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS One. 2008;3(10):e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plantinga TS, et al. Crohn’s disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut. 2011;60(9):1229–1235. doi: 10.1136/gut.2010.228908. [DOI] [PubMed] [Google Scholar]
- 46.Kaufmann AM, Krise JP. Lysosomal sequestration of amine-containing drugs: Analysis and therapeutic implications. J Pharm Sci. 2007;96(4):729–746. doi: 10.1002/jps.20792. [DOI] [PubMed] [Google Scholar]
- 47.Kornhuber J, et al. Identification of novel functional inhibitors of acid sphingomyelinase. PLoS One. 2011;6(8):e23852. doi: 10.1371/journal.pone.0023852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halliwell WH. Cationic amphiphilic drug-induced phospholipidosis. Toxicol Pathol. 1997;25(1):53–60. doi: 10.1177/019262339702500111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.