Significance
Heat shock protein 70 (Hsp70) molecular chaperones play key roles in protein folding and other cellular processes. The effect of Hsp70 on the conformation of its substrate proteins is still largely unknown. This study unveils, for the first time to our knowledge, the effect of the bacterial Hsp70 chaperone DnaK on the structure of the full-length substrate protein SRC homology 3 domain (SH3). We show that multiple largely unstructured conformations of SH3, distinct from the protein’s unfolded state, interact with DnaK. The bound client protein shares a flexible N terminus and multiple slowly interconverting conformations in different parts of the sequence. In all, there is significant structural and dynamical heterogeneity. This result is important because it reveals that proteins may undergo conformational sampling while chaperone-bound.
Keywords: molecular chaperone, protein folding, triple-resonance NMR, Hsp70, DnaK
Abstract
The molecular chaperone heat shock protein 70 (Hsp70) plays a vital role in cellular processes, including protein folding and assembly, and helps prevent aggregation under physiological and stress-related conditions. Although the structural changes undergone by full-length client proteins upon interaction with DnaK (i.e., Escherichia coli Hsp70) are fundamental to understand chaperone-mediated protein folding, these changes are still largely unexplored. Here, we show that multiple conformations of the SRC homology 3 domain (SH3) client protein interact with the ADP-bound form of the DnaK chaperone. Chaperone-bound SH3 is largely unstructured yet distinct from the unfolded state in the absence of DnaK. The bound client protein shares a highly flexible N terminus and multiple slowly interconverting conformations in different parts of the sequence. In all, there is significant structural and dynamical heterogeneity in the DnaK-bound client protein, revealing that proteins may undergo some conformational sampling while chaperone-bound. This result is important because it shows that the surface of the Hsp70 chaperone provides an aggregation-free environment able to support part of the search for the native state.
Proteins attain their most stable native conformation upon refolding in vitro, under given environmental conditions (1). However, ribosome-bound, newly synthesized, stress-unfolded, large or highly concentrated proteins often cannot fold efficiently in vitro and/or are particularly prone to aggregation. Remarkably, in the cellular environment, molecular chaperones and the ribosome help ensure that all proteins attain their native state (2–5). Molecular chaperones are overexpressed under stress-related conditions and help maintain a population of correctly folded proteins. Some chaperones also interact with cellular proteins cotranslationally and posttranslationally under nonstress conditions (6).
The heat shock protein 70 (Hsp70; ∼70 kDa) class of molecular chaperones (7) are single-chain proteins comprising a nucleotide-binding domain (NBD; ca. 42 kDa) capable of hydrolyzing ATP, a substrate-binding domain (SBD; ca. 27 kDa) devoted to interacting with the substrate, and a flexible linker connecting the two domains (6, 8) (Fig. 1 A–C). Hsp70 is involved in a variety of protein folding and trafficking processes (9), and it plays major yet diverse roles in disease. For instance, overexpression of Hsp70 is related to cancer (10), and Hsp70 overexpression in transgenic mice leads to spontaneous development of T-cell lymphoma (10, 11). In addition, Hsp70 plays a protective role against a number of neurodegenerative diseases by preventing protein aggregation (12–14).
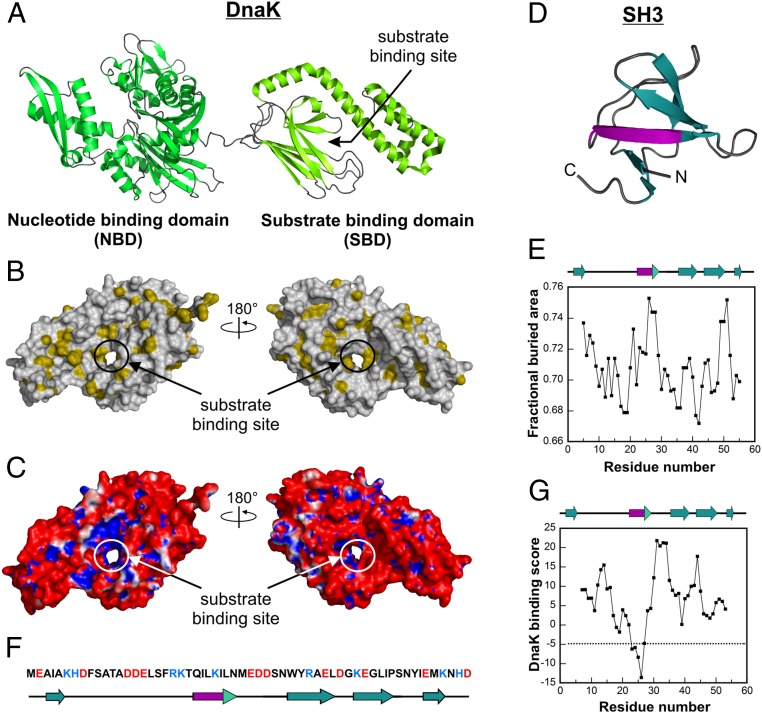
Fig. 1.
Structures and properties of DnaK and SH3. (A) Cartoon representation of DnaK, a multidomain chaperone with an ∼42-kDa NBD (dark green) and an ∼27-kDa SBD (light green) consisting of an α-helical “lid” subdomain and a β-sheet subdomain [Protein Data Bank (PDB) ID code 2KHO (42)]. (B) Space-filling representation of the SBD of DnaK [PDB ID code 1DKX (18)]. Hydrophobic side chains with a fractional buried area >0.74 (43) are shown in gold. The image on the right has the same orientation as the SBD of panel A. (C) Electrostatic surface potential map of the SBD of DnaK [PDB ID code 1DKX (18)] showing the charge of the surface accessible to solvent with a radius of 1.4 Å. Blue denotes regions of positive potential (>+1 kBT/e, where kB and T denote the Boltzmann constant and the temperature, respectively), and red denotes regions of negative potential (<−1 kBT/e). Electrostatic surface potentials were generated with the PyMOL software package and the APBS algorithm (Materials and Methods). (D) Cartoon representation of SH3 [PDB ID code 2A36 (44)]. The putative binding site for DnaK (purple) comprises most of one strand of a β-sheet. “N” and “C” denote the N and C termini, respectively. (E) Fractional buried area according to Rose et al. (43) on a per-residue basis for SH3. (F) Amino acid sequence of SH3. Positively and negatively charged residues are colored blue and red, respectively. (G) DnaK binding scores on a per-residue basis for SH3 based on the algorithm by Bukau and coworkers (29). Scores less than −5 indicate significant binding to DnaK.
Due to the central importance of Hsp70, significant efforts have been devoted to understand its function, including how ATP hydrolysis modulates substrate affinity, how this chaperone interacts with its cochaperones, and how Hsp70 undergoes allosteric regulation during the different stages of its functional cycle (8, 15).
On the other hand, a crucial piece of information for understanding the role of Hsp70 in the mechanism of protein folding (i.e., how the conformation of the client protein is affected by its interaction with Hsp70) is still poorly understood (16, 17). Most previous studies used small peptides (ca. 5–13 residues) as Hsp70 clients, instead of full-length proteins. Peptide substrates comprising primarily the Hsp70 binding site region undergo a conformational change upon binding Hsp70 by transitioning from a disordered to a fully extended conformation (18–20), or from a weakly helical state to a less ordered nonhelical state (21). A folding-incompetent larger peptide comprising a protein fragment (the 77-residue N-terminal region of apomyoglobin) binds the SBD of Hsp70 in a globally unfolded state (22). A specialized periplasmic chaperone (HscA) that shares some similarities with Hsp70 causes depletion in the native-state population of its only substrate IscU (23).
Hence, although it is slowly becoming clear that small polypeptides interacting with Hsp70 are generally not folded, there is still a severe lack of information on full-length protein clients. Importantly, an assessment of the conformation of Hsp70-bound proteins is lacking. This gap of knowledge is particularly severe in view of the role of Hsp70 in protein folding. It was proposed that Hsp70 is either a holdase, capable of preventing protein aggregation by kinetic partitioning (24, 25), or an (un)foldase [i.e., a molecular machine capable of actively folding and/or unfolding (26, 27) proteins by binding to their native, misfolded, or partially folded state].
Here, we investigate the conformational changes experienced by the SRC homology 3 domain (SH3) client protein upon binding the Hsp70 molecular chaperone. We use full-length Escherichia coli Hsp70, also known as DnaK, and focus on its ADP-bound state (ADP-DnaK), given its known high affinity for substrate proteins. The N-terminal domain of the Drosophila melanogaster adaptor protein drk, denoted here as drkN SH3 or simply SH3 (Fig. 1D), serves as a model substrate. The synergistic use of NMR chemical shifts and volumes, NMR-detected translational diffusion, and native gels shows that several globally unstructured SH3 states interact with Hsp70. These states share a common highly flexible N terminus and multiple globally disordered conformations in other regions of the protein. Our data provide the first high-resolution evidence, to our knowledge, for the fact that DnaK-bound client proteins populate a variety of conformational states, some of which are explicitly detectable and some of which are not. We show that the detectable bound states are unfolded or incompletely folded and lack much of the native protein’s well-defined secondary and tertiary structure. This result highlights the role of structural heterogeneity in client protein-Hsp70 binding, and provides a glimpse at the fact that Hsp70 may allow the client protein to sample conformational space even while chaperone-bound.
Results
SH3 Interacts Reversibly with DnaK.
The chosen Hsp70 client for our studies, SH3 (Fig. 1 D–G), is a well-characterized and marginally stable protein with comparably populated folded and unfolded states. Conveniently, given that SH3 exchanges slowly on the NMR chemical shift time scale, both native and unfolded forms are concurrently detected at atomic resolution (28) in the absence and presence of the DnaK chaperone. Further, the lack of any SH3 aggregation propensity greatly facilitates the analysis. Prior experimental studies based on the peptide-array scanning of many protein clients for their interactions with DnaK showed that specific protein regions (5–8 residues long) comprising nonpolar stretches usually surrounded by positively charged residues have specific affinity for DnaK (29). This work led to the generation of a computational tool to predict the local affinity of any polypeptide or protein sequence for DnaK (29). Application of this tool to a variety of proteins resulted in computational predictions in excellent agreement with experiments (30–32). A similar approach was used for SH3 (33), leading to the identification of one putative binding site for DnaK comprising SH3 residues 23–27 (Fig. 1G).
Our SH3 NMR backbone assignments in the absence of DnaK, pertaining to amide protons (HN) and nitrogens (N), as well as to carbonyl (C′), and α- and β-carbons (Cα and Cβ) of the13C-15N–enriched client protein in the absence of chaperone, are consistent with published data collected under slightly different conditions (28). Comparably intense resonances for both the native (N) and unfolded (U) states are clearly visible by 2D NMR in the absence of chaperone (Fig. 2B), confirming that SH3 is thermodynamically unstable and exchanges slowly on the NMR chemical shift time scale.
Fig. 2.
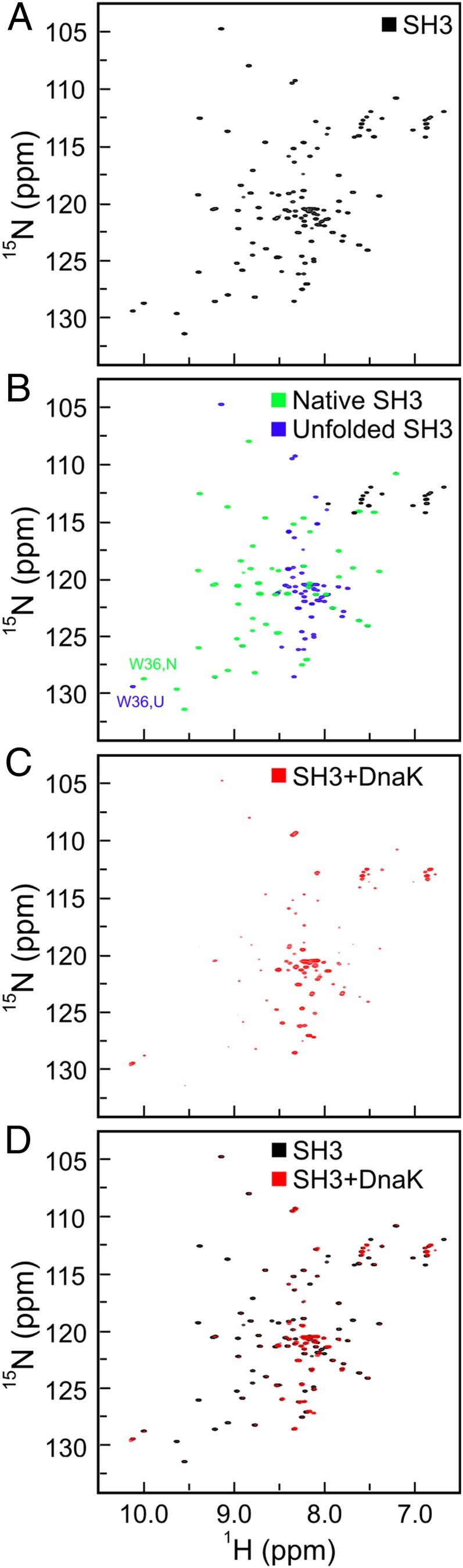
Addition of an excess of ADP-DnaK leads to dramatic spectral changes. (A) 1H-15N HSQC spectrum of 15N-13C–labeled SH3. (B) 1H-15N HSQC spectrum of SH3 as in A, with resonances corresponding to the native and unfolded species in green and purple, respectively. Side-chain resonances of Asp and Gln were not colored. (C) 1H-15N HSQC spectrum of a 1:4 ratio of SH3/ADP-DnaK, where only SH3 is isotopically enriched. (D) Overlaid spectra from both experiments. All samples were prepared in 50 mM Tris⋅HCl, 5 mM MgCl2, 50 mM KCl, 5 mM ADP, and 15 mM DTT in 95%:5% H2O/D2O. All spectral data were collected at 25 °C and pH 7.2, with eight scans and 256 increments over a sweep width of 1,937 Hz in the indirect dimension.
Upon addition of a fourfold excess of DnaK under conditions where DnaK is ADP-bound, a dramatic variation in the 2D spectral features is observed (Fig. 2). The resonances due to native SH3 undergo a significant decrease in intensity, whereas the resonances due to unfolded SH3 persist in the spectrum and undergo either an increase or decrease in intensity (Fig. 2 C and D). Negligible to moderate line width variations are observed (SI Appendix and SI Appendix, Fig. S4), after correcting the data for the changes in solution viscosity due to addition of DnaK to the medium. These spectral characteristics, including the intensity and width of the side-chain indole NH resonances of the only Trp (Trp36), suggest variations in the relative populations of the N and U states upon addition of DnaK. We carried out NMR assignments for a variety of SH3 nuclei in the presence of DnaK and deduced that the resonances present in the absence of chaperone experience negligible chemical shift changes (SI Appendix, Figs. S1 and S2).
The above spectral changes are entirely reversible and highly dependent on the [SH3]/[DnaK] ratio. For instance, addition of an excess of unlabeled SH3 reverts the features of the 1H-15N sensitivity-enhanced (SE) heteronuclear single quantum coherence (HSQC) NMR spectrum of SH3/DnaK to the features of chaperone-free SH3 (Fig. 3 A and B). The above observations suggest a mere shift in the N and U equilibrium populations upon addition of the chaperone but do not provide any conclusive evidence for the binding of SH3 to DnaK. Once additional observations are taken into account, however (discussed below), including native-gel analysis, diffusion-ordered NMR, and the appearance of new NMR resonances in the HSQC spectrum, it becomes evident that SH3 and DnaK interact with each other extensively and form a highly heterogeneous bound state.
Fig. 3.
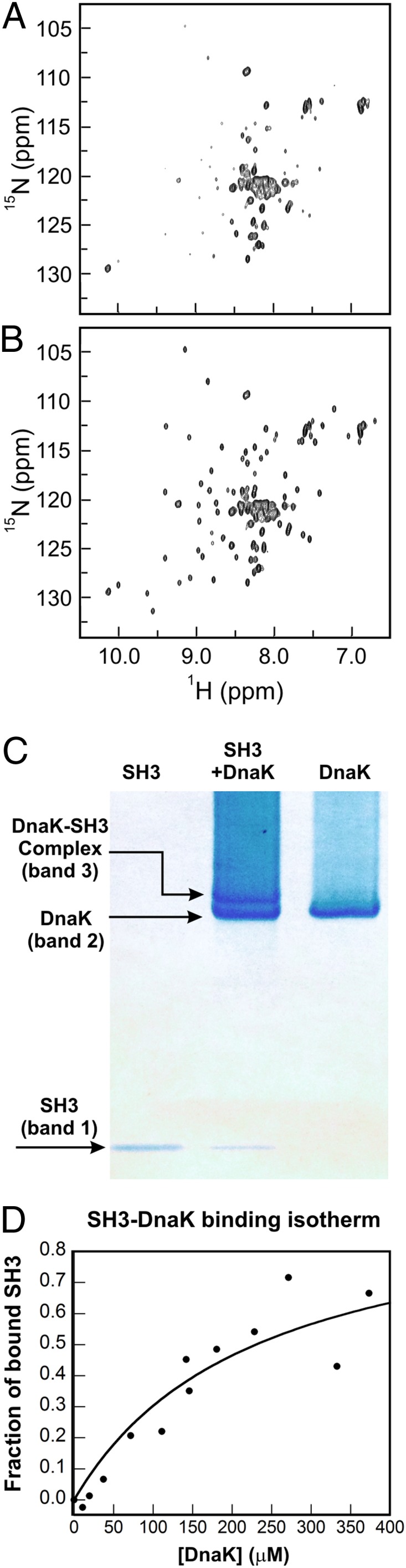
ADP-DnaK binds SH3 reversibly. SH3/ADP-DnaK samples of Fig. 2 were incubated for 2 wk (A) followed by (B) threefold dilution by addition of an excess amount of unlabeled SH3 (1 mM). (A) 1H-15N HSQC spectrum of 100 μM 13C-15N–labeled SH3 and 400 μM ADP-DnaK. (B) 1H-15N HSQC spectrum of 100 μM 13C-15N–labeled SH3, 400 μM ADP-DnaK, and 1 mM SH3. All samples were prepared in 50 mM Tris⋅HCl, 5 mM MgCl2, 50 mM KCl, 5 mM ADP, and 15 mM DTT in 95%:5% H2O/D2O. All spectra were acquired at 25 °C and pH 7.2, with eight scans and 64 increments over a sweep width of 1,900 Hz. (C) Native-gel image showing lanes for SH3, SH3/ADP-DnaK, and ADP-DnaK. The putative complex is shown as band 3. (D) Binding isotherm illustrating the interaction of SH3 with ADP-DnaK. The best-fit curve is shown as a solid line. The average Kd for two independent experiments is 243 ± 13 μM, with the listed uncertainty denoting the SE.
First, the native-gel and MS data analysis outlined below prove that SH3 and DnaK form a complex. Specifically, gel shift assays show that addition of DnaK to the medium containing client protein gives rise to a new well-defined band (band 3 in Fig. 3C). In addition, liquid chromatography followed by electrospray MS (SI Appendix, Fig. S3) of the extracted native-gel band 3 unequivocally indicates that the new native-gel band comprises both SH3 and DnaK. This result rules out the possibility that band 3 may have resulted from SH3 or DnaK conformational changes or from SH3–SH3 or DnaK–DnaK interactions. Hence, SH3 and DnaK form a complex in solution. Although the native-gel band 3 is consistent with the presence of one single complex, the NMR data analysis presented below shows that the bound population comprises more than one client protein conformation. Given that the chaperone-bound client protein conformers are not sufficiently different to run as distinct bands in the native gel, we will allude here to a single class of substrate protein–DnaK complex.
The binding isotherm generated by gel shift titrations, shown in Fig. 3D, is consistent with saturation behavior, and it yields an apparent dissociation constant (Kd) of 243 ± 13 μM. Hence, the data support the presence of a relatively weak specific class of binding sites upon interaction of SH3 with the DnaK chaperone.
Chaperone-Free Unfolded SH3 Is in Dynamic Equilibrium with Chaperone-Bound SH3.
Additional insights into the nature of the client protein–DnaK interactions are gained from translational diffusion NMR, via 1H 1D and 1H-13C–correlated 2D diffusion-ordered spectroscopy (DOSY). The Ala13 resonance was chosen for the DOSY data analysis because of its high intensity (Ala5 and Ala11 were also monitored). Fig. 4 shows the translational diffusion coefficient of DnaK and compares it with the corresponding values for SH3 in the absence and presence of DnaK (4:1 ratio of DnaK to SH3). The observed diffusion coefficients were corrected for viscosity changes (Materials and Methods and SI Appendix) and are reported as the values of a hypothetical solution bearing the same viscosity as plain buffer. Hence, any observed variations in the viscosity-corrected diffusion coefficients upon introduction of DnaK to the medium must be due to the interaction between SH3 and DnaK.
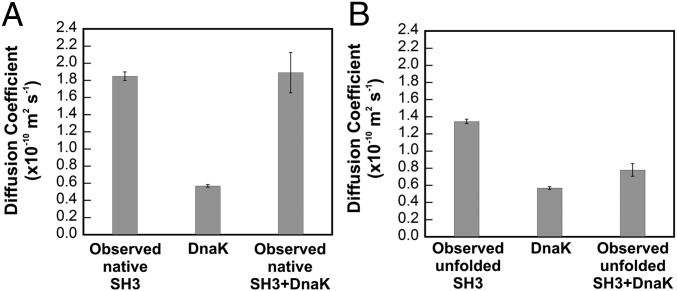
Fig. 4.
Unfolded state of SH3 interacts with ADP-DnaK. Viscosity-corrected translational diffusion coefficients of native (A) and unfolded (B) SH3 in the absence and presence of a fourfold excess of ADP-DnaK. Data were obtained from proton-carbon convection-compensated DOSY HSQC (DOSY-HC-HSQC) experiments (refs. 45 and 46 and SI Appendix).
Given that the corrected translational diffusion coefficient derived from DOSY for the native SH3 resonances is identical within error to the corresponding value in the absence of chaperone (Fig. 4A), we conclude that there is no evidence that the SH3 native state binds DnaK. The overall conformational equilibrium between the chaperone-free and chaperone-bound native conformations is likely in the slow exchange regime on the DOSY translational diffusion NMR time scale (34) (i.e., ca. 100 ms). Regardless, we monitored the DOSY behavior via the intense resonances of side-chain methyl groups in the N-terminal region whose detectable chemical shifts are virtually unchanged upon complex formation. Hence a conformational equilibrium between the native state of chaperone-free SH3 and its corresponding native bound form bearing the same (or very similar) chemical shifts, and on the fast exchange on the NMR chemical shift time scale, would have resulted in a decrease in the observed diffusion coefficient, which we did not detect. Hence, our study shows no explicit evidence for a chaperone-bound native state. Of course this state could still exist (and possibly be one of the invisible states identified in the following sections) in case the DnaK-bound native state were to be spectroscopically invisible, as well as in slow exchange on both the NMR chemical shift and translational diffusion time scales. In all, we were unable to detect any direct thermodynamic paths linking the chaperone-free native state to the chaperone-bound ensemble.
In contrast, in the case of unfolded SH3, it is clear that the observed translational diffusion coefficient for the unfolded SH3 resonances in the presence of DnaK is considerably smaller than the value observed for pure SH3, consistent with slower translational diffusion in the presence of chaperone. The observed diffusion coefficient is similar to the value observed for the large and slow-diffusing pure DnaK chaperone (Fig. 4B). Hence, the DOSY data show that the unfolded state is in dynamic exchange with a DnaK-bound state.
A few additional technical considerations ought to be kept in mind. First, the viscosity-corrected translational diffusion coefficient of pure ADP-DnaK is consistent with the chaperone behaving as a large globular protein (apparent molecular mass >70 kDa), according to simple calculations based on the Stokes–Einstein equation and assuming, for simplicity, a spherical shape (SI Appendix). Interestingly, this solution behavior, leading DnaK to behave as one large molecule, applies even though the smaller ADP-DnaK NBD and SBD are known to interact only loosely with each other (35). Second, at least some of the intense side-chain Ala methyl groups of unfolded SH3 used in the DOSY measurements are in fast exchange on the NMR chemical shift time scale but in slow exchange on the DOSY time scale. The latter is true because at least some of the bound complex exchanges with free unfolded SH3 more slowly (due to new resonances in slow exchange on the chemical shift time scale; Fig. 5 B–D) than the DOSY experiment diffusion time (34) (i.e., 100 ms). The double-exponential DOSY profile expected for this scenario was not observed simply because of the relatively small differences in diffusion coefficients of free and bound states. This statement is supported by a collection of 1,000 simulations of expected DOSY profile decays that we ran in MATLAB (MathWorks), assuming a 5% experimental error. Each simulation generated data points for free and bound client protein assuming their ratios from NMR volume analysis data (discussed below). Single-exponential NMR intensity decays adequately fit the simulated data and yielded expected translational diffusion coefficients differing by less than 8% from our experimental results.
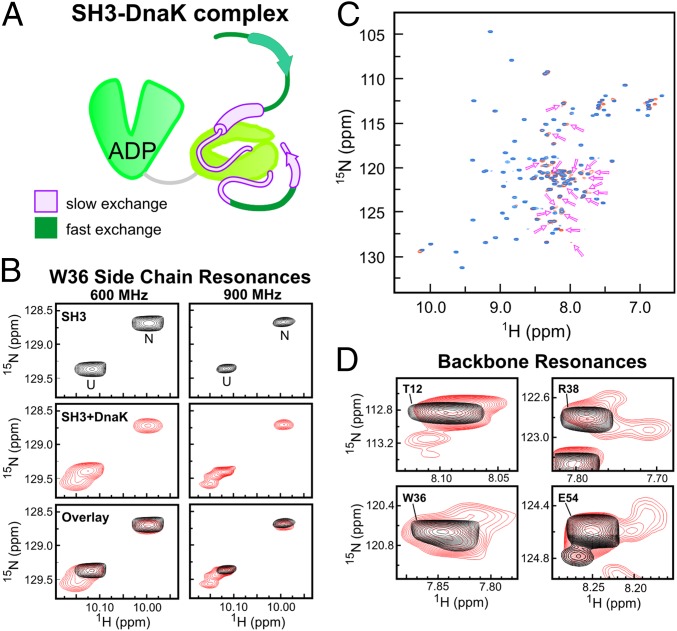
Fig. 5.
Hsp70-bound state of SH3 is heterogeneous. (A) Schematic representation of proposed SH3 binding mode to Hsp70. The SH3 regions directly interacting with the chaperone (pink) are in slow exchange on the NMR chemical shift time scale, whereas the SH3 regions not interacting with Hsp70 (dark green, including N-terminal residues ca. 3–11 and residues 43–46) are in fast exchange. The residual α-helical and β-strand secondary structure is shown as solid cylinders and arrows, respectively. (B) Expanded 1H-15N HSQC spectral region of SH3 showing the Trp (W36) side-chain indole NH resonances for the native and unfolded states in the absence (Top) and presence (Middle) of a fourfold excess of ADP-DnaK, together with an overlay of both spectra (Bottom) collected on 600-MHz (Left) and 900-MHz (Right) NMR spectrometers, respectively. (C) Overlay of 1H-15N HSQC of SH3 in the absence (blue) and presence (dark orange) of ADP-DnaK. The arrows denote new backbone resonances that appear only upon addition of DnaK. These resonances reversibly disappear upon addition of an excess of unlabeled SH3. (D) Expanded spectral regions of the 1H-15N HSQC in the absence (black) and presence (red) of ADP-DnaK, showing the appearance of multiple new conformational states for each of the residues upon addition of DnaK.
In all, the results of the DOSY analysis are important because they unequivocally show that the chaperone-free U state is linked to chaperone-bound SH3 via one or more explicit thermodynamic pathways. Conversely, our data were unable to detect evidence for pathways directly connecting native chaperone-free SH3 to chaperone-bound states.
Exploring Different Hypotheses to Explain the NMR Spectral Features of SH3 in the Presence of DnaK.
The absence of any significant amide 1H, 15N, and C′ SH3 chemical shift variations upon addition of chaperone (SI Appendix, Figs. S2 and S3) means that each SH3 resonance must comply with one of the following three scenarios: (i) fast exchange between free and bound species on the NMR chemical shift time scale, with only a small fraction of the total SH3 population bound to DnaK, so that chemical shift perturbations are undetectable; (ii) fast exchange on the NMR chemical shift time scale, with DnaK-bound conformations and chemical environment of the bound resonances virtually identical to the values of the respective free states, causing no variations in chemical shift; and (iii) slow exchange on the NMR chemical shift time scale and distinct resonances from those due to the chaperone-free state. Resonances due to the bound state may either be detectable in different regions of the spectrum or too broad to be observed.
Hypothesis i is disproved by the fact that the native-gel data (Fig. 3C), supported by DOSY, indicate that overall a large fraction of SH3 is DnaK-bound. Hence, the chaperone-bound species must be significantly populated.
For any rigidly bound state, hypothesis ii is disproven by line broadening arguments. A large population of fast-exchanging bound species would, in fact, have significantly broadened resonances (SI Appendix). This possibility is discarded because all resonances are still detectable after addition of DnaK and, most importantly, negligible to moderate line broadening is observed after correcting for the small line broadening imparted by the viscosity increase due to addition of DnaK to the medium (SI Appendix and SI Appendix, Fig. S4). However, hypothesis ii can be valid for any resonances that remain sufficiently dynamic upon binding. This scenario is plausible for residues, such as residues 3–11 or residues 43–46, that correspond to highly dynamic portions of the client protein sequence away from the contact region with the chaperone, in analogy to what was previously observed in the case of the complex of the apomyoglobin1–77 fragment with the DnaK SBD (22).
Finally, some bound-state SH3 residues may undergo slow conformational exchange on the NMR chemical shift time scale upon chaperone binding/release, as described by hypothesis iii. A simple back-of-the-envelope calculation (SI Appendix) shows that resonances from rigidly held DnaK-bound SH3 residues are expected to experience a decrease in intensity by at least 100-fold. This value would almost certainly lead to line broadening beyond detection.
The scenario we propose for SH3 binding to DnaK involves a highly heterogeneous bound form, involving cases ii and iii applying to different regions of the client protein sequence. Specifically, the SH3 core-binding region comprises residues in slow exchange on the NMR chemical shift time scale (hypothesis iii) and residues further removed from the core-binding region that are in fast exchange with the unbound state and remain highly dynamic, as described in the cartoon in Fig. 5A. Consistent with this model, the observed resonances in the spectrum are due to the free unbound client protein population, to some highly dynamic bound residues experiencing negligible interactions with the chaperone (hence leading to negligible chemical shift perturbations), and to some new distinct resonances in slow exchange on the NMR chemical shift time scale (discussed in the following section).
DnaK-Bound State of the SH3 Client Protein Is Heterogeneous and Comprises an Ensemble of Globally Disordered Conformations.
Important insights into the binding interaction are gained upon additional inspection of the 1H-15N HSQC spectra. Starting from the analysis of the Trp side-chain data (Fig. 5B), there are, as expected, two distinct resonances for the Trp36 indole NH due to the N and U states in the absence of the chaperone. Upon addition of ADP-DnaK, the resonances corresponding to folded and unfolded SH3 experience a significant loss in intensity and volume. In addition, a clear third class of resonances appears in the spectrum (Fig. 5B). Data collected at 900 MHz (Fig. 5C) are particularly revealing in that they show the presence of multiple Trp36-bound conformations with a chemical shift similar to the chemical shift of the unfolded state. Shifting the equilibrium so that the entire SH3 population is chaperone-bound was not possible due to aggregation of DnaK at high concentrations. Given the chemical shift proximity with the unfolded Trp36 resonance, the new spectral features are tentatively ascribed to globally unstructured chaperone-bound Trp36 conformations. Analogous data on backbone resonances confirm this idea (discussed below).
A close inspection of the entire 1H-15N HSQC spectrum reveals that several additional new resonances, about 28, due to backbone amides are also observed upon addition of ADP-DnaK to the client protein solution (Fig. 5C). The new resonances reflect a reversible phenomenon, because they disappear when an excess of unlabeled SH3 is added to the solution. The new resonances are close to the parent peaks in the absence of the chaperone. We ascribe these new spectral features to multiple DnaK-bound SH3 conformations in slow exchange with free SH3 on the NMR chemical shift time scale. Importantly, most relevant residues have multiple bound conformations (red resonances in Fig. 5D) showing that there is significant conformational heterogeneity in the chaperone-bound SH3 population. Further, all of the new backbone resonances are clustered within a narrow proton chemical shift range (Fig. 5C, pink arrows and SI Appendix, Fig. S7), showing that all of the bound conformations are globally unstructured. Analysis of triple-resonance data was rendered complicated by weak intensities and spectral overlaps. However, inspection of selected detectable HNCA resonances revealed that the secondary chemical shifts (from inspection of Cα carbons) of the resonances appearing upon addition of DnaK are similar to the chemical shifts of the chaperone-free unfolded state (SI Appendix, Fig. S2). Hence the residual secondary structure of the bound conformations is similar to the secondary structure of unfolded SH3 in the absence of chaperone. The cartoon in Fig. 8B, discussed in detail later, illustrates the peculiar features of the client protein-bound state, with emphasis on its conformational heterogeneity.
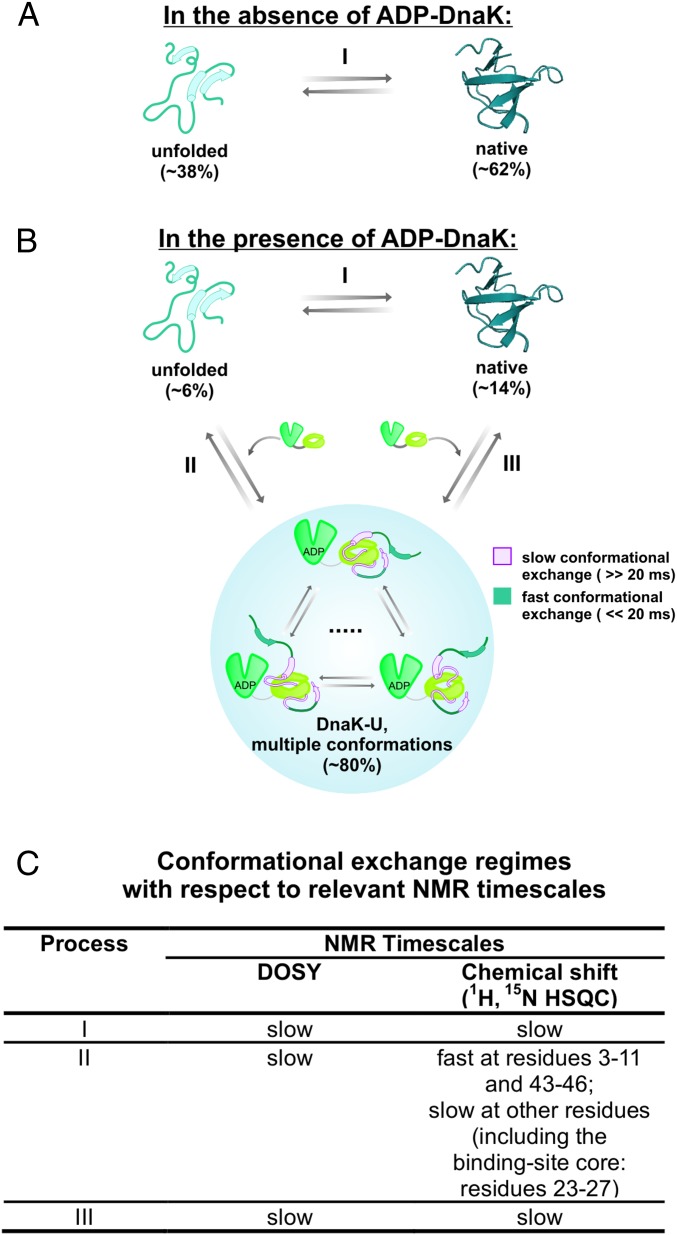
Fig. 8.
Model illustrating the conformational changes experienced by the SH3 substrate protein upon interaction with ADP-DnaK. (A) Cartoon representation illustrating the equilibrium of chaperone-free SH3 between its unfolded and native states in the absence of chaperones. Percent values denote relative populations assessed by NMR resonance volume analysis. (B) Cartoon representation of the proposed model for the interaction of SH3 with ADP-DnaK. The percent values in parentheses denote the estimated fraction of each major species with respect to the total SH3 population, as assessed by NMR under the conditions of our experiments, in the presence of a fourfold excess of ADP-DnaK. The residual α-helical and β-strand secondary structure is shown as solid cylinders and arrows, respectively. In addition to the representative bound species shown in the cartoon, a spectroscopically invisible chaperone-bound SH3 state (accounting for a loss of ca. 43% in the total client protein population) is present and is shown in the cartoon as contiguous dots. Reversible process III is shown for illustration purposes, although we were not able to gather evidence for its existence in this work. (C) Table illustrating the time scale regime for each major equilibrium process in B with respect to NMR-relevant parameters (translational diffusion monitored via DOSY and NMR chemical shifts).
NMR Analysis of Resonance Volumes Reveals Additional Details.
The volume of the 1H-15N HSQC resonances in the absence and presence of ADP-DnaK reveals additional information. Resonance volumes for HSQC data collected with a long relaxation delay and a favorable digital resolution in both the direct and indirect dimensions and processed with no window functions can be taken as a measure of the observable free and bound populations. Resonance volumes are independent of tumbling rates, in the absence of any experimental artifacts. Therefore, in the presence of binding events leading to resonance line broadening, the total HSQC volume due to a specific nucleus in all pertinent environments should remain constant regardless of the exchange rate regime (on the chemical shift and spin–spin relaxation time R2 time scales). Any fraction of the total volume that remains unaccounted for is, in principle, diagnostic of populations whose intrinsic line width is too broad to be detected. However, these theoretical considerations are complicated by the fact that slow-tumbling species with fast transverse relaxation may undergo selective relaxation losses during NMR pulse sequence, causing artifactual imbalances in volume values.
The volume analysis of the Trp side-chain resonances reveals a pattern of significant volume loss. Given that the resonance volume is proportional to the corresponding population, the populations of each species can be compared before and after addition of DnaK. In the case of the Trp side-chain resonances, the sum of the volumes of the three resonances in the presence of DnaK (corresponding to N, U, and U-type bound) represents only 67% of the sum of the N and U resonance volumes in the absence of DnaK. This 33% volume loss must be linked to either relaxation losses during the HSQC pulse sequence or a loss of corresponding magnitude due to a theoretically detectable population whose resonances are broadened beyond detection due to extremely slow tumbling.
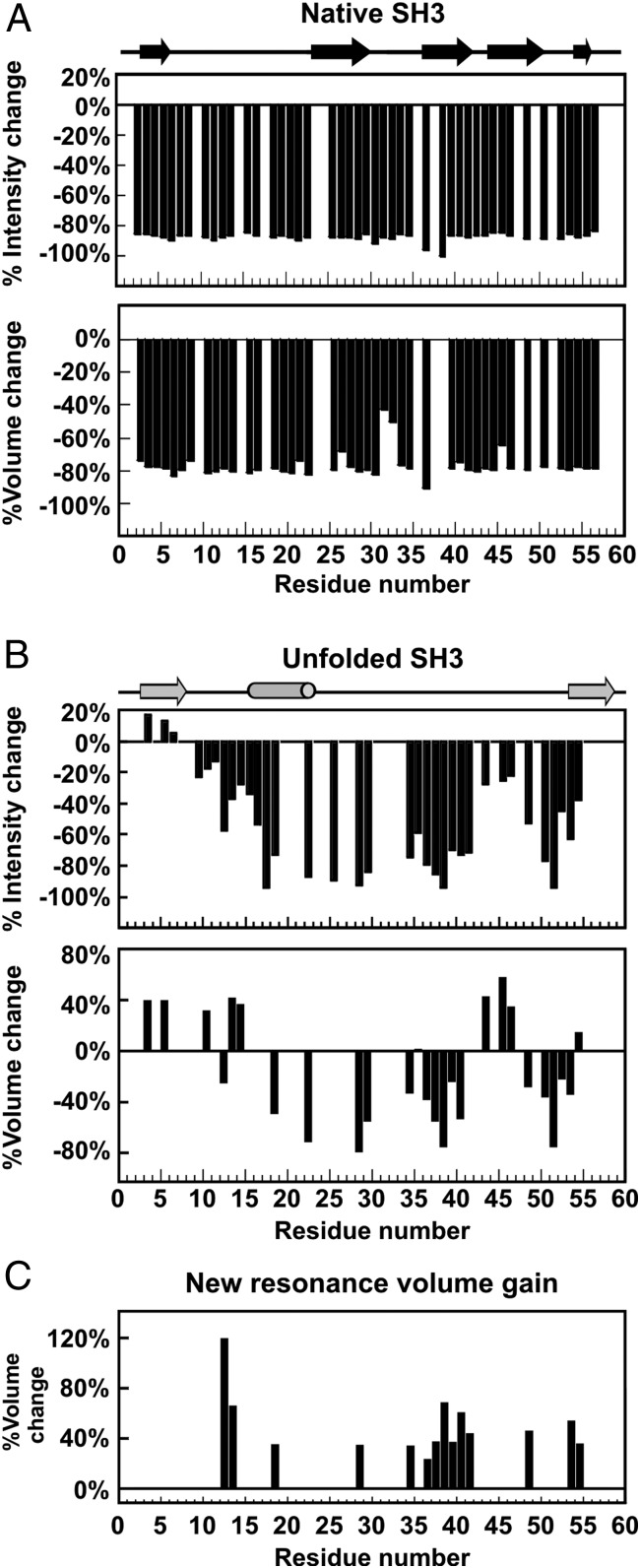
A resonance-specific analysis of volume loss across all backbone residues shows similar trends. Volume changes of the N and U 1H-15N SE-HSQC resonances of SH3 upon addition of ADP-DnaK are shown in Fig. 6. Changes in intensity are also displayed, although they are less diagnostic. As shown in Fig. 6A, the N state of SH3 undergoes a uniform volume loss, ca. 80%, upon addition of DnaK. This result illustrates the fact that about 80% of the N population shifts to a different state in the presence of the chaperone.
Fig. 6.
Addition of ADP-DnaK to pure SH3 leads to dramatic variations in resonance intensities and volumes. (A) Percent changes in 1H-15N HSQC intensities and volumes of resonances corresponding to native SH3 upon addition of ADP-DnaK chaperone. (B) Percent changes in 1H-15N HSQC intensities and volumes of resonances corresponding to unfolded SH3 upon addition of ADP-DnaK chaperone. Missing bars denote residues whose intensities and volumes could not be accurately assessed due to resonance overlapping. (C) Percent volume recovery due to new peaks relative to the volume of corresponding unfolded resonances in the absence of ADP-DnaK.
Given that addition of a fourfold excess of DnaK leads to a spectrum dominated by U-type resonances (Fig. 2C), that there is no evidence from DOSY for the presence of chaperone-bound species exchanging with the N state (Fig. 4A), and that most of the SH3 population becomes chaperone-bound upon addition of DnaK (Fig. 3C), it is important to gather additional information on the nature of the chaperone-bound population. If the lost N population were converted into U, and U were to be mostly chaperone-bound upon addition of DnaK, the 80% volume loss from the N resonances would be recovered by an increase of ca. 80% in the U state resonance volume. However, the U state does not show a uniform 80% increase in volume upon addition of DnaK (Fig. 6B).
Instead, the central region of the protein (resonances 17–42) experiences a decrease in U-state resonance volume. We interpret this result as evidence for binding of some of the SH3 population to DnaK on the slow NMR chemical shift time scale. Some of the expected bound-state resonances are explicitly detected (Figs. 5 B–D and 6C), whereas others are broadened beyond detection. The observable unshifted U-type resonances for residues 17–42 (and also residues 47–54) in the presence of DnaK are assigned to the free (i.e., non–chaperone-bound) U state. This scenario explains the lack of any observable chemical shift variations upon addition of DnaK and the corresponding appearance of some new nearby resonances for residues 17–42. Other portions of the SH3 sequence show a different behavior.
In contrast, the backbone U-state NH resonances 3–11 (and resonances 43–46) experience a volume gain upon addition of DnaK (Fig. 6B). Hence, we deduce that these resonances correspond to conformations in fast exchange on the NMR chemical shift time scale. These conformations must include both the free and bound SH3 unfolded states. Therefore, by subtracting an average volume for resonances 17–42 (which nominally represent only the free U state, as discussed above) from an average volume for resonances 3–11 (which reflect free and bound states) and comparing the resulting value with the volumes in the chaperone-free state, we obtain an estimate of the populations of free and bound U-type species that are 19% and 24% of the total SH3 population, respectively. A similar analysis performed on the native state yields 14% of the total SH3 population in the free N state. Importantly, resonances 3–11 (and resonances 43–46) in the free and bound states share virtually identical chemical shifts, and hence a similar chemical environment, indicating that these unfolded-like residues do not appreciably interact with DnaK within the bound complex. These residues are in fast exchange on the NMR chemical shift time scale and fit scenario ii described above.
Interestingly, the sum of these percentages does not equal 100% but, rather, only 57% of the total SH3 population. In addition, no new resonances are detected for these residues. We therefore conclude that there is a volume loss of ca. 43% due to the presence of one or more additional slow-tumbling spectroscopically invisible chaperone-bound states, with unknown conformational features, amounting to roughly 43% of the total SH3 population. The resonances belonging to this spectroscopically invisible bound species must be in slow exchange on the chemical shift NMR time scale, and must be undetectable due to either broadening beyond detection (resulting from extremely slow tumbling during the acquisition time) or selective relaxation losses by the slow-tumbling complex during the multidimensional NMR pulse sequences. In addition, the conformational heterogeneity of this invisible population may be so extreme that the several broad resonances are individually too small in volume for explicit detection. This analysis is in agreement with the volume analysis of the Trp side chains discussed above and with line width estimations discussed in SI Appendix. The invisible nature of this population suggests that the corresponding bound SH3 population is likely more rigidly held to DnaK than the detectable SH3 resonances in slow exchange. Back-of-the-envelope line width calculations also show that the latter resonances (visible in slow exchange) must be preserving a significant degree of residual local dynamics (SI Appendix).
The resulting scenario leads to the identification of an additional population of a spectroscopically invisible class of chaperone-bound states that is likely extremely heterogeneous and/or poorly dynamic. The structural features of this bound state are unknown, and its population must be added to the population of the visible bound states.
As discussed in earlier sections, the main, visible, globally unstructured bound SH3 state is in reversible thermodynamic equilibrium with chaperone-free unfolded SH3 (Fig. 3 A and B). In addition, zz exchange experiments performed at high fields (600 and 900 MHz) with variable mixing times displayed no evidence for cross-peaks between any of the visible bound states and the chaperone-free native state, suggesting the lack of a direct path between these states [up to the approximate second (∼s) time scale] in the case of ADP-DnaK. Cross-peaks between free-unfolded and chaperone-bound states, and among spectroscopically visible bound states in slow exchange, may be present, although they were not explicitly discernible due to poor spectral resolution or very slow exchange rates (<1 s−1).
Careful data analysis shows that one additional correction to the free and bound SH3 populations is required as follows. A direct comparison with the percentage of bound SH3 assessed by native-gel analysis shows that the observed Kd imposes the requirement that the total chaperone-bound SH3 population under our NMR experimental conditions has to be 80%. Further, we noticed that the apparently unbound unfolded-state population Uf derived from the SH3 residues in the middle of the sequence (which has identical HN and N chemical shifts in the absence and presence of DnaK) shows a moderate line broadening even after correcting for the increase in viscosity resulting from the addition of DnaK to the sample (SI Appendix, Fig. S4B). The combination of these observations led us to conclude that some of the volume assigned to the 100% unbound unfolded population must also include some chaperone-bound species. The extent of this additional bound state was determined by imposing that 80% of the total SH3 population is chaperone-bound, as independently dictated by the native-gel analysis. This operation led to the conclusion that there is a 6% truly chaperone-free unfolded SH3 population and that the chaperone-bound population must include an additional 13% of an unconventional bound population of unspecified origin, which shows no observable chemical shift variations (relative to the free species) and weak line broadening.
Core of the DnaK-Bound SH3 Client Protein Is Buried from the Solvent.
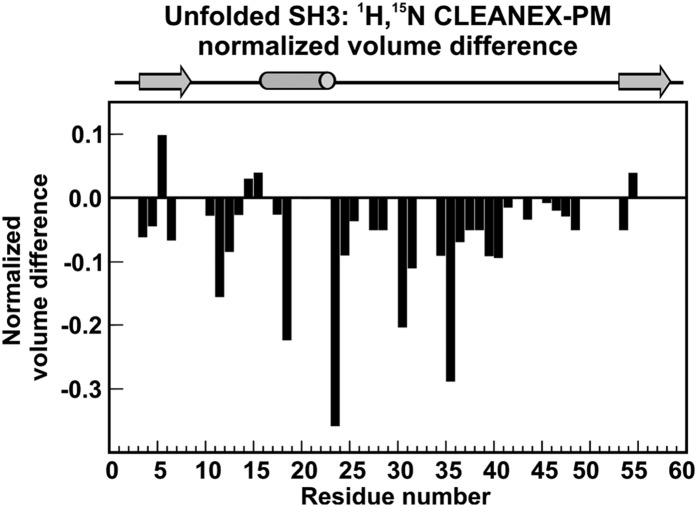
The preceding description of protein binding is supported by results of the clean solvent-exposed amides HSQC (Clean SEA-HSQC) experiment (36) performed on SH3 in the absence and presence of DnaK. The clean SEA-HSQC sequence is artifact-free, and it employs the phase-modulated CLEAN chemical exchange (CLEANEX-PM) (37, 38) pulse train to provide information about the degree of solvent exposure of specific protein regions. The normalized CLEANEX-PM volume difference with respect to HSQC upon addition of unlabeled DnaK to the solution (Fig. 7 and SI Appendix, Figs. S5 and S6) is particularly interesting for the U state. The data show that the central region of the protein becomes less solvent-exposed in the presence of the chaperone. This result is consistent with the previous description, and, in addition, it shows that chaperone-bound states have, on average, a solvent-buried core and floppy dynamic ends, including the N terminus.
Fig. 7.
Central portion of the chaperone-bound substrate protein sequence is protected from exchange with the solvent. SH3 backbone amide proton exchange with water is slowed down in the presence of DnaK. Normalized volume difference of unfolded SH3 resonances (calculated according to ), where VCLNX,S+D and VCLNX,S are the 1H-15N HSQC CLEANEX-PM resonance volumes for the SH3 sample in the absence (S) and presence of DnaK (S+D). VHSQC,S+D and VHSQC,S are the corresponding volumes in the 1H-15N HSQC. All volumes were normalized to 64 scans. The missing bars denote residues whose volumes could not be assessed accurately due to resonance overlapping. Negative normalized volume differences denote backbone amide protons buried from the solvent.
Hence, the interaction of SH3 with the DnaK chaperone affects a broad region of the central portion of the client protein rather than only the much narrower computationally predicted binding site region. This finding on the nature of the DnaK-bound protein state, supported by volume analysis and by Clean SEA-HSQC, is likely important to protect a large number of aggregation-prone residues. In summary, the region spanning the DnaK-protected residues is much larger than the one corresponding to the much narrower DnaK binding site predicted for small peptides spanning the SH3 sequence (Fig. 1G).
Discussion
The effect of Hsp70 chaperones on the conformation of their substrate proteins and on protein folding has been largely unexplored. Whereas previous work focused mainly on the effect of Hsp70 on small peptides, this study offers a first glimpse at the effect of the bacterial Hsp70 chaperone DnaK on the conformation of a full-length substrate protein.
Model Illustrating the Conformational Changes Undergone by SH3 upon Interaction with the DnaK Chaperone.
The data presented in this work lead to the model illustrated in Fig. 8 A and B for SH3 in the absence and presence of DnaK, respectively. The NMR time scale regimes corresponding to each of the proposed steps are shown in Fig. 8C. In essence, the free client protein is in equilibrium with a variety of mutually interconverting bound species. The detectable bound conformations are globally unstructured. The N-terminal portion of the sequence (amino acids ca. 3–11) and residues 43–46 are highly flexible, interact negligibly with the chaperone, and undergo fast conformational interconversion on a time scale <<20 ms. The rest of the client protein sequence is bound more tightly to the chaperone and is composed of a variety of globally unstructured states that undergo mutual interconversion on a time scale >>20 ms. Some of these residues are explicitly detectable, whereas others, likely interacting with the chaperone more tightly, are broadened beyond detection. We were able to identify 14 amino acids (showing new resonances in slow exchange) that experience a variety of different chemical environments upon addition of DnaK. These residues have either slightly different conformations or interact with different regions of the chaperone surface. Importantly, all detectable regions of the bound species retain the same small residual secondary structure as the unfolded state in the absence of chaperone.
In addition to this finding, there is another fully spectroscopically invisible bound population (dots in Fig. 8B) where all residues interact tightly with Hsp70 and have such slow tumbling rates that they are either broadened beyond detection in the NMR spectra or experience relaxation losses during the pulse sequence. Further investigations on this additional “invisible” state are necessary and will likely shed light on its potential role in protein folding and aggregation/disaggregation.
We propose that these multiple bound states play an integral role in the folding pathway of SH3 in the cellular environment, given the slow folding rates of this protein (33). In addition, we hypothesize that the heterogeneous chaperone-binding pattern identified in this study may underscore a general aspect of client protein conformational changes induced by interaction with DnaK. Specifically, the observed conformational heterogeneity suggests that conformational sampling takes place on the chaperone surface, implying that one of the roles of the DnaK chaperone may be to promote some of the search for the native state in an environment free from the risk of deleterious aggregation.
Although the conformational search proceeds successfully even in the absence of the chaperone for a small substrate protein, such as SH3, a heterogeneous bound state may be essential for the folding of larger proteins, which are more prone to populate kinetically trapped and aggregation-prone misfolded states in the absence of Hsp70, yet harder to study in the presence of chaperones via NMR or X-ray crystallography. Interactions such as that of SH3 with DnaK are likely important when DnaK serves as an unfoldase (39). Under such circumstances, ATP hydrolysis prompts partial unfolding of misfolded states, thus enabling one or more unfolded or partially unfolded states to be generated and leading to the ADP-bound state of the substrate–DnaK complex. We propose that the sampling of conformational space while ADP-DnaK–bound is a competitively advantageous process for misfolding-prone proteins (e.g., luciferase), in that nonpolar groups of these client proteins are capable of weakly and reversibly binding/unbinding DnaK while sampling different conformational states, thus minimizing the likelihood of falling into misfolded kinetically trapped conformations. As noted by Sharma et al. (39) for proteins such as luciferase, multiple cycles of ATP-hydrolysis/binding/unbinding are necessary to unfold misfolded conformations fully and reach a fully folded state. Hence, the conformational sampling that the client protein experiences in the ADP-bound state of DnaK, as studied in this work, is likely insufficient to disentangle misfolded states that fall into deep free-energy wells. However, the fact that the major DnaK binding sites are typically evenly spread across the protein primary structure (29) (e.g., the middle of the sequence as we saw here in the case of SH3) suggests that DnaK binding has the ability to unravel compact conformations and may enable the exploration of new regions of conformational space. In support of this idea, DnaK binding has recently been shown to cause unfolded-state expansion (40).
Our native-gel analysis shows that the overall dissociation constant for the binding of SH3 to DnaK exceeds 200 μM. Such a surprisingly weak interaction is compatible with conformational sampling on the ADP-DnaK chaperone’s surface, which requires client protein plasticity. Relatively weak binding is likely also helpful to maximize DnaK’s capacity to assist the folding of a large population of cellular proteins, although it has been proposed that the whole Hsp70 ATPase cycle increases the effective affinity of client proteins below the Kd value for binding to ADP-Hsp70 alone (41).
Finally, we determined that DnaK affects a wider region of SH3 sequence than formerly thought, which represents an aspect of its activity that may be critical to facilitate conformational sampling and maximize the prevention of aggregation across a wide range of residues and proteins.
Concluding Remarks.
In summary, we used a variety of multidimensional NMR experiments, native-gel analysis, and MS to identify a major class of heterogeneous chaperone-bound states of the drkN SH3 model protein. The bound client protein is highly heterogeneous, and it consists of multiple poorly structured states sharing a uniquely floppy N terminus, effectively not interacting with the chaperone, and a variety of more tightly bound core conformations. This mode of binding highlights the ability of DnaK to host a variety of protein conformational states. This result is important because it reveals that proteins undergo conformational sampling while chaperone-bound, highlighting the possibility of chaperone surface-mediated protein folding, a process free from complications arising from competing aggregation. Elucidating the presence and characteristics of Hsp70-chaperone–bound states of full-length proteins, presented here for the first time to our knowledge, crucially advances our understanding of chaperone-mediated protein folding.
Materials and Methods
Sample Preparation.
Unlabeled and isotopically labeled SH3 was expressed and purified according to known procedures, as described in further detail in SI Appendix. Stock solutions of pure SH3 were prepared in 50 mM Tris (pH 7.2), 5 mM MgCl2, and 50 mM KCl; flash-frozen; and stored at −80 °C. DnaK was prepared either according to published procedures or according to a streamlined procedure described in detail in SI Appendix. Stock solutions of pure DnaK were prepared and stored in 50 mM Tris, 5 mM MgCl2, and 50 mM KCl (pH 7.2).
NMR Sample Preparation, Data Collection, and Analysis.
NMR samples containing only 13C-15N SH3 consisted of a solution of pure SH3 in storage buffer diluted to a final concentration of 300 μM in 50 mM Tris (pH 7.2), 5 mM MgCl2, 50 mM KCl, 5 mM ADP, and 15 mM DTT in 5% (vol/vol) D2O. SH3 samples containing ADP-DnaK were identical except for the presence of 1.2 mM DnaK. Unless otherwise noted, all NMR experiments were performed using an Agilent 600-MHz (14.1-T) NMR spectrometer equipped with a 50-mm z-axis pulse field gradient (PFG) triple-resonance cryogenically cooled probe. All NMR experiments were carried out at 25 °C. All data were processed with NMRPipe (version 9.0.0-b108) and analyzed with NMRviewJ (version 2009.015.15.35). Volume measurements were carried out using the AutoFit script of NMRPipe. Additional details are described in SI Appendix.
Other Experimental Procedures.
Details on native-gel analysis, MS, viscosity measurements, and electrostatic surface potential calculations are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Hon Nam (Ken) Lam for the gift of unlabeled SH3; Charlie Fry, Tom Record, and Marco Tonelli for helpful discussions; and Ivan Akhremitchev (Rheosense, Inc.) for technical assistance. This work was supported by the National Science Foundation (Grant MCB-0951209) and by University of Wisconsin–Madison Bridge Funds from the College of Letters and Sciences (to S.C.). C.H. and Y.O. were awarded NIH Chemistry–Biology Interface (Grant T32 GM008505) and NIH Molecular Biophysics (Grant T32 GM08293) predoctoral training fellowships, respectively. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH National Institute of General Medical Sciences Grant P41GM103399 (old Grant P41RR002301). Equipment was purchased with funds from the University of Wisconsin–Madison; NIH Grants P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, and S10RR029220; NSF Grants DMB-8415048, OIA-9977486, and BIR-9214394; and the US Department of Agriculture.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession nos. 25500 and 25501).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505173112/-/DCSupplemental.
References
- 1.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14(10):630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82(82):323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 4.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92(3):351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 5.Fedyukina DV, Cavagnero S. Protein folding at the exit tunnel. Annu Rev Biophys. 2011;40:337–359. doi: 10.1146/annurev-biophys-042910-155338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 7.Georgopoulos C, Ang DKL, Zylic M. Properties of the Escherichia coli heat shock proteins and their role in bacteriophage lambda growth. In: Morimoto R, Tissieres A, Georgopoulos C, editors. Biology and Medicine. Cold Spring Harbor Laboratory Press; Plainview, NY: 1990. pp. 191–221. [Google Scholar]
- 8.Zuiderweg ER, et al. Allostery in the Hsp70 chaperone proteins. Top Curr Chem. 2013;328:99–153. doi: 10.1007/128_2012_323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young JC, Barral JM, Ulrich Hartl F. More than folding: Localized functions of cytosolic chaperones. Trends Biochem Sci. 2003;28(10):541–547. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Murphy ME. The HSP70 family and cancer. Carcinogenesis. 2013;34(6):1181–1188. doi: 10.1093/carcin/bgt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo JS, et al. T cell lymphoma in transgenic mice expressing the human Hsp70 gene. Biochem Biophys Res Commun. 1996;218(2):582–587. doi: 10.1006/bbrc.1996.0103. [DOI] [PubMed] [Google Scholar]
- 12.Evans CG, Wisén S, Gestwicki JE. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1-42) aggregation in vitro. J Biol Chem. 2006;281(44):33182–33191. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- 13.Muchowski PJ. Protein misfolding, amyloid formation, and neurodegeneration: A critical role for molecular chaperones? Neuron. 2002;35(1):9–12. doi: 10.1016/s0896-6273(02)00761-4. [DOI] [PubMed] [Google Scholar]
- 14.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6(1):11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 15.Zhuravleva A, Clerico EM, Gierasch LM. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012;151(6):1296–1307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clerico EM, Tilitsky JM, Meng W, Gierasch LM. How Hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J Mol Biol. 2015;42(7):1575–1588. doi: 10.1016/j.jmb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trend Biochem Sci. 2013;38(10):507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Zhu X, et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272(5268):1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landry SJ, Jordan R, McMacken R, Gierasch LM. Different conformations for the same polypeptide bound to chaperones DnaK and GroEL. Nature. 1992;355(6359):455–457. doi: 10.1038/355455a0. [DOI] [PubMed] [Google Scholar]
- 20.Popp S, et al. Structural dynamics of the DnaK-peptide complex. J Mol Biol. 2005;347(5):1039–1052. doi: 10.1016/j.jmb.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z, Kurt N, Rajagopalan S, Cavagnero S. Secondary structure mapping of DnaK-bound protein fragments: Chain helicity and local helix unwinding at the binding site. Biochemistry. 2006;45(40):12325–12333. doi: 10.1021/bi0612263. [DOI] [PubMed] [Google Scholar]
- 22.Kurt N, Cavagnero S. Nonnative helical motif in a chaperone-bound protein fragment. Biophys J. 2008;94(7):L48–L50. doi: 10.1529/biophysj.107.127647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JH, Tonelli M, Frederick RO, Chow DC-F, Markley JL. Specialized Hsp70 chaperone (HscA) binds preferentially to the disordered form, whereas J-protein (HscB) binds preferentially to the structured form of the iron-sulfur cluster scaffold protein (IscU) J Biol Chem. 2012;287(37):31406–31413. doi: 10.1074/jbc.M112.352617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seckler R, Jaenicke R. Protein folding and protein refolding. FASEB J. 1992;6(8):2545–2552. doi: 10.1096/fasebj.6.8.1592207. [DOI] [PubMed] [Google Scholar]
- 25.Sekhar A, Santiago M, Lam HN, Lee JH, Cavagnero S. Transient interactions of a slow-folding protein with the Hsp70 chaperone machinery. Protein Sci. 2012;21(7):1042–1055. doi: 10.1002/pro.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priya S, Sharma SK, Goloubinoff P. Molecular chaperones as enzymes that catalytically unfold misfolded polypeptides. FEBS Lett. 2013;587(13):1981–1987. doi: 10.1016/j.febslet.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Slepenkov SV, Witt SN. The unfolding story of the Escherichia coli Hsp70 DnaK: Is DnaK a holdase or an unfoldase? Mol Microbiol. 2002;45(5):1197–1206. doi: 10.1046/j.1365-2958.2002.03093.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang O, Kay LE, Olivier JP, Forman-Kay JD. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J Biomol NMR. 1994;4(6):845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]
- 29.Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16(7):1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vega CA, Kurt N, Chen Z, Rüdiger S, Cavagnero S. Binding specificity of an alpha-helical protein sequence to a full-length Hsp70 chaperone and its minimal substrate-binding domain. Biochemistry. 2006;45(46):13835–13846. doi: 10.1021/bi061432a. [DOI] [PubMed] [Google Scholar]
- 31. Van Durme J, et al. (2009) Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLoS Comput Biol 5(8):e1000475. [DOI] [PMC free article] [PubMed]
- 32.Ivey RA, 3rd, Subramanian C, Bruce BD. Identification of a Hsp70 recognition domain within the rubisco small subunit transit peptide. Plant Physiol. 2000;122(4):1289–1299. doi: 10.1104/pp.122.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekhar A, Lam HN, Cavagnero S. Protein folding rates and thermodynamic stability are key determinants for interaction with the Hsp70 chaperone system. Protein Sci. 2012;21(10):1489–1502. doi: 10.1002/pro.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson CS. Effects of chemical-exchange in diffusion-ordered 2D NMR-spectra. J Magn Reson A. 1993;102(2):214–218. [Google Scholar]
- 35.Swain JF, et al. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26(1):27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin D, Sze KH, Cui Y, Zhu G. Clean SEA-HSQC: A method to map solvent exposed amides in large non-deuterated proteins with gradient-enhanced HSQC. J Biomol NMR. 2002;23(4):317–322. doi: 10.1023/a:1020225206644. [DOI] [PubMed] [Google Scholar]
- 37.Hwang TL, van Zijl PCM, Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J Biomol NMR. 1998;11(2):221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 38.Hwang TL, Mori S, Shaka AJ, vanZijl PCM. Application of phase-modulated CLEAN chemical EXchange spectroscopy (CLEANEX-PM) to detect water-protein proton exchange and intermolecular NOEs. J Am Chem Soc. 1997;119(26):6203–6204. [Google Scholar]
- 39.Sharma SK, De los Rios P, Christen P, Lustig A, Goloubinoff P. The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat Chem Biol. 2010;6(12):914–920. doi: 10.1038/nchembio.455. [DOI] [PubMed] [Google Scholar]
- 40.Kellner R, et al. Single-molecule spectroscopy reveals chaperone-mediated expansion of substrate protein. Proc Natl Acad Sci USA. 2014;111(37):13355–13360. doi: 10.1073/pnas.1407086111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Los Rios P, Barducci A. Hsp70 chaperones are non-equilibrium machines that achieve ultra-affinity by energy consumption. eLife. 2014;3:e02218. doi: 10.7554/eLife.02218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ERP. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci USA. 2009;106(21):8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose GD, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229(4716):834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- 44.Bezsonova I, Singer A, Choy WY, Tollinger M, Forman-Kay JD. Structural comparison of the unstable drkN SH3 domain and a stable mutant. Biochemistry. 2005;44(47):15550–15560. doi: 10.1021/bi0512795. [DOI] [PubMed] [Google Scholar]
- 45.Jerschow A, Muller N. Suppression of convection artifacts in stimulated-echo diffusion experiments. Double-stimulated-echo experiments. J Magn Reson. 1997;125(2):372–375. [Google Scholar]
- 46.Pelta MD, Morris GA, Stchedroff MJ, Hammond SJ. A one-shot sequence for high-resolution diffusion-ordered spectroscopy. Magn Reson Chem. 2002;40(13):S147–S152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.