Significance
The propensity for weight gain is detrimental to modern human health. However, under environmental conditions where nutrients are limiting, this trait can be highly adaptive. Currently, the genetic basis of population level differences in appetite control and metabolism is still largely mysterious. Here, we describe changes in metabolism that evolved in the small tetra Astyanax mexicanus as it adapted from surface rivers to the nutrient-poor environment found in caves. We identified coding mutations in melanocortin 4 receptor responsible for an increase in appetite and starvation resistance of cavefish compared with surface fish populations. These results provide important genetic insights into metabolic evolution and show that mutations in a single gene can have profound effects on multiple physiological adaptations.
Keywords: Astyanax mexicanus, cavefish, MC4R, metabolic evolution, hyperphagia
Abstract
Despite recent advances in the understanding of morphological evolution, the genetic underpinnings of behavioral and physiological evolution remain largely unknown. Here, we study the metabolic changes that evolved in independently derived populations of the Mexican cavefish, Astyanax mexicanus. A hallmark of cave environments is scarcity of food. Cavefish populations rely almost entirely on sporadic food input from outside of the caves. To survive under these conditions, cavefish have evolved a range of adaptations, including starvation resistance and binge eating when food becomes available. The use of these adaptive strategies differs among independently derived cave populations. Although all cavefish populations tested lose weight more slowly than their surface conspecifics during restricted rations, only a subset of cavefish populations consume more food than their surface counterparts. A candidate gene-based screen led to the identification of coding mutations in conserved residues of the melanocortin 4 receptor (MC4R) gene, contributing to the insatiable appetite found in some populations of cavefish. Intriguingly, one of the mutated residues has been shown to be linked to obesity in humans. We demonstrate that the allele results in both reduced maximal response and reduced basal activity of the receptor in vitro. We further validate in vivo that the mutated allele contributes to elevated appetite, growth, and starvation resistance. The allele appears to be fixed in cave populations in which the overeating phenotype is present. The presence of the same allele in multiple caves appears to be due to selection from standing genetic variation present in surface populations.
The dark and relatively nutrient-poor environment of caves imposes strong selective pressures on colonizing species. As a consequence of the pitch-black environment, no photosynthetic primary producers exist in the caves. Cave inhabitants therefore rely entirely on food chains originating outside of the caves. The external food input can be introduced into the cave environments by bats living in the caves or through seasonal flooding. As a consequence, the food supply is limited and infrequent (1). To deal with this challenge, obligate cave species converge on similar metabolic adaptations, such as reduced metabolic rate, increased metabolic efficiency (weight gain/food consumed), starvation resistance (reduced weight loss during fasting), and increased body fat composition (2). To understand the underlying genetic basis of metabolic evolution better, we have focused on Astyanax mexicanus, the Mexican cavefish. There are two distinct forms of this species, a surface form and a cave form that displays a reduction or absence of melanin pigmentation and is eyeless (reviewed in 3). Although these morphs exhibit numerous morphological and behavioral differences, they remain interfertile. Furthermore, there are multiple independently evolved cave populations that share similar traits, allowing for the study of parallelism in evolution (4). Previous work has focused on understanding the genetic architecture of morphological and behavioral traits, such as pigmentation (5–7), eye size (6), schooling (8), and feeding angle (9); however, relatively little work has gone into understanding physiological traits (10, 11). Protas et al. (6) mapped sensitivity to dissolved amino acids, weight loss on sustained fasts, and condition factor using quantitative trait loci (QTL) analysis, but the genetic underpinning has not been identified for any of these traits. The genetic basis of metabolic variation, in particular, has recently undergone a resurgence in interest (12–14), but it still remains poorly understood in vertebrates.
Cave populations of A. mexicanus, like other obligate cave species, are well adapted to nutrient-poor environments. For example, a recent study has shown that cavefish have a slower metabolic rate, although they lack a circadian rhythm (15). Paradoxically, despite their lower metabolic rate, cavefish are actually more active. On this basis, Beale et al. (11) proposed that the cavefish experience “constant light” rather than perpetual darkness. The distinct metabolic rates between cave and surface populations provide a unique opportunity to uncover the genetic architecture underlying metabolic variation in vertebrates and how cave animals can cope with limited and infrequent food supply.
Results
We initially studied starvation resistance in individuals of similar body length and body weight taken from laboratory-bred surface and cave populations of A. mexicanus. After a 2-mo fast, all of the cave populations tested fared better under fasting conditions. Individuals from each of these caves lost only half as much weight as individuals from the surface population during a 2-mo fast [Fig. 1A; Tinaja-surface P value = 3.99 × 10−8, Pachón-surface P value = 2.69 × 10−9, and Molino-surface P value = 2.55 × 10−10 by Tukey honest significant difference (HSD) test]. Consistent with this observation, adult regularly fed Tinaja cavefish had a higher triglyceride composition than surface fish (Fig. 1B; Tinaja: 0.78 ± 0.018 nmol/μg vs. surface: 0.15 ± 0.05 nmol/μg; P value = 0.0016 by Tukey HSD test). Similar results in triglyceride levels were observed with individuals descended from another independently derived cave population, Pachón (Fig. 1B; 0.4 ± 0.025 nmol/μg; P value = 0.021 by Tukey HSD test). Although individuals of the two cave populations showed the same trend, those individuals from the Tinaja cave displayed by far the highest fat levels. In addition, the Tinaja cavefish are distinct in the location of fat storage within their bodies. The Tinaja cavefish had greatly enlarged livers not seen in fish from other caves (Pachon or Molino) that, moreover, stained strongly for the presence of lipids (Fig. 1 C and D). Despite developing fatty livers under high nutrient conditions, in preliminary studies, we observed no obvious difference in the life span or health of these fish, suggesting that they have evolved mechanisms to use the liver as a fat storage organ without the deleterious consequences seen in other species (16). This analysis thus reveals previously unidentified differences in metabolism not only between cave and surface populations but also among cave populations.
Fig. 1.
Cavefish are adapted to nutrient-poor environments. (A) Percentage of weight loss after a 2-mo fast. Eleven to 13 fish per population were used. Each cave population was significantly different from the surface population. ***P < 0.0001 (Tukey HSD test). (B) Total triglyceride content/protein in adult (>1-y-old) fish. Each population was significantly different from one another. *P < 0.05 (Tukey HSD test). (C) Pictomicrograph of livers. (Scale bars: 2 mm.) (D) Oil red O staining of liver sections. (Scale bars: 250 μm.) Red symbols (Pachón, Tinaja, and Molino) represent cavefish, and blue symbols represent surface fish. M, Molino; P, Pachón; S, surface; T, Tinaja.
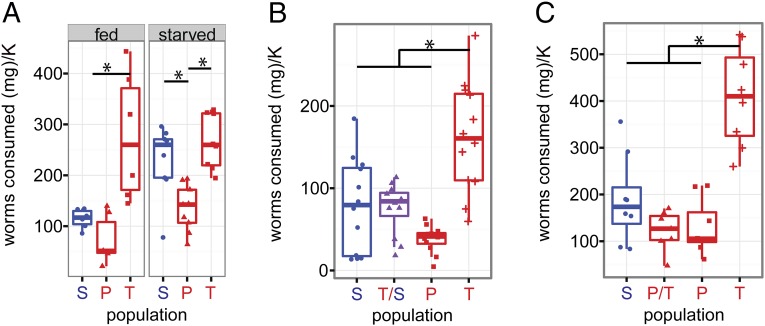
To study how cavefish acquire increased fat storage when reared under identical conditions as surface fish, we analyzed food consumption of individuals from the cave and surface populations. To detect differences in appetite, we measured the amount of black worms consumed over 1 to 2 d using ∼3- to 4-cm fish (<1-y-old). Tinaja cavefish allowed to feed ad libitum displayed greater appetite than the surface fish over a 24-h period (eating 276 ± 51 and 114 ± 7.7 mg per 100 g⋅cm−3) of worms, respectively; Fig. 2A; P value = 0.0018 by Tukey HSD test). The appetite of well-fed Tinaja cavefish was not significantly different from the appetite that surface fish reach after 1 mo of fasting (Fig. 2A; P value = 0.79 by Tukey HSD test). Although Pachón cave fish reach higher fat levels than surface fish (Fig. 1B; P value = 0.021 by Tukey HSD test), we did not detect a difference in their appetite from their surface counterparts (Fig. 2A; P value = 0.88 by Tukey HSD test), suggesting a divergent mechanism of weight gain. Neither Pachón nor Tinaja cavefish appetites were affected by a 1-mo fast, whereas surface fish that have fasted display a significant elevation in appetite (Fig. 2A; P value = 0.021 by Tukey HSD test). These data suggest that there are significant differences in appetite regulation between populations. To investigate the genetic basis of these phenotypes, we analyzed F1 hybrids between Pachón and Tinaja cavefish and between surface fish and Tinaja cavefish. Both classes of F1 hybrids had appetites similar to Pachón and surface fish during a fed state, which suggests that the excessive Tinaja appetite is a recessive trait (Fig. 2 B and C; Tinaja/surface F1 − Pachón: P value = 0.245, Tinaja/surface F1 − surface: P value = 0.99, Pachón/Tinaja F1 − Pachón: P value = 0.99, Pachón/Tinaja F1 − surface: P value = 0.48; Tinaja/surface F1 − Tinaja: P value = 0.00029, and Tinaja/Pachón F1 − Tinaja: P value = 1.34 × 10−6 by Tukey HSD test).
Fig. 2.
Differential appetite regulation. (A) Appetite comparison between fed and starved surface and cavefish (3 wk). Six fish were used for each condition, and population combination and the amount of worms consumed were recorded over a 24-h assay period. (B) Appetite comparison among fed Tinaja, Pachón, surface, and Tinaja/surface F1 hybrids. Twelve fish were used for each population and tested over a 36-h assay period. (C) Appetite comparison with fed Tinaja, Pachón, surface, and Pachón/Tinaja F1 hybrids. Eight fish were used for each population and tested over a 48-h period. Red symbols (Pachón, Tinaja, and Molino) represent cavefish, and blue symbols represent surface fish. P/T, Pachón/Tinaja F1 hybrid; T/s, Tinaja/surface F1 hybrid. *P < 0.05 (Tukey HSD test). K = condition factor (100 g⋅cm−3, cm = body length from nose to base of tail).
To identify genes contributing to the Tinaja cavefish hyperphagia phenotype, we took a candidate gene approach, taking advantage of the newly available genome assembly for A. mexicanus (17). We focused on the genes in the leptin pathway, because leptin levels are correlated with adiposity in other species (a list of genes, primers, and sequences is provided in Dataset S1). One of the key targets of leptin is melanocortin 4 receptor (MC4R), a G protein-coupled receptor that integrates leptin and insulin levels in the paraventricular nucleus in the hypothalamus (18). MC4R neurons are known to induce anorexia when there are high leptin and insulin levels (19). Comparison of the MC4R sequence found in the surface and Tinaja cave genomes revealed three nonsynonymous sequence differences in highly conserved residues (Fig. 3A and Fig. S1). One difference, in particular, resulting in a Gly-to-Ser shift in the encoded MC4R protein, is particularly interesting because mutations in this highly conserved residue are associated with obesity in human patients. Mutations of this residue in humans have been characterized as having reduced signaling efficiency and lower basal activity (20), although it also must be noted that there is an Ala at this position in all other vertebrate genomes; thus, even the surface fish differ from other vertebrates (Fig. S1).
Fig. 3.
Hypomorphic mutations in MC4R cosegregate with hyperphagia. (A) Protein sequences of A. mexicanus MC4R [adapted after Tao (18)] shown in red are Tinaja-specific mutations. (B) Dose–response curve of Tinaja and surface alleles in vitro using 293T cells. Tinaja cavefish have a lower maximal response and basal activity than surface fish (P < 0.05). Each data point represents the mean of three wells being independently transfected and measured. (C) Appetite comparison between Pachón cavefish crosses carrying either the surface or cave allele of MC4R. C, Tinaja cave allele; S, surface allele. Twenty-two homozygous fish and 38 heterozygous fish (3-mo-old) were used, and their appetite was assayed over a 1-wk assay period. **P < 0.005 (two-tailed t test). (D) Body length comparison among Pachón cavefish MC4R genotypes. The same fish shown as in Fig. 3C, but 6 mo older. At the end of the experiment, the fish were 9 mo old. (E) Percentage of weight loss comparison among Pachón cavefish MC4R genotypes during a 3-wk fast. Same fish as in Fig. 3D were used, but they were starved after measuring length. *P < 0.05 (two-tailed t test).
Fig. S1.
Conservation of the amino acids affected by coding changes in MC4R within A. mexicanus populations among other vertebrate genomes, including the mutated allele in a case of human obesity (20). Red asterisks highlight the residue changes in the Tinaja allele.
The Gly-to-Ser change we identified affects a residue in the G protein interaction domain of MC4R (Fig. 3A) that could potentially result in less efficient binding between the receptor and the G protein. To determine whether the observed substitutions are indeed significant for the activity of the MC4R protein, we directly compared the signaling efficiencies of the surface and Tinaja cavefish alleles. Utilizing an in vitro approach previously used in studies of the piscine (21) and mammalian MC4R (20), we transfected MC4R surface and Tinaja alleles into 293T cells and generated a dose–response curve to the melanocortin receptor agonist, [Nle4, D-Phe7]-α-melanocyte stimulating hormone (NDP-α-MSH). Similar to the analogous human A154D MC4R variant (20), the Tinaja MC4R allele had a significantly lower maximal response and basal activity (45% P value = 0.0029 and 83% P value = 0.016, respectively) but an equivalent EC50 to the surface form (Fig. 3B), consistent with a deficiency in transduction but not in ligand binding.
Carrying a variant of MC4R with decreased activity could have given a selective advantage to fish in the nutrient-poor Tinaja cave environment. One of the advantages of the A. mexicanus cavefish system is that multiple caves, with similar ecological conditions, were independently invaded by surface populations of the same species, providing the opportunity to study parallel evolution. To see whether the A. mexicanus populations in other caves also carry this alteration or other alterations in the MC4R gene, we sequenced this gene from multiple individuals of eight additional cavefish populations (Pachón, Sabinos, Micos, Yerbaniz, Piedras, Molino, Arroyo, and Japonese) (Fig. 4A and Table 1) of which at least three (Micos, Pachón, and Molino) are known to represent independent and geographically separated colonization events from the Tinaja cave and from each other (compare also Fig. 4A).
Fig. 4.
Parallelism of hyperphagia in Astyanax mexicanus. (A) Geographic distribution of different cave populations. (B) Appetite comparison between fed young (<1-y-old) surface fish and another independently derived cave population (Molino) carrying a derived MC4R allele over a 72-h period. ***P < 0.001 (two-tailed t test).
Table 1.
Derived Tinaja MC4R mutations are prevalent in independently derived cave populations and present in surface fish populations
| Cave population | Genotype | ||
| s/s | c/s | c/c | |
| Pachón | 2 | 3 | 4 |
| Molino* | 0 | 0 | 9 |
| Arroyo | 0 | 0 | 7 |
| Yerbaniz | 0 | 0 | 9 |
| Piedras | 0 | 0 | 1 |
| Micos | 0 | 0 | 1 |
| Japonese | 0 | 0 | 5 |
| Sabinos | 0 | 0 | 1 |
| Tinaja | 0 | 0 | 9 |
| Surface | 107 | 6 | 1 |
We found that in six of these populations, the same mutations in MC4R were detected in 100% of the fish tested (Table 1). One exception is Molino, which had only two of the three mutations fixed (Table 1), with the other being Pachón, which showed the surface allele in our laboratory stocks. Further genotyping of wild-caught Pachón DNA revealed that both alleles are present in the Pachón cave (Table 1). Additional sampling will be necessary to establish the frequency of this mutation in the Pachón cave. The presence of the exact same mutations (at least the most conserved two mutations) in four independently derived cave populations is intriguing and points toward the possibility of selection from standing variation in the ancestral surface populations. To see if standing genetic variation is present in the current surface population, we sampled 114 wild-caught surface fish from different locations in the Sierra de el Abra (Figs. S1–S3 and Dataset S1). Of these fish, we found six that were heterozygous and one that was homozygous for the derived allele (Table 1). These frequencies are in line with previous results of standing genetic variation in cavefish (4) and in marine stickleback populations for alleles responsible for reduced body armor in fresh water populations (14), supporting the possibility that there was selection from standing genetic variation.
Fig. S3.
(A) Liver oil red O staining of fed Pachón (>1-y-old fish) (Scale bars: 250 μm.) (B) Comparisons of body length of (>1-y-old) Pachón fish among MC4R genotypes. *P < 0.05. (C) Appetite comparison among MC4R genotypes in old (>1-y old) fed Pachón fish during a 2-wk period. MC4R genotypes have an age-dependent association with appetite, with older fish (>1-y-old) not showing a difference in appetite among genotypes.
The fact that the Tinaja allele is present to some extent in the Pachón population allowed us to investigate the extent to which the allele of MC4R with reduced signaling activity contributes to the hyperphagic phenotype within this population. To that end, we tested crosses between Pachón cavefish that were carriers and noncarriers of the derived MC4R allele in the appetite assay described above. We observed that 3-mo-old Pachón cavefish that were homozygous for the derived allele had a greater appetite than heterozygotes of the same cross [Fig. 3B; 309 mg/K vs. 258 mg/K; P value = 0.0018 by two-tailed t test; K = condition factor (100 g⋅cm−3) where cm = body length]. However, older fish (>>1-y-old) did not show this difference (Figs. S2B and S3C), potentially as a result of reduced growth rates exhibited by bigger fish due to space constraints. Moreover, the difference in appetite we did observe in younger animals with the two genotypes was less pronounced than the difference in appetite between Tinaja and Pachón cavefish. Indeed, observing the variation between the genetic classes of Pachón cavefish required extending the appetite assay from 2 d to 1 wk. These results demonstrate that the observed mutations in MC4R contribute to a hyperphagic phenotype but that additional genes affecting appetite must also be present in the Tinaja cave population. Further substantiating that the derived MC4R allele is functionally significant, we observed that Pachón cavefish homozygous for the derived allele appeared larger than their clutch mates on average; however, they also displayed a rather large variation [Fig. 3D, heterozygotes (cave [c] and surface [s]): 33.8 ± 0.55 mm and homozygotes (cc): 36.3 ± 1.14 mm, P value = 0.06 by two-tailed t test; and Fig. S3B, cc-cs P value = 0.032 and cc-ss P value = 0.098 by two-tailed t test]. Furthermore when we tested for starvation resistance, Pachón cavefish that were homozygous for the derived allele were more starvation-resistant than heterozygous fish (Fig. 3E; P value = 0.018 by two-tailed t test). This finding shows that even such a small increase in appetite can have substantial effects on the growth rates and starvation resistance of these fish, potentially contributing to the adaptive metabolic changes cavefish have experienced. However, the changes were not enough to induce a fatty liver phenotype (Fig. S3A), suggesting that other genes are mainly responsible for this phenotype. As additional evidence to support the ecological and evolutionary significance of the MC4R mutations, Molino cavefish, which are homozygous for a derived allele of MC4R, are also hyperphagic relative to their surface counterparts (Fig. 4B; P value < 0.001 by two tailed t test) and starvation-resistant (Fig. 1A; P value = 2.55 × 10−10 by Tukey HSD test).
Fig. S2.
(A) Comparison of appetite between fed old (>1-y-old) and young (<1-y-old) Tinaja fish over a 72-h period. ***P < 0.0005 (two-tailed t test). (B) Appetite comparison among MC4R genotypes of older surface/Tinaja F2 fish over a 5-d period. ns, not significant ANOVA.
Discussion
In this study, we explored the physiological adaptations that cave populations of A. mexicanus have made to survive and thrive in their unique nutrient-poor environment. These adaptations include, to a varying extent from cave to cave, an increased efficiency in using food, increased appetite when food is available, increase in stored body fat, and decrease in the speed of weight loss in the face of starvation. All of these metabolic and behavioral changes work together to increase the fitness of fish facing only sporadic periods of food availability, punctuating general nutrient deprivation. Although the cave populations from Tinaja, Molino, and Pachón all lose weight more slowly than surface fish, the Tinaja and Molino cavefish ate more than their relatives from the Pachón cave. This finding could reflect differences in the severity of nutrient limitation in these respective caves, or, alternatively, it could indicate that the fish in the Pachón cave have adapted in other ways not analyzed here that give them a similar ability to cope with starvation without hyperphagia. Further ecological and metabolic studies will be needed to explore this possibility. Additionally, we identified coding mutations in MC4R that may contribute to some of the physiological adaptations of A. mexicanus described here, at least to the increase in appetite, growth, and starvation resistance. Other traits, such as the fatty liver phenotype, appear to be independent of MC4R and hyperphagia. In line with this observation is the finding that Molino cavefish, despite having a greater appetite and the derived MC4R allele, do not display the fatty liver phenotype.
We provide evidence that the mutations in cavefish MC4R lead to reduced signaling efficiency and basal activity of MC4R, similar to an analogous mutation in human MC4R. In vivo data demonstrate that the cave allele of MC4R cosegregates with higher appetites. Interestingly, the hyperphagia cosegregating with the derived MC4R allele was age-dependent, because older fish did not show an increase in appetite (Figs. S2 and S3), although this finding could be explained, in part, by reduced growth rates of bigger fish due to tank size constraints. It is intriguing to note that certain human MC4R mutants have also been reported to affect childhood obesity specifically, with the impact being counteracted in older patients (22). Other genes might act to modulate appetite in older cavefish. It is also plausible that higher appetite is particularly important during early stages of fish development (<1-y-old) to obtain higher growth rates to the point where the fish can store sufficient reserves to make it through the starvation periods. It is worth mentioning that in some species of platyfish, MC4R copy number variation with both functional and nonfunctional alleles has been shown to play a role in energy balance and reproductive maturity (21). In any event, it is clear that MC4R is only one of a number of genes modulating feeding activity in the Tinaja and Molino cavefish, because the difference in level of appetite between Pachón individuals carrying the derived MC4R allele and those individuals homozygous for the WT allele is far less than the difference in level of appetite between the Tinaja or Molino fish and their surface conspecifics.
The same derived MC4R allele, with decreased signaling efficiency was found in all eight of the cave populations sampled, and, moreover, it appears to be fixed in all but one of them. Four of these caves are known to be independently colonized. This suggests that the mutations likely entered the caves as standing variations present in the surface populations at the times of the invasions. Indeed, we detected the derived MC4R allele at low frequency in the current surface population. The level of standing variation we observed is consistent with the level of standing variation seen for various traits selected during fresh water invasion by sticklebacks (23).
Although the trajectory of evolutionary change in A. mexicanus is clear (the cavefish are the derived phenotype relative to the surface fish), there is some low level of escape from certain caves back to the surface population (24, 25). Thus, in principle, the migration from the caves could contribute to the present day standing variation, although it must be noted that not only are the escape events uncommon but surface/cave matings are believed to be rare in the river environment due to surface fish aggression, as well as cavefish being maladapted to the surface environment (24). Intriguingly, however, the derived allele of MC4R appears to be fixed in the cavefish populations despite the fact that there is significant gene flow in the opposite direction from the outside into the caves (24). This finding suggests a strong selective advantage of the allele in the seven caves where we found the mutations to predominate. Of course, we cannot exclude that natural selection is actually acting on a nearby locus and that the derived allele of MC4R is carried along by linkage disequilibrium. However, our in vitro data clearly show a significant decrease in the function of MC4R similar to human obesity studies, making the cave allele of MC4R a compelling candidate for phenotypic selection. In principle, all three observed coding changes could work in concert to reduce signaling of MC4R; however, given that Molino fish carry only two of them (G145S and M259I) and mutations in amino acid 145 alone are associated with obesity in humans, we favor the hypothesis that the first mutation (G145S) is responsible for the observed phenotype. Further studies will be needed to determine the interplay of these mutations and the potential epistatic effects with other genes and signaling pathways in the endocrine signaling of the fish. This selective pressure seems to be reduced in the Pachón cave, however, where, in addition to the derived allele, we found the ancestral allele to be present. Relatively lower levels of gene flow into the Pachón cave further support this notion of reduced selective pressure (24), but it would be interesting to study whether the relaxed selection for this allele is related to the general absence of hyperphagia in the population and to what extent this adaptation is connected to the ecological constraints in the Pachón cave. It has been speculated that the Pachón cave has a relatively large food input due to bat colonies located above the cave water and from the influx of plant debris that is carried into the system from some unknown source(s) (26).
The unique aspects of the cave environment create conditions where metabolic traits, such as obesity and hyperphagia, which would be maladaptive in other settings, give a strong survival advantage. Further genetic analysis of this fascinating species will give further insight into the process by which metabolism evolves in the face of environmental challenge.
Materials and Methods
Animal Husbandry.
All animal procedures were conducted in accordance with the guidelines of National Institutes of Health and were approved by the Institutional Animal Care and Use Committee at Harvard Medical School. To generate starved animals, we housed animals individually and fasted them for a maximum of 2 mo. Unfasted animals were fed in excess three times daily with brine shrimp, pellet, black worms, and other live food. Cave and surface F1 hybrids were generated through dark matings, because surface fish tend to attack their cave counterparts in the light. Animals were housed at around 25–30 fish per tank to reduce variation caused by differential housing density.
Triglyceride Quantification.
Four older male fish from each population (>1-y-old) with a similar body length were flash-frozen with liquid nitrogen and ground to powder with a ceramic mortar and pestle under liquid nitrogen. The powder was stored at −80 °C until processing. Fifty milligrams of powder from each fish was resuspended and homogenized in 1 mL of 0.5% IGEPAL CA-630 (I8896; Sigma). A 100-μL subsample of homogenized tissue was used to measure total protein using a Pierce BCA Protein Assay Kit (23225; Thermo Scientific). The rest of the sample was used for triglyceride quantification utilizing a colorimetric Triglyceride Quantification Kit (ab65336; Abcam). Triglyceride levels (micromoles) were normalized to total protein levels (micrograms).
Candidate Gene Sequencing.
RNA and DNA were isolated from tail fin clips and extracted following phenol or proteinase K treatment, respectively. RNA was reverse-transcribed using the cloned AMV First-Strand Synthesis Kit (12328-032; Invitrogen). Primers were designed using the Ensembl version of the A. mexicanus genome and tested in different populations (Dataset S1). For MC4R, all genotyping was performed using genomic DNA because MC4R has no introns (primers and sequences are provided in Dataset S1).
Appetite Assay.
Appetite was assessed by measuring the amount of black worms, Lumbriculus variegatus, consumed after 24–72 h. When comparing appetite between MC4R genotypes in Pachón fish, the assay was extended up to 2 wk. Animals assayed were of similar body length (∼3.5 ± 0.5 cm) and of similar age (<1-y-old). Fish were said to be “fed” if they were fed ad libitum three times daily for 1 wk. Before measuring appetite, animals were housed individually in 800-mL cups and acclimatized for 24 h. Before starting the assay, the water was changed and individuals were then weighed and measured before preweighed black worms (∼1 g) were added. Following the testing interval, animals were removed into a fresh 800-mL cup and were reweighed and measured following 24 h. Black worms were separated from fecal matter and reweighed. Appetite measurements were normalized to condition factor (K = 100 g⋅cm−3, cm = body length from nose to base of tail).
In Vitro Functional Characterization.
MC4R alleles were functionally characterized from a protocol adapted from Tao and Segaloff (20). HEK 293T cells were transiently transfected with constructs expressing MC4R cavefish alleles. MC4R alleles were amplified from cDNA and cloned into pcDNA3 using Gibson assembly. Cells were seeded on six-well poly-d-lysine–coated plates with DMEM + 10% FBS + 1% penicillin/streptomycin the night before transfection. The media used is standard growth media for 293T cells. Six microliters of polyethylemine was used to transfect 1.5 μg of pcDNA-MC4R and 0.5 μg of CAG-GFP (transfection control) per well for 24 h. Media were changed to DMEM (serum-free) for 12 h. Transfection efficiency was assessed via expression of GFP. Following serum starvation, media were changed to DMEM + 0.5 mM isobutyl methylxanthine. After a 15-min incubation, buffer alone or different concentrations of NDP-αMSH (M8764; Sigma) were added to cells and incubated for 1 h at 37 °C. cAMP was measured using a cAMP Direct Immunoassay Kit (ab65355; Abcam). Measurements were normalized by number of GFP-positive cells. The P value was derived from at least three independent transfections for each condition, and maximal and basal activity was assessed by two independent experiments.
Statistics.
Dose–response curves were generated using a four-parameter log–logistic function in the drm function in the drc package in R. Pairwise comparisons were done with Student’s t tests. Multiple comparisons were done with ANOVA and post hoc Tukey HSD tests in R (27).
Supplementary Material
Acknowledgments
We thank F. Miller, C. Normand, S. Roberts, R. Jonas-Closs, A. Webster, and M. Vitali for assistance in the appetite experiments and for weighing, measuring, and general maintenance of fish. We are grateful to Y. Chinchore, Y. Kamberov, and B. Rabe for their technical expertise and advice for the in vitro experiments. This work was supported by Grant HD047360 from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510802112/-/DCSupplemental.
References
- 1.Culver D, Pipan T. The Biology of Caves and Other Subterranean Habitats. Oxford Univ Press; Oxford: 2009. [Google Scholar]
- 2.Hüppop K. Phänomene und Bedeutung der Energieersparnis bei dem Höhlenfisch Astyanax fasciatus (Characidae) Universität Hamburg; Hamburg, Germany: 1988. German. [Google Scholar]
- 3.Jeffery WR. Cavefish as a model system in evolutionary developmental biology. Dev Biol. 2001;231(1):1–12. doi: 10.1006/dbio.2000.0121. [DOI] [PubMed] [Google Scholar]
- 4.Bradic M, Teotónio H, Borowsky RL. The population genomics of repeated evolution in the blind cavefish Astyanax mexicanus. Mol Biol Evol. 2013;30(11):2383–2400. doi: 10.1093/molbev/mst136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Protas M, Conrad M, Gross JB, Tabin C, Borowsky R. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr Biol. 2007;17(5):452–454. doi: 10.1016/j.cub.2007.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Protas M, et al. Multi-trait evolution in a cave fish, Astyanax mexicanus. Evol Dev. 2008;10(2):196–209. doi: 10.1111/j.1525-142X.2008.00227.x. [DOI] [PubMed] [Google Scholar]
- 7.Protas ME, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38(1):107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- 8.Kowalko JE, et al. Loss of schooling behavior in cavefish through sight-dependent and sight-independent mechanisms. Curr Biol. 2013;23(19):1874–1883. doi: 10.1016/j.cub.2013.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowalko JE, et al. Convergence in feeding posture occurs through different genetic loci in independently evolved cave populations of Astyanax mexicanus. Proc Natl Acad Sci USA. 2013;110(42):16933–16938. doi: 10.1073/pnas.1317192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salin K, Voituron Y, Mourin J, Hervant F. Cave colonization without fasting capacities: An example with the fish Astyanax fasciatus mexicanus. Comp Biochem Physiol A Mol Integr Physiol. 2010;156(4):451–457. doi: 10.1016/j.cbpa.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Beale A, et al. Circadian rhythms in Mexican blind cavefish Astyanax mexicanus in the lab and in the field. Nat Commun. 2013;4:2769. doi: 10.1038/ncomms3769. [DOI] [PubMed] [Google Scholar]
- 12.Carreno-Quintero N, Bouwmeester HJ, Keurentjes JJ. Genetic analysis of metabolome-phenotype interactions: From model to crop species. Trends Genet. 2013;29(1):41–50. doi: 10.1016/j.tig.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Chan EK, Rowe HC, Hansen BG, Kliebenstein DJ. The complex genetic architecture of the metabolome. PLoS Genet. 2010;6(11):e1001198. doi: 10.1371/journal.pgen.1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed LK, et al. Systems genomics of metabolic phenotypes in wild-type Drosophila melanogaster. Genetics. 2014;197(2):781–793. doi: 10.1534/genetics.114.163857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran D, Softley R, Warrant EJ. Eyeless Mexican cavefish save energy by eliminating the circadian rhythm in metabolism. PLoS ONE. 2014;9(9):e107877. doi: 10.1371/journal.pone.0107877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchesini G, et al. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology. 2003;37(4):917–923. doi: 10.1053/jhep.2003.50161. [DOI] [PubMed] [Google Scholar]
- 17.McGaugh SE, et al. The cavefish genome reveals candidate genes for eye loss. Nat Commun. 2014;5:5307. doi: 10.1038/ncomms6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao YX. The melanocortin-4 receptor: Physiology, pharmacology, and pathophysiology. Endocr Rev. 2010;31(4):506–543. doi: 10.1210/er.2009-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz MW, Woods SC, Porte D, Jr, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404(6778):661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- 20.Tao YX, Segaloff DL. Functional analyses of melanocortin-4 receptor mutations identified from patients with binge eating disorder and nonobese or obese subjects. J Clin Endocrinol Metab. 2005;90(10):5632–5638. doi: 10.1210/jc.2005-0519. [DOI] [PubMed] [Google Scholar]
- 21.Lampert KP, et al. Determination of onset of sexual maturation and mating behavior by melanocortin receptor 4 polymorphisms. Curr Biol. 2010;20(19):1729–1734. doi: 10.1016/j.cub.2010.08.029. [DOI] [PubMed] [Google Scholar]
- 22.Farooqi IS, et al. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. 2003;348(12):1085–1095. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- 23.Colosimo PF, et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307(5717):1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 24.Bradic M, Beerli P, García-de León FJ, Esquivel-Bobadilla S, Borowsky RL. Gene flow and population structure in the Mexican blind cavefish complex (Astyanax mexicanus) BMC Evol Biol. 2012;12:9. doi: 10.1186/1471-2148-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panaram K, Borowsky R. Gene flow and genetic variability in cave and surface populations of the Mexican Tetra, Astyanax mexicanus (Telcostei: Characidae) Copeia 2005. 2005;(2):409–416. [Google Scholar]
- 26. Mitchell RW, Russell WH, Elliott WR (1997) Mexican Eyeless Characin Fishes, Genus Astyanax: Environment, Distribution, and Evolution (Texas Tech Univ Press, Lubbock, TX)
- 27. R Development Core Team (2010) R: A Language and Environment for statistical computing R Foundation for Statistical Computing Vienna. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.