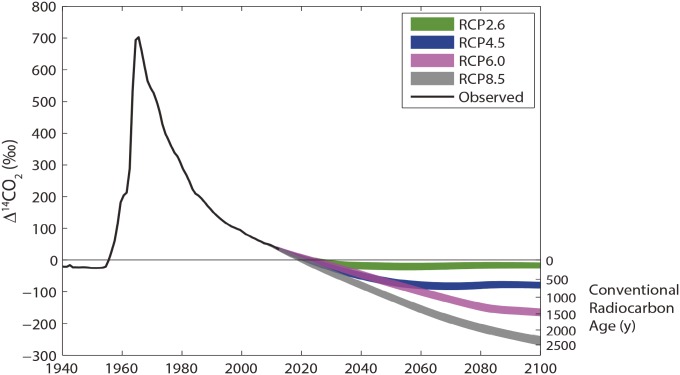
Changes in atmospheric radiocarbon
Radiocarbon content in atmospheric CO2 projected through 2100 using representative concentration pathway carbon emission scenarios.
The amount of radioactive carbon-14 in atmospheric CO2 has varied since 1890 due to fossil fuel emissions and nuclear weapons testing. Heather Graven (pp. 9542–9545) calculated the effect of fossil fuel consumption and resultant CO2 emissions on the ratio of radiocarbon to stable carbon in the atmosphere over the 21st century. Because fossil fuels have lost all radiocarbon through radioactive decay, the radiocarbon concentration in the atmosphere can be diluted by fossil fuel-generated CO2. The dilution is currently increasing the radiocarbon age of the atmosphere by 30 years per year. According to the author, ambitious emission reductions could hold atmospheric radiocarbon at preindustrial levels, while business-as-usual emission scenarios suggest significant depletion of atmospheric radiocarbon equivalent to more than 2,000 years of radioactive decay. By 2050, under a business-as-usual scenario, fresh organic material may be indistinguishable by radiocarbon dating techniques from material from 1050. The results suggest that fossil fuel use may decrease atmospheric radiocarbon content more rapidly than previously expected and that such changes may strongly affect the viability of techniques that use radiocarbon in fields such as earth science, archaeology, and forensics, according to the author. — P.G.
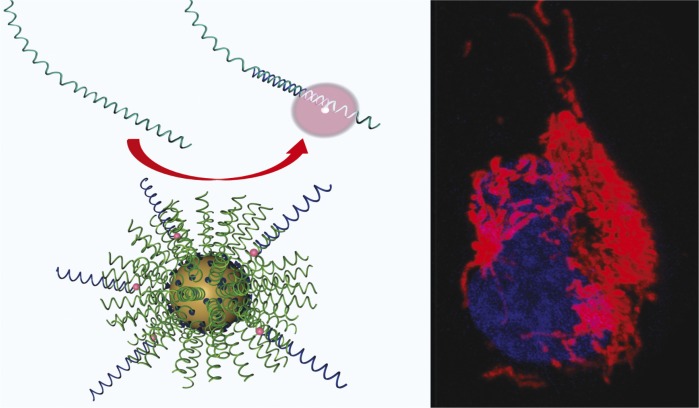
Quantifying and tracking RNA in live cells
Sticky-flares label endogenous RNA, allowing RNA tracking in live cells.
The transport and localization of messenger RNA (mRNA) are critical for cellular health, and understanding RNA misregulation can aid the diagnosis and treatment of numerous disorders. However, current techniques for RNA analysis have limitations, including the inability of many methods to report on intracellular RNA localization. William Briley et al. (pp. 9591–9595) developed spherical nucleic acid gold nanoparticle conjugates termed Sticky-flares, which can report on both expression levels of RNA transcripts and their intracellular localization. Upon recognition of a target RNA, the Sticky-flare transfers a fluorescent reporter to the transcript, thereby labeling the transcript’s movements throughout the cell to be tracked via fluorescence microscopy. Extending previous work with the Nanoflare, the authors rendered the fluorophore-conjugated reporter strand long and complementary to the targeted RNA transcript, thus allowing the Sticky-flare to determine the spatial distribution of RNA within cells, a capability that the Nanoflare lacks. The authors used Sticky-flares to quantify β–actin mRNA in HeLa cells as well as to follow the real-time transport of β–actin mRNA in mouse embryonic fibroblasts. Future studies to develop additional Sticky-flare constructs might allow simultaneous targeting of multiple transcripts, according to the authors. — L.G.
Misreading the genetic code
Successful translation of the genetic code depends on the ability of transfer RNAs (tRNAs) to discriminate between correct codons and mismatches, as measured by parameters known as d-values. Using a cell-free system, Jingji Zhang et al. (pp. 9602–9607) estimated the d-values of seven tRNAs for nine close but mismatched codons per tRNA, representing about 15% of all such possible codon–tRNA combinations. During protein synthesis, tRNA binds to the ribosome as part of a complex involving the molecule GTP, which is hydrolyzed upon codon recognition. The authors measured the rates of GTP hydrolysis at varying Mg2+concentrations, and determined the catalytic efficiency of codon recognition. Accuracy was calculated as the ratio of the efficiencies for correct and incorrect recognitions, and increased linearly with decreasing efficiency of correct recognitions. Extrapolating to zero efficiency of correct recognition yielded the d-value for each tRNA-mismatch combination. The d-values varied 400-fold, from 200 to 80,000, depending on the type of mismatch, mismatch position in the codon, and tRNA type. Particularly frequent errors were recognition of the codons GGA, GAU, or GAC instead of GAA by tRNAGlu, and recognition of the codons CGC and CAA instead of CAC by tRNAHis. The authors suggest that the proofreading mechanism in protein synthesis may have evolved in response to frequent errors from low-d mismatches. — B.D.
Infectious diseases at the wildlife–livestock interface
Direct contact between a horse and a bat entangled in barbed wire in Queensland, Australia.
The transfer of infectious diseases between wildlife and livestock poses a threat to the livestock sector, the ecosystem, and human populations. However, global studies of the diseases and animals involved are lacking. Anke Wiethoelter et al. (pp. 9662–9667) analyzed almost 16,000 scientific publications on infectious diseases at the wildlife–livestock interface from 1912 to 2013 to characterize the types of diseases and animal species of greatest interest and to identify trends. The number of publications per year increased continuously with a shift in focus from parasitic diseases to viral diseases over time. Ten diseases, most of which are capable of afflicting humans, accounted for half of the published research. The most frequently studied interfaces were those between species that are closely related or that share the same habitat, with the interface between wild birds and poultry being cited most frequently worldwide. The relative importance of other interfaces varied among geographic regions. The authors suggest that the results could be used as a starting point for analyses of specific diseases, animal species, and geographic regions, potentially helping to identify effective surveillance and research strategies for controlling diseases at the wildlife–livestock interface. — B.D.
How cavefish survive food scarcity
Surface-dwelling (Top) and cave-dwelling (Bottom) Mexican cavefish.
Animals living in the relative obscurity of caves rely on decaying animal matter, flotsam from seasonal floods, and occasional bat droppings for nutrition, displaying efficient metabolism, resistance to starvation, and large reserves of body fat when food is available. To examine the genetic basis of such adaptations, Ariel Aspiras et al. (pp. 9668–9673) compared surface- and cave-dwelling forms of Mexican cavefish (Astyanax mexicanus), and found that cave dwellers had a larger appetite and more triglyceride fats than surface dwellers upon regular feeding, and lost half as much body weight as surface dwellers after two months of fasting. Analysis of genome sequences of surface- and cave-dwelling populations known to have evolved independently uncovered three changes in the signaling protein melanocortin 4 receptor (MC4R), linked to anorexia, in cave dwellers, suggesting that the receptor may be associated with overfeeding. One of these changes replaces the amino acid glycine with serine in the receptor, and the variant’s human counterpart has been linked to obesity. Cell-based assays revealed less efficient signaling by the cave dwellers’ variant MC4R, compared with the glycine-containing receptor. Genetic analysis suggested a role for other appetite-regulating genes in cave dwellers. Though a neighboring gene linked to MC4R could possibly underlie the observed effects, the authors suggest that the receptor is a strong candidate for natural selection in cavefish. — P.N.
Nocturnal redolence of petunias
Petunia flowers attract nocturnal pollinators like hawkmoths. Image courtesy of Kiley Riffell Photography.
The swan-white flowers of common garden petunias (Petunia hybrida cv. Mitchell) turn fragrant at night, luring nocturnal pollinators with a scent laden with volatile organic compounds. Because genes involved in the synthesis of the odorants and the switches that control them both contain sequences targeted by a circadian clock protein called LATE ELONGATED HYPOCOTYL (LHY), Myles Fenske et al. (pp. 9775–9780) tested the role of LHY in timing scent emission in petunias. Experiments in continuous light and dark conditions over 3 days revealed that the emission of four of the plant’s main odorants in the dark followed a circadian pattern, controlled by the protein P. hybrida LHY (PhLHY), the petunia equivalent of LHY. Altering PhLHY levels influenced the expression levels of other clock genes as well as genes implicated in floral odorant metabolism. Continuous PhLHY expression abolished floral odorant emission, whereas suppression of PhLHY shifted emission from late afternoon to morning. By restricting the expression of a master regulator and fragrance-associated genes to the evening, the clock protein helps increase the plant’s odds of pollination. Unraveling the genetic control of the timing and composition of floral fragrance might help engineer crops with scent emission patterns tailored to the routines and preferences of local pollinators, according to the authors. — P.N.