Significance
The protein α-synuclein (αS) forms neuronal aggregates in Parkinson’s disease. Its normal folding and assembly state are unsettled. Cross-linking αS in living neurons reveals a major soluble 60-kDa form sizing as a tetramer, but the specificity of the cross-linking has been questioned. Here, we find that certain amino acid substitutions in the conserved αS “KTKEGV” repeat motifs specifically abolish the tetrameric form, indicating that cross-linking of normal αS into tetramers is not artifactual and models of monomeric αS as the major normal form should be reconsidered. An independent method, Venus-YFP complementation, confirms the cross-linking data. Importantly, tetramer abrogation causes αS insolubility, inclusions, and neurotoxicity, suggesting that the adoption of a folded tetrameric form is indispensible for normal αS homeostasis.
Keywords: alpha-synuclein, multimer, tetramer, Parkinson's disease, neurotoxicity
Abstract
α-Synuclein (αS) is a highly abundant neuronal protein that aggregates into β-sheet–rich inclusions in Parkinson’s disease (PD). αS was long thought to occur as a natively unfolded monomer, but recent work suggests it also occurs normally in α-helix–rich tetramers and related multimers. To elucidate the fundamental relationship between αS multimers and monomers in living neurons, we performed systematic mutagenesis to abolish self-interactions and learn which structural determinants underlie native multimerization. Unexpectedly, tetramers/multimers still formed in cells expressing each of 14 sequential 10-residue deletions across the 140-residue polypeptide. We postulated compensatory effects among the six highly conserved and one to three additional αS repeat motifs (consensus: KTKEGV), consistent with αS and its homologs β- and γ-synuclein all forming tetramers while sharing only the repeats. Upon inserting in-register missense mutations into six or more αS repeats, certain mutations abolished tetramer formation, shown by intact-cell cross-linking and independently by fluorescent-protein complementation. For example, altered repeat motifs KLKEGV, KTKKGV, KTKEIV, or KTKEGW did not support tetramerization, indicating the importance of charged or small residues. When we expressed numerous different in-register repeat mutants in human neural cells, all multimer-abolishing but no multimer-neutral mutants caused frank neurotoxicity akin to the proapoptotic protein Bax. The multimer-abolishing variants became enriched in buffer-insoluble cell fractions and formed round cytoplasmic inclusions in primary cortical neurons. We conclude that the αS repeat motifs mediate physiological tetramerization, and perturbing them causes PD-like neurotoxicity. Moreover, the mutants we describe are valuable tools for studying normal and pathological properties of αS and screening for tetramer-stabilizing therapeutics.
Alpha-synuclein (αS) is an abundant neuronal protein that may function in synaptic vesicle trafficking (1–3). A portion of αS forms insoluble neuronal aggregates (Lewy bodies and neurites) during aging and in some neurodegenerative diseases, such as PD, dementia with Lewy bodies, multiple system atrophy, and Alzheimer’s disease (4). Genetic evidence increasingly implicates αS dyshomeostasis as a cause of PD, via missense mutations, copy number variants, or up-regulated expression (5–8). Two decades of research suggest that αS occurs principally as a natively unfolded monomer in neurons, and this assumption is commonly stated in articles and reviews of αS. In the last 4 y, unexpected findings from our (9–11) and other (3, 12–14) laboratories have provided evidence that αS forms physiological multimers, principally tetramers, that have α-helical conformation. Several criteria indicate that the tetramers/multimers present in intact, healthy neurons are physiological and we thus refer to these species as “multimers” to emphasize their distinction from pathological, β-sheet–rich aggregates traditionally called oligomers.
We performed chemical cross-linking of endogenous αS in intact cells, including primary neurons, that support the existence of soluble, low-n multimers (10). We trapped abundant αS in ∼60-kDa species, the size of four monomers (4 × 14,502 Da = 58,010 Da). Known monomeric and multimeric proteins were trapped by cross-linking in their expected monomeric or multimeric states. Multiple controls ruled out artifactual induction of multimers, abnormally migrating monomers or hetero-multimers (10). If cells were lysed before cross-linking, multimers appeared depolymerized unless cross-linking was done at high protein concentration, suggesting that molecular crowding may stabilize native tetramers/multimers. Several endogenous multimeric proteins did not show this cell lysis sensitivity, except αS homolog β-synuclein (βS). These findings suggested to us that a dynamic intracellular population of metastable αS multimers and monomers coexists normally, and others have proposed similar models of dynamic/metastable tetramers (12), multimers (15), or “conformers” that may represent multimers (16). Analogous dynamic equilibria have been proposed for well-known tetrameric proteins such as hemoglobin (17) and p53 (18). An older study, from when αS was assumed to be solely monomeric, showed cross-linked low-n αS multimers in intact cells, and the authors discussed the possibility that synuclein normally exists in cells as low molecular mass oligomers, primarily dimers and trimers, that are in equilibrium with monomeric synuclein and are stabilized by experimentally induced covalent association (19).
After our initial report (9), two laboratories published data supporting the earlier model of αS existing as natively unfolded monomers. These studies either did not use cross-linking of intact cells (20) or considered any cross-linked multimeric αS to be nonspecific (21). One of the reports suggested that a small and variable portion of brain αS was helically folded (20) and, subsequently, this group reported multimers (including abundant tetramers) on membranes/vesicles (14). The unresolved but central debate about the existence and biological relevance of native αS tetramers/multimers led us to perform systematic mutagenesis of αS in intact neural cells to learn whether there are discrete sequence requirements that could mediate tetramer/multimer formation and, if so, what the cellular consequences of fully depolymerizing αS might be. To quantify multimers in these experiments, we used two independent intact-cell methods: cell-penetrant covalent cross-linking and fluorescent protein complementation. We hypothesized that if mutating αS residues abolished multimers and led to detection of only monomers in cells, then αS multimers are not induced by the two intact-cell detection methods we use. Moreover, mutants that make αS solely monomeric might be toxic to cells and lead to αS-rich inclusions. Finally, solely monomeric αS mutants could be useful tools for future studies to identify whether tetramers or monomers (or both) are the functional form of αS in neurons.
Results
Native Multimers Are the Result of Overall Synuclein Structure.
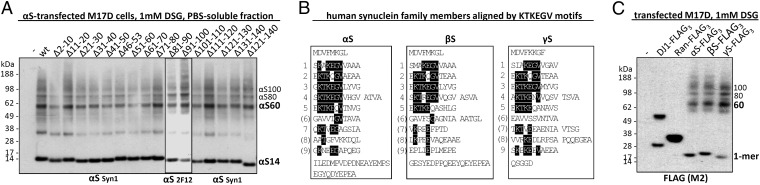
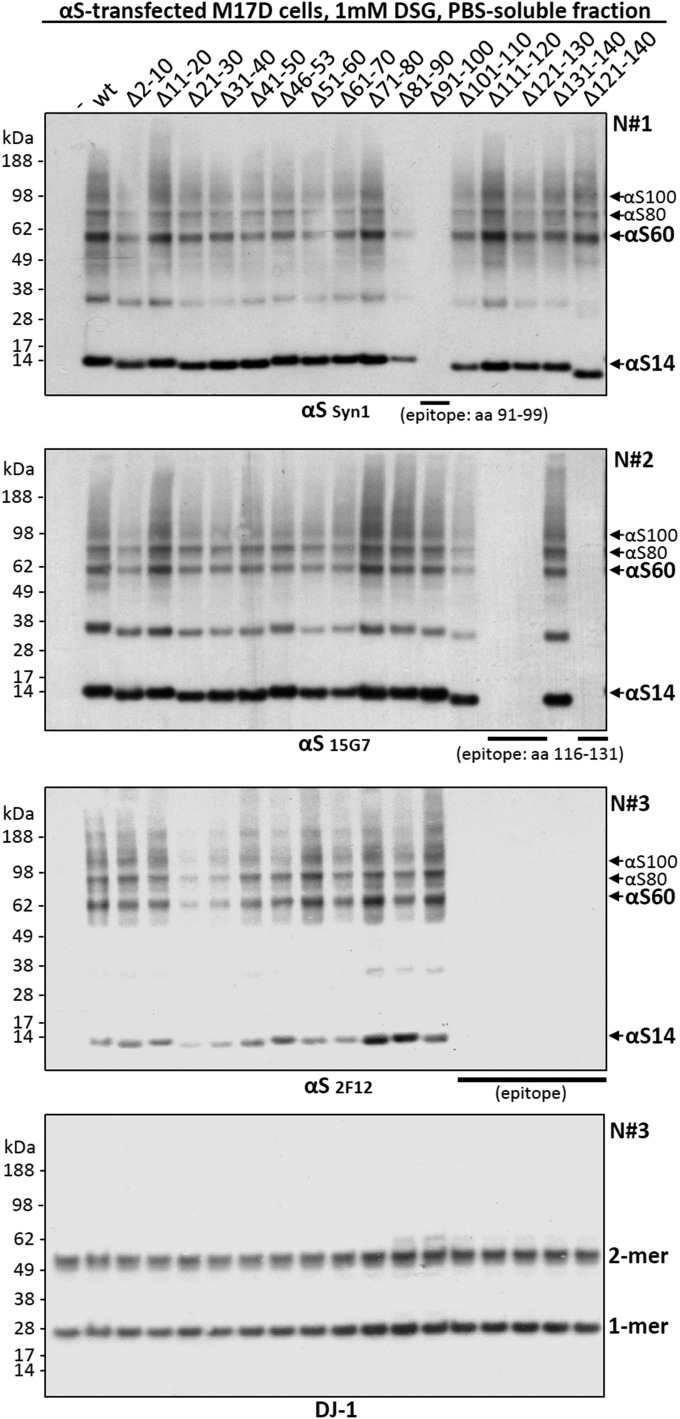
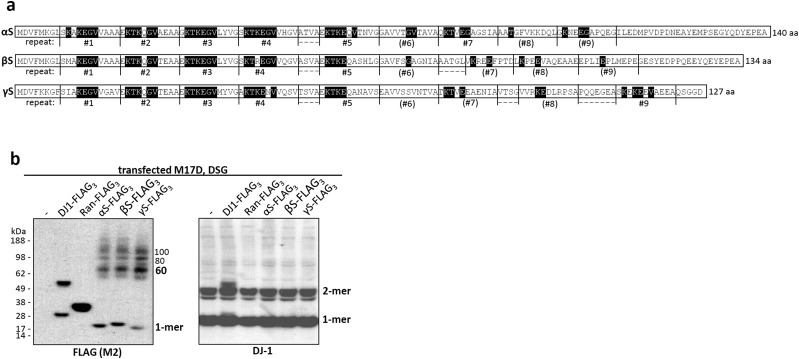
We previously reported that the cross-linking pattern of αS in living cells using cell-permeant cross-linkers such as disuccinimidyl glutarate (DSG) or dithiobis[succinimidyl propionate] (DSP) differed from the diffusion-controlled cross-linking pattern (mono- > di- > tri- > tetramers) of recombinant monomeric αS in vitro, in that it strongly favored tetramers (10). On αS overexpression, cross-linking produced a similar pattern but often with additional dimers and high-molecular-weight smears that rarely occur with just endogenous αS (figures 3a and d and 6d in ref. 10). As an unbiased approach to identify regions required for multimerization, we now sought to apply DSG cross-linking to analyze a complete set of sequential 10-amino acid (aa) deletions (9 aa for Δ2–10) across the 140-residue protein (Fig. 1A and Fig. S1 for more details and dimeric PARK7 protein DJ-1 as a control). In addition, we tested a construct lacking the amino acids 46–53, where four PD-linked point mutations cluster, and one lacking amino acids 121–140, a truncation that enhanced pathological aggregation in a PD mouse model (22). To our surprise, all 16 deletion mutants were cross-linked in live human M17D neural cells to the familiar multimeric pattern we had reported (10): 60-, 80-, and 100-kDa species (αS60, αS80, αS100; slight differences in gel migration due to deletions) (Fig. 1A). Although relatively subtle effects on multimer levels occurred with some deletion mutants, we found no key regions indispensible for multimerization. While initially surprising, this finding was consistent with our evidence that βS forms multimers in a pattern similar to that of αS (10). Despite their generally different amino acid sequences, the members of the synuclein family share conserved repeat motifs (consensus: KTKEGV) that occur at least six times in αS (nine if less conserved repeats are included), five times in βS, and six times in γ-synuclein (γS) (Fig. 2B and Fig. S2A). Accordingly, we next tested the relative multimerization propensities of all three homologs by expression with C-terminal FLAG3 tags in human M17D neuroblastoma cells. Upon cross-linking intact cells, anti-FLAG Western blots (WBs) of the cytosols revealed that each synuclein has a similar pattern of intracellular multimers (Fig. 2C and Fig. S2B for cross-linking/loading control DJ-1).
Fig. 1.
Intact-cell cross-linking analysis of αS deletion mutants and FLAG-tagged αS, βS, and γS. (A) Post-20,000g cytosols of cross-linked (PBS/1 mM DSG) M17D cells transfected with empty vector (-), wt αS, or αS deletion mutants. To detect all αS variants by WB, mAb 2F12 was used in addition to mAb Syn1 (epitope: amino acids 91–100). (B) Sequences of αS, βS, and γS aligned by their KTKEGV repeats. (amino acids that match this motif are shaded in black). KTKTEGV repeats are labeled 1–9, motifs with less than 4 aa conservation are in parentheses. (C) Triple-FLAG (FLAG3)-variants of all three human synucleins, the control proteins DJ-1 and Ras-related nuclear protein (Ran), and empty vector (-) analyzed analogous to A. Shown is a WB for the FLAG3-tag (M2 antibody). All WBs represent at least four independent experiments.
Fig. S1.
Intact-cell cross-linking analysis of αS deletion mutants. M17D neuroblastoma cells were transfected with empty vector (-), wt αS, or the indicated αS deletion mutants. The cell-permeant cross-linker DSG was added at a 1 mM final concentration to live cells in PBS followed by 30′ incubation at 37 °C and quenching (50 mM Tris). Post-20,000g cytosols were prepared after lysing cells in PBS plus protease inhibitors by sonication. To detect all variants by WB, we had to use three different mAbs to αS: Syn1 (Top), 15G7 (Upper Middle) and 2F12 (Lower Middle). The three WBs represent independent experiments done on different days (N#1, N#2, N#3). Equal loading as well as equal and specific cross-linking was confirmed by trapping the endogenous cytosolic protein DJ-1 partly in its native dimeric form (Bottom, run with N#3).
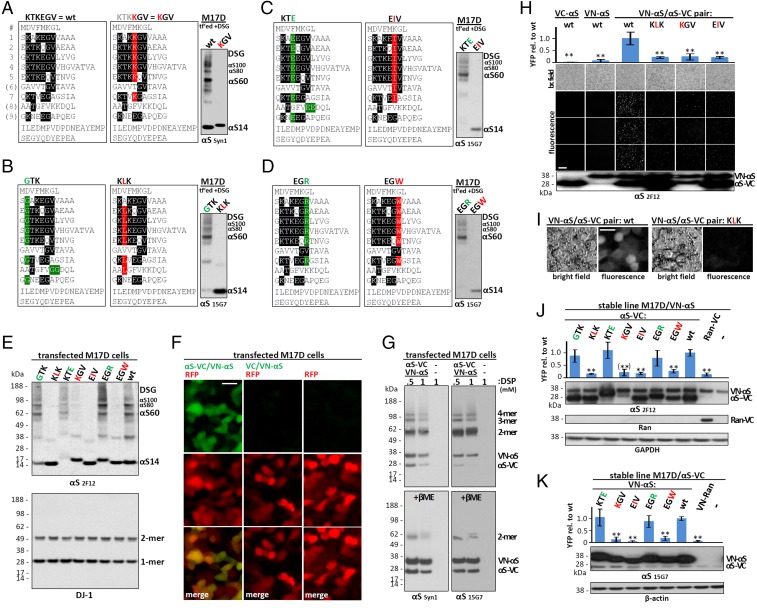
Fig. 2.
Multimerization propensity of αS repeat motif mutants. (A–D) Schematics of wt αS and repeat-motif mutants by aligning amino acid sequences via the repeat consensus sequence KTKEGV. Far right: WBs of the respective DSG–cross-linked M17D transfectants (cytosols; mAbs Syn1 and 15G7); WBs represent at least four independent experiments. (E) DSG–cross-linked M17D transfectants (cytosols; mAb 2F12). Endogenous DJ-1 is the control for cross-linking and loading. (F) Fluorescence microscopy of M17D cells transfected with RFP (red) plus VN-αS/αS-VC, VN-αS/VC or empty vector (green), and merge below. (G) VN-αS/αS-VC in M17D cells cross-linked with 0.5 or 1 mM DSP, plus vector-only control (-); WBs of cytosols (mAbs Syn1 and 15G7). Top, DSP; Bottom, DSP+5% βME (reduction of crosslink). (H) Venus-YFP complementation: bright field (top row) and YFP fluorescence images; WB (mAb 2F12) at bottom. αS-VC alone, VN-αS alone, or the indicated complementation pairs expressed in M17D cells. Graph (Top) quantifies n = 3 independent experiments, 4 fields for each. (I) Venus-YFP complementation assay: higher magnification bright-field and YFP fluorescence images for wt and KLK complementation pairs. (J) Venus-YFP complementation by automated fluorescence plate reading. M17D/VN-αS stable cells were transiently transfected with wt αS-VC or the indicated KTKEGV mutants (or Ran-VC as negative control). (Top) YFP fluorescence relative to wt αS-VC (n = 6). (Bottom) Representative WBs (total lysates in PBS/1% TX-100) for αS (mAb 2F12) and loading control glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Note that low KGV expression prevented a definite conclusion, indicated by parentheses. (K) Analogous experiment to J using low-expressing M17D/αS-VC cells transfected with the indicated VN-tagged αS variants (n = 6). WBs, αS (mAb 15G7) and loading control β-actin. One-way ANOVA for all statistical analyses. Bars are means ± SD. Criteria for significance: *P < 0.05; **P < 0.01. (Scale bars: F and I, 10 μm; H, 100 μm.)
Fig. S2.
Partially conserved KTKEGV motifs are shared between all three human, multimer-forming synuclein homologs. (A) Amino acid sequences of human αS, βS, and γS emphasizing the conservation of an 11-aa repeat structure having the core consensus motif KTKEGV (amino acids that match this motif are shaded in black). KTKTEGV repeats are labeled 1–9, but motifs with less than 4 aa conservation are in parentheses. (B) Triple-FLAG (FLAG3) variants of all three human synucleins, the control proteins DJ-1 and Ran, and empty vector (-) were each expressed in M17D cells followed by intact-cell DSG-cross-linking of the cytosols. WB for the FLAG3-tag (M2 antibody; Left) and DJ-1 (Right). Presence of DJ-1 dimers (natively dimeric) and absence of multimeric bands for Ran (natively monomeric) confirm specificity of the FLAG-tagged multimers trapped by cross-linking. WBs are representative of four independent experiments.
Native Multimerization Arises from Conserved Repeat Motifs.
Based on the above findings, we asked whether the overall repeat structure (Fig. 1B), rather than specific regions, mediates αS multimerization. We postulated that mutations simultaneously introduced into six or seven KTKEGV repeats might disrupt multimers, and our approach was to “reverse” the nature of the respective residue. A first example was to change the negatively charged amino acid glutamate (E) to positively charged lysine (K). This mutant is compared with wild-type (wt) αS in Fig. 2A by aligning amino acid sequences via the repeat motifs. DSG cross-linking of live M17D cells expressing αS with the altered sequence KTKKGV six times (termed “KGV”) revealed the complete absence of both αS60 tetramers and related αS80 and αS100 multimers (Fig. 2A, WB on the right). We thus created an extensive set of αS repeat-motif mutants, continuing our strategy, e.g., by altering charge (KTKEGV→KTEEGV; Fig. 2C, Left) or size (KTKEGV→KTKEIV; Fig. 2C, Middle). Using live-cell DSG cross-linking, we established that KLK (T→L substitution in all relevant repeats: 2–5, 7, 8), but not GTK (K→G in repeats 1–5, 7, 9 plus two additional changes in 8) could abolish cytosolic tetramers (Fig. 2B, WB). Similarly, EIV (repeats 1–7) but not KTE (repeats 1–5, 7, 9, plus two changes in 8), and EGW but not EGR (both in repeats 1–5 and 7) abrogated tetramer formation (Fig. 2 C and D) (mAb 15G7 can still detect all of these mutants). Fig. 2E summarizes these intact-cell cross-linking results by comparing all variants to wt αS in one gel (WB with mAb 2F12, which also detects all variants). As a second intact-cell method, Venus-YFP complementation has recently been shown to detect native, functional αS multimers in primary neurons (3). The assay is based on the split Venus-YFP protein whose two halves (VN and VC) have negligible affinity (3) but reassemble when fused to proteins that interact, producing YFP fluorescence in living cells. We found that coexpression of fusion proteins VN-αS and αS-VC led to strong complementation that colocalized with the soluble, cytoplasmic red fluorescent protein (RFP), whereas signals were weak for VN-αS and VC alone (Fig. 2F), excluding intrinsic affinity of VN and VC, as expected from prior work (23). When we coexpressed and cross-linked VN-αS and αS-VC in intact cells, we found a pattern consistent with monomers, dimers, trimers, and tetramers (Fig. 2G, Upper, DSP; Lower, DSP followed by βME reductive cleavage of the cross-linker). Compared with untagged αS, dimers were more prominent, suggesting that steric hindrance by the tags prevents further assembly or efficient cross-linking. We examined wt αS, KLK, KGV, and EIV complementation pairs with live-cell microscopy (Fig. 2H, four images below graph). Complementation was strong for wt αS but weak for the three tetramer-abolishing mutants (Fig. 2H, graph), despite high expression levels (vs. wt) for KLK and EIV and modestly reduced levels for KGV (Fig. 2H, Bottom). Fig. 2I shows representative bright field and fluorescent images for wt and KLK complementation pairs at a higher magnification. To achieve better consistency (more even expression) for quantitative analyses, we next expressed VC-tagged versions of all αS variants tested by cross-linking (Fig. 2 A–E) in a cell line that stably expressed VN-αS wt (Fig. 2J) and quantified YFP fluorescence with an automated plate reader. Strong fluorescence occurred for all those KTKEGV mutants shown by cross-linking to preserve tetramers, whereas for all of the tetramer-abolishing mutants, complementation was reduced to the level of the control monomeric protein Ran-VC (Fig. 2J, graph). These highly significant differences were observed despite robust expression (Fig. 2J, top WB image), except for KGV-VC, which showed a trend toward lower expression, precluding definite conclusions about KGV in this experimental setting. We therefore expressed VN-tagged KGV and, for comparison, VN-tagged wt, KTE, EIV, EGR, and EGW in an M17D cell line stably expressing low levels of wt αS-VC (Fig. 2K). Even with excess VN-tagged variants, we again found strong YFP complementation only for the multimer-sparing KTE, EGR, and wt cells, whereas EIV, EGW, and KGV (well-expressed when VN-tagged) produced insignificant fluorescence, similar to VN-Ran.
Loss of αS Tetramers Is Neurotoxic and Causes Inclusions.
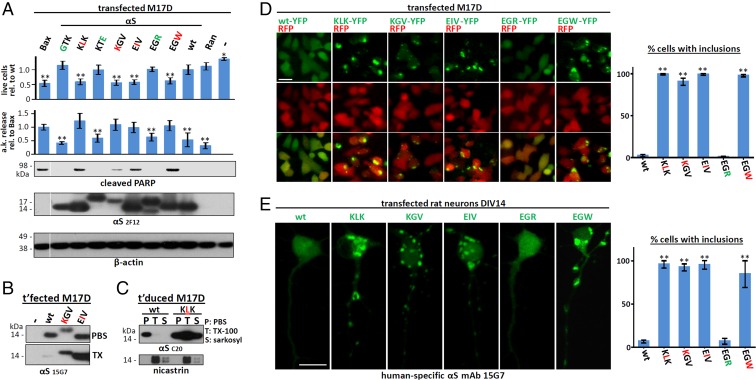
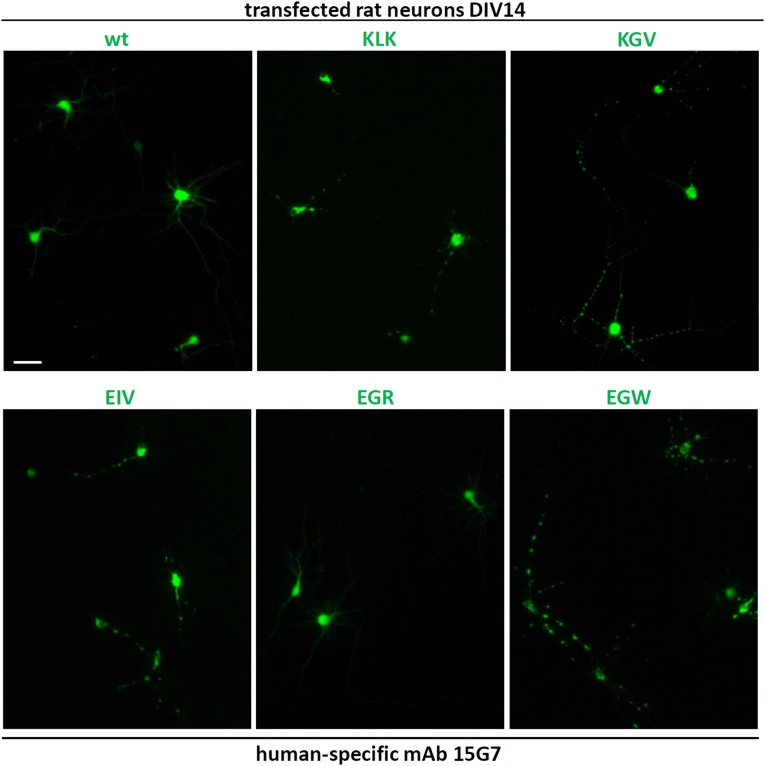
Having identified αS residues that abolish native tetramers in intact cells with two independent methods, we asked how this shift to monomers affected neuronal health. We correlated tetramer presence or absence with neurotoxicity by using a range of assays. When expressed in human M17D neural cells, all tetramer-abolishing KTKEGV variants were cytotoxic compared with wt αS by Trypan blue exclusion assay (Fig. 3A, top graph). Trypan blue penetration due to tetramer-lacking mutants mirrored that of the major proapoptotic factor Bcl2-associated X protein (Bax). Those KTKEGV mutants that did not reduce tetramers by intact-cell cross-linking and fluorescence complementation (GTK, KTE, EGR) were indistinguishable from wt-expressing cells, providing a key negative control throughout these mutagenesis experiments. A second assay for neurotoxicity, adenylate kinase release, showed significantly higher cytotoxicity for each of the tetramer-abolishing mutants, again in the same range as the proapoptotic Bax protein, whereas all tetramer-sparing variants produced little or no toxicity, i.e., were similar to both wt αS and the negative control protein, Ran (Fig. 3A, second graph). As a third indicator of cytotoxicity, we assayed Poly(ADP-ribose) polymerase (PARP) cleavage as a marker for activated apoptosis; PARP cleavage was clearly detectable only with tetramer-abolishing mutants, not tetramer-sparing mutants, producing a binary pattern (Fig. 3A, third row). The similar or lower (for KGV) expression levels of all tetramer-abolishing mutants relative to wt αS (using β-actin as a loading control) ruled out that these results were based on expression levels (Fig. 3A, lowest two rows). We then asked whether lack of tetramerization is associated with decreased αS solubility in cells and found substantially increased αS levels (vs. wt) in PBS-insoluble extracts, as seen with KGV and EIV in the sequential TX-100 fraction (1% TX-100 in PBS) of transiently transfected M17D cells (Fig. 3B) and with KLK in both the TX-100 and the sequential sarkosyl (2% sarkosyl in PBS) fractions of stably transduced M17D cells (Fig. 3C). Strikingly, the absence of tetramers and reduced aqueous solubility of total αS was accompanied by formation of round, dense, cytoplasmic αS inclusions, as seen with YFP-tagged tetramer-abolishing variants KLK, KGV, EIV, and EGW (inclusions in >90% of cells), but not tetramer-sparing EGR or wt αS (small inclusions in <6% of cells) (Fig. 3D). This highly consistent difference was reproduced with untagged αS constructs in primary rat neurons, where the tetramer-abolishing variants (but not wt or EGR) readily led to round inclusions in the somata and, strikingly, altered neurites (Fig. 3E). Whereas wt αS and EGR had a smooth, diffuse cytoplasmic distribution, the KLK, KGV, EIV, and EGW variants showed multiple round inclusions of varying size in the somata and an abnormal “beaded” neurite pattern due to varicosities (Fig. 3E; quantifications on the right). Fig. S3 shows further examples for wt αS and the five variants in a wider field with lower magnification.
Fig. 3.
Cytotoxicity and inclusion formation of αS repeat-motif mutants. (A) Cytotoxicity assays: Trypan-blue exclusion for live-cell count (n = 18) relative to wt αS. Toxilight assay for adenylate-kinase (a.k.) release relative to Bax (n = 12). WB for cleaved PARP (n = 6). M17D cells were transiently transfected as indicated or mock (-). WBs shown are for cleaved PARP, αS (2F12), and β-actin (total lysates in PBS/1% TX-100). Membrane cut once, as indicated by white line. (B) WB for αS (15G7) in the PBS- or TX-100–soluble fractions of the indicated M17D transfectants; representative of three independent experiments. (C) WB for αS (15G7) in the PBS-, 1% TX-100– or 2% sarkosyl-soluble fractions of M17D cells stably transduced with wt or KLK αS; representative of three independent experiments. (D, Left) Fluorescence microscopy of M17D cells transfected with RFP and the indicated αS variants as YFP fusion proteins. (Right) Percentage of cells with αS inclusions was counted in a blinded fashion (n = 3; 100 cells each). (E, Left) Fluorescence microscopy of DIV14 rat primary neurons transfected with the indicated untagged αS variants: human-specific αS mAb 15G7. (Scale bar: 20 μm.) (Right) Percentage of cells with inclusions was counted in a blinded fashion (n = 3; 100 cells each). Note that differences in morphology reflect cell-to-cell variability, not differences caused by the respective variants. One-way ANOVA for all statistical analyses. Bars are means ± SD. Criteria for significance: *P < 0.05; **P < 0.01.
Fig. S3.
Fluorescence microscopy of DIV14 rat primary neurons transfected with the indicated untagged αS variants. Human-specific αS mAb 15G7 was used. Lower magnification images of the data shown in Fig. 3E. Three to four transfected cells are shown for each condition; note that because of the application of human-specific mAb 15G7, only transfected human αS is visible, and transfection efficiency was relatively low. (Scale bar: 50 μm.)
Discussion
A common principle in molecular biology is to test the relevance of protein characteristics and activities by introducing defined point mutations. If one or a minimal set of such mutations abolishes a proposed property, the property is commonly regarded as biologically true and specific. An example is the catalytic subunit of γ-secretase, presenilin: Proteolysis carried out by a polytopic transmembrane protein lacking major cytosolic or luminal stretches was considered highly unlikely. This opinion changed when mutagenesis showed two intramembrane aspartates in presenilin to be absolutely required for γ-secretase cleavage activity (24). Analogously, we demonstrate here that a set of 6–7 defined amino acid substitutions in the 140-aa αS protein can fully abolish low-n multimers (principally tetramers) of αS in intact cells. The resulting proteins are monomeric in the cytosol, as judged by two independent intact-cell methods, and they accumulate to some extent in buffer-insoluble fractions, form abnormal cytoplasmic inclusions, and are associated with frank neurotoxicity. These highly consistent findings provide key validation of the recent and still controversial concept of native αS multimerization in cells (3, 9, 10, 12–15). They also support the physiological importance of native, cytosolic αS multimers in cells: Without normal quaternary assembly, αS becomes a neurotoxic protein. Despite the findings of many in vitro studies, αS is not particularly aggregation-prone in the context of a whole organism: Only a small percentage of humans develop αS aggregates in a small fraction of their αS-expressing neurons, and this process usually occurs later in life in relatively infrequent (considering all humans) synucleinopathies such as PD and dementia with Lewy bodies. Based on the findings of this study, we propose that these neurodegenerative diseases are linked in part to a chronic inability of neurons to properly assemble and maintain physiological αS multimers.
We first tried to disrupt multimerization by eliminating 10-aa stretches (we could not eliminate the N-terminal methionine in intact cells) throughout αS. For each such modification, cytosolic 60-kDa αS tetramers and related 80- and 100-kDa multimers (10) could still be trapped. Thus, we conclude that no single amino acid is indispensible for multimer formation. Instead, these results suggest redundancy that compensates for the loss of a 10- or even a 20-aa stretch (121–140). Notably, even eliminating amino acids 41–50, amino acids 51–60, or amino acids 46–53 still permitted αS60/80/100 formation. This region harbors most known familial PD-causing mutations in αS and has been proposed to form a loop in the protein (25), a property that may be taken over by neighboring sequences when this stretch is disrupted. The likelihood that multimerization propensity of αS results from overall structure is further supported by direct comparison of αS, βS, and γS. To detect all three homologs on the same WB, we fused them to C-terminal FLAG3 tags and trapped 60-, 80-, and 100-kDa multimers to a similar degree as FLAG3-tagged DJ-1 in its physiological dimeric form, whereas monomeric Ran-FLAG3 remained monomeric (Fig. 1C). This finding is of particular interest, as only αS is thought of as an “aggregation-prone” protein, further indicating that the cytosolic multimers cross-linked in intact cells are distinct from pathological oligomers and are instead a feature of normal synuclein biology. Our findings agree with trapping both αS and βS at tetrameric (10) and a report of physiological γS tetramerization (26). αS (140 aa), βS (134 aa), and γS (127 aa) share general protein structure, and their N-terminal halves containing the repeat motifs are considerably conserved, although the exact sequences within the 11-aa repeats vary, as do the positions and lengths of intervening sequences not assigned to a repeat (Fig. 1B). In all three homologs, repeats 1–5 are highly conserved (four or more aa of KTKEGV are identical), and additional conserved repeat motifs occur in αS (repeat 7) and γS (repeat 9). The length of the C terminus after the last repeat is 28 aa in αS, 19 in βS, and 5 aa in γS, and as all three homologs multimerize to a similar extent, we assume the C terminus is unlikely to affect multimerization propensity.
Both of these broad findings—the inability to disrupt multimers by deletions and similar multimerization of all three synuclein homologs—led us to focus on the partially conserved αS repeats. We expressed in-register missense mutations in as many as six or seven of the imperfect repeat motifs. Among the seven different amino acid substitutions we chose, four abolished αS multimers: KTKEGV→KLKEGV, KTKKGV, KTKEIV, or KTKEGW. Neutral mutations were GTKEGV, KTEEGV, and KTKEGR. Strikingly, all four tetramer-abolishing variants caused signficant cytoxicity in neural cells by three independent assays and led to αS-immunoreactive cytoplasmic inclusions and abnormal αS distribution in neurites. Conversely, the three repeat-motif mutations that preserved tetramers/multimers caused neither cytotoxicity nor abnormal αS patterns. The correlation between tetramer abolition and neurotoxicity in this dataset provides strong support for our hypothesis that αS tetramers/multimers are both physiological and neuroprotective. Accordingly, we postulate that shifts away from a normal complement of cytosolic tetramers/multimers are adverse to neuronal health. In this regard, a repeat-motif mutant KTKEGV→KKKEGV—similar to our “KLK” mutant—was recently reported to prevent normal αS-YFP complementation in neurons and abrogate the proper tethering of synaptic vesicles by αS (3), consistent with our data. And an earlier publication (27) studied a KLK-like αS variant and concluded that general membrane binding was elevated, consistent with our sequential extraction data for KLK (Fig. 3C).
Interestingly, despite the apparent aggregation propensity of the four tetramer-abolishing mutants (Fig. 3), they produced low levels of YFP complementation compared with wt αS, suggesting that the complementation assay (23) might favor the detection of physiological α-helical multimers, not pathological β-sheet–rich αS aggregates, consistent with a recent YFP study of physiological αS in intact neurons by Wang et al. (3). Alternatively, αS in these inclusions may not be fibrillar, but instead bound to other subcellular components such as vesicles/membranes. Secondary events like impaired vesicle trafficking could then contribute to decreased cell viability, as observed in patient-derived neurons (28). Although membrane association of αS is considered a physiological aspect of αS biology (29, 30) and can be associated with decreased amyloid formation (31), it has also been shown to trigger pathological aggregation under certain conditions, due to increased local αS concentration on membranes (32). In earlier work, we showed that purified tetrameric αS is aggregation-resistant (9), and others subsequently proposed that tetrameric αS is a safe “storage form” (15). Additional work should focus on the identification of cellular factors involved in the formation and stabilization of αS tetramers/multimers and place multimerization of this abundant neuronal protein into a functional context. Is tetrameric/multimeric αS the active form, as suggested by in situ vesicle tafficking experiments (3), or is it an inert storage form, or does αS function derive from its ability to transition between different assembly forms in cells? In any event, our mutagenesis data strongly imply that cells tightly regulate the homeostatic equlibrium between αS monomeric forms and tetramers/related multimers.
Materials and Methods
Materials were purchased from Invitrogen unless stated otherwise.
Cell Culture, Transfection, and Primary Rat Neurons.
Human BE (2)-M17 neuroblastoma cells (called M17D, ATCC number CRL-2267) were cultured as published (10) and Lipofectamine 2000 transfected according to the manufacturer’s instructions (unsupplemented neurobasal medium instead of Optimem for primary neurons). Monoclonal cell lines M17D/VN-αS and M17D/αS-VC were generated by transfection of M17D cells with pcDNA3/VN-αS or pcDNA3/αS-VC, followed by G418 selection for clones. Primary neurons were cultured from embryonic day 18 Sprague–Dawley rats (Charles River) as described (10), acquired under protocol number 05022, approved by the appropriate Institutional Animal Care and Use Committee, the Harvard Medical Area Standing Committee on Animals.
cDNA Cloning.
Deletion and repeat-motive mutant constructs were synthesized as GeneArt Strings DNA fragments (GeneArt/Life Technologies) and inserted into pcDNA4/TO/myc-His A (pcDNA4) with the In-Fusion HD Cloning Kit (Clontech). FLAG3-tagged αS (10) analogous βS and γS constructs were generated from cDNA libraries by using specific primers and inserted into pcDNA4. pcDNA3/VN-αS and pcDNA3/αS-VC were subcloned into pcDNA4 and αS cDNA was replaced by In-Fusion to generate tagged variants. The VC construct was amplified from pcDNA3/αS-VC with a forward primer adding a start codon. Full-length YFP was reconstituted from the VN and VC tags and cloned into pcDNA4; YFP-tagged constructs were subsequently created by insertion of cDNA upstream of the tag. pCAX/dsRed and pcDNA3.1/Bax were the kind gifts of T. Young-Pearse, Brigham and Women's Hospital, and M. LaVoie, Brigham and Women's Hospital, respectively.
Protein Biochemistry and Immunocytochemistry.
Cross-linking and immunoblotting have been described (10, 11). To sequential extractions (10), we added a 1-h 2% sarkosyl incubation of TX-100–insoluble pellets. Immunocytochemistry was performed as described (33), except that cells were rinsed twice with HBSS with divalent cations before fixing, and cells were imaged directly in the dish instead of after mounting.
Primary Antibodies.
We used mAbs Syn1 to αS (Clone 42; Becton-Dickinson), 2F12 to αS (10), 15G7 to αS (34), 71.1 to GAPDH (Sigma), M2 to the FLAG-tag (Sigma), and pAbs C20 to αS (Santa Cruz), ab8227 to β-actin (Abcam), D64E10 to cleaved PARP (Asp214) (Cell Signaling), anti-Ran (4462; Cell Signaling), and anti-DJ-1 (35).
Statistical Analyses.
We performed one-way ANOVA by using GraphPad Prism Version 6.05 following the program’s guidelines (Tukey’s multiple comparison’s test, calculation of adjusted P values, “repeated measures” correction where applicable). Normal distribution and similar variance were observed for all values. Graphs are means ± SD. Criteria for significance, routinely determined relative to wt αS: *P < 0.05; **P < 0.01. Sufficient experiments and replicates were analyzed to achieve statistical significance.
Cell Toxicity Assays.
The ToxiLight Nondestructive Cytotoxicity BioAssay Kit (Lonza) was used following the manufacturer’s directions to measure adenylate kinase release, signals from untransfected cells were subtracted for normalization. Cell suspensions were mixed 1:1 with 0.4% Trypan blue (Sigma) and counted on a TC10 automated cell counter (Bio-Rad).
YFP Complementation Assay and Microscopy.
Cell lines M17D/VN-αS and M17D/αS-VC were transfected for 40 h with VC- and VN-tagged plasmids, respectively, followed by fluorescence detection on Synergy H1 HybridReader (BioTek; excitation 505 nM, emission 535 nm). Alternatively, we coexpressed αS-VC (4 μg per 6-cm dish) and VN-αS plasmids (2 μg) for 40 h followed by bright field and fluorescence microscopy of live cells (AxioVert 200 microscope, AxioCam MRm camera, AxioVision Release 4.8.2; Zeiss). YFP images were collected by using a GFP/FITC filter cube. Blinded analyses were performed by assigning random numbers to dishes by one investigator before representative images were taken or cells (with or without inclusions) were counted by another investigator. Confocal images were obtained on a Zeiss LSM710 system.
Acknowledgments
We thank N. Exner and C. Haass (Munich) for mAb 15G7; T. F. Outeiro (Goettingen) and P. McLean (Jacksonville) for wt VN-αS and αS-VC plasmids; T. Young-Pearse for the RFP plasmid; M. LaVoie for the Bax plasmid; E. Luth, S. Nuber, and all other members of the D.S., LaVoie, and Young-Pearse laboratories in the Ann Romney Center for Neurologic Diseases for many helpful discussions. This work was supported by NIH Grant R01 NSNS083845 (to D.S.), a research grant from the Fidelity Biosciences Research Initiative (www.fidelitybiosciences.com) (to D.S.), and a research grant from the Michael J. Fox Foundation (https://www.michaeljfox.org) (to U.D.).
Footnotes
Conflict of interest statement: D.S. is a director and consultant to Prothena Biosciences.
This article is a PNAS Direct Submission.
See Commentary on page 9502.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505953112/-/DCSupplemental.
References
- 1.Chandra S, et al. Double-knockout mice for alpha- and beta-synucleins: Effect on synaptic functions. Proc Natl Acad Sci USA. 2004;101(41):14966–14971. doi: 10.1073/pnas.0406283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott D, Roy S. α-Synuclein inhibits intersynaptic vesicle mobility and maintains recycling-pool homeostasis. J Neurosci. 2012;32(30):10129–10135. doi: 10.1523/JNEUROSCI.0535-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, et al. α-synuclein multimers cluster synaptic vesicles and attenuate recycling. Curr Biol. 2014;24(19):2319–2326. doi: 10.1016/j.cub.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spillantini MG, et al. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 5.Krüger R, et al. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18(2):106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 6.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 7.Zarranz JJ, et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55(2):164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 8.Singleton AB, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 9.Bartels T, Choi JG, Selkoe DJ. α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477(7362):107–110. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dettmer U, Newman AJ, Luth ES, Bartels T, Selkoe D. In vivo cross-linking reveals principally oligomeric forms of α-synuclein and β-synuclein in neurons and non-neural cells. J Biol Chem. 2013;288(9):6371–6385. doi: 10.1074/jbc.M112.403311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman AJ, Selkoe D, Dettmer U. A new method for quantitative immunoblotting of endogenous α-synuclein. PLoS One. 2013;8(11):e81314. doi: 10.1371/journal.pone.0081314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, et al. A soluble α-synuclein construct forms a dynamic tetramer. Proc Natl Acad Sci USA. 2011;108(43):17797–17802. doi: 10.1073/pnas.1113260108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westphal CH, Chandra SS. Monomeric synucleins generate membrane curvature. J Biol Chem. 2013;288(3):1829–1840. doi: 10.1074/jbc.M112.418871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burré J, Sharma M, Südhof TC. α-Synuclein assembles into higher-order multimers upon membrane binding to promote SNARE complex formation. Proc Natl Acad Sci USA. 2014;111(40):E4274–E4283. doi: 10.1073/pnas.1416598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurry T, et al. The dynamic structure of α-synuclein multimers. J Am Chem Soc. 2013;135(10):3865–3872. doi: 10.1021/ja310518p. [DOI] [PubMed] [Google Scholar]
- 16.Gould N, et al. Evidence of native α-synuclein conformers in the human brain. J Biol Chem. 2014;289(11):7929–7934. doi: 10.1074/jbc.C113.538249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manning LR, et al. Human embryonic, fetal, and adult hemoglobins have different subunit interface strengths. Correlation with lifespan in the red cell. Protein Sci. 2007;16(8):1641–1658. doi: 10.1110/ps.072891007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaglia G, Guan Y, Shah JV, Lahav G. Activation and control of p53 tetramerization in individual living cells. Proc Natl Acad Sci USA. 2013;110(38):15497–15501. doi: 10.1073/pnas.1311126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole NB, et al. Lipid droplet binding and oligomerization properties of the Parkinson’s disease protein alpha-synuclein. J Biol Chem. 2002;277(8):6344–6352. doi: 10.1074/jbc.M108414200. [DOI] [PubMed] [Google Scholar]
- 20.Burré J, et al. Properties of native brain α-synuclein. Nature. 2013;498(7453):E4–E6; discussion E6–E7. doi: 10.1038/nature12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fauvet B, et al. α-Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J Biol Chem. 2012;287(19):15345–15364. doi: 10.1074/jbc.M111.318949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michell AW, et al. The effect of truncated human alpha-synuclein (1-120) on dopaminergic cells in a transgenic mouse model of Parkinson’s disease. Cell Transplant. 2007;16(5):461–474. doi: 10.3727/000000007783464911. [DOI] [PubMed] [Google Scholar]
- 23.Outeiro TF, et al. Formation of toxic oligomeric alpha-synuclein species in living cells. PloS One. 2008;3(4):e1867. doi: 10.1371/journal.pone.0001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfe MS, et al. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398(6727):513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 25.Kara E, et al. α-Synuclein mutations cluster around a putative protein loop. Neurosci Lett. 2013;546:67–70. doi: 10.1016/j.neulet.2013.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golebiewska U, Zurawsky C, Scarlata S. Defining the oligomerization state of γ-synuclein in solution and in cells. Biochemistry. 2014;53(2):293–299. doi: 10.1021/bi401479a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pranke IM, et al. α-Synuclein and ALPS motifs are membrane curvature sensors whose contrasting chemistry mediates selective vesicle binding. J Cell Biol. 2011;194(1):89–103. doi: 10.1083/jcb.201011118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung CY, et al. Identification and rescue of α-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013;342(6161):983–987. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273(16):9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 30.Burré J, Sharma M, Südhof TC. Definition of a molecular pathway mediating α-synuclein neurotoxicity. J Neurosci. 2015;35(13):5221–5232. doi: 10.1523/JNEUROSCI.4650-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fonseca-Ornelas L, et al. Small molecule-mediated stabilization of vesicle-associated helical α-synuclein inhibits pathogenic misfolding and aggregation. Nat Commun. 2014;5:5857. doi: 10.1038/ncomms6857. [DOI] [PubMed] [Google Scholar]
- 32.Galvagnion C, et al. Lipid vesicles trigger α-synuclein aggregation by stimulating primary nucleation. Nat Chem Biol. 2015;11(3):229–234. doi: 10.1038/nchembio.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dettmer U, et al. Transmembrane protein 147 (TMEM147) is a novel component of the Nicalin-NOMO protein complex. J Biol Chem. 2010;285(34):26174–26181. doi: 10.1074/jbc.M110.132548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kahle PJ, et al. Subcellular localization of wild-type and Parkinson’s disease-associated mutant alpha -synuclein in human and transgenic mouse brain. J Neurosci. 2000;20(17):6365–6373. doi: 10.1523/JNEUROSCI.20-17-06365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baulac S, LaVoie MJ, Strahle J, Schlossmacher MG, Xia W. Dimerization of Parkinson’s disease-causing DJ-1 and formation of high molecular weight complexes in human brain. Mol Cell Neurosci. 2004;27(3):236–246. doi: 10.1016/j.mcn.2004.06.014. [DOI] [PubMed] [Google Scholar]