Significance
Acquired thrombotic thrombocytopenic purpura (TTP) is primarily caused by autoantibodies that inhibit the ability of ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13) to proteolyze von Willebrand factor (VWF). The molecular mechanism of inhibition is not known. We used a hydrogen–deuterium exchange mass spectrometry (HX MS) method to determine at near–single-residue resolution the epitope of three monoclonal anti-ADAMTS13 autoantibodies isolated from TTP patients. Additional results show that the same autoantibody-binding epitope is responsible for ADAMTS13 binding to VWF to manage VWF proteolysis. These observations reveal the mechanism of autoimmune TTP and, together with the epitope determination, suggest a knowledge-based approach for finding a novel therapeutic.
Keywords: ADAMTS13, von Willebrand factor, thrombotic thrombocytopenic purpura, hydrogen exchange, autoimmunity
Abstract
Acquired thrombotic thrombocytopenic purpura (TTP), a thrombotic disorder that is fatal in almost all cases if not treated promptly, is primarily caused by IgG-type autoantibodies that inhibit the ability of the ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13) metalloprotease to cleave von Willebrand factor (VWF). Because the mechanism of autoantibody-mediated inhibition of ADAMTS13 activity is not known, the only effective therapy so far is repeated whole-body plasma exchange. We used hydrogen–deuterium exchange mass spectrometry (HX MS) to determine the ADAMTS13 binding epitope for three representative human monoclonal autoantibodies, isolated from TTP patients by phage display as tethered single-chain fragments of the variable regions (scFvs). All three scFvs bind the same conformationally discontinuous epitopic region on five small solvent-exposed loops in the spacer domain of ADAMTS13. The same epitopic region is also bound by most polyclonal IgG autoantibodies in 23 TTP patients that we tested. The ability of ADAMTS13 to proteolyze VWF is impaired by the binding of autoantibodies at the epitopic loops in the spacer domain, by the deletion of individual epitopic loops, and by some local mutations. Structural considerations and HX MS results rule out any disruptive structure change effect in the distant ADAMTS13 metalloprotease domain. Instead, it appears that the same ADAMTS13 loop segments that bind the autoantibodies are also responsible for correct binding to the VWF substrate. If so, the autoantibodies must prevent VWF proteolysis simply by physically blocking normal ADAMTS13 to VWF interaction. These results point to the mechanism for autoantibody action and an avenue for therapeutic intervention.
Acquired thrombotic thrombocytopenic purpura (TTP) is characterized by severe thrombocytopenia and microangiopathic hemolytic anemia, resulting from disseminated microvascular thrombosis. Patients with TTP may suffer from widespread organ damage, resulting in death if not aggressively treated (1, 2). In most patients, thrombotic microangiopathy is caused by IgG-type autoantibodies against the plasma metalloprotease ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13) (3–5). ADAMTS13 regulates the function of the multidomain von Willebrand factor (VWF) (∼600 to ∼20,000 kDa) by cleaving it at the central A2 domain to regulate platelet-induced blood clot formation (3, 4).
Immunological studies show that many acquired TTP patients with low plasma ADAMTS13 activity (less than 10%) harbor IgG autoantibodies that bind ADAMTS13 especially at the Cys-rich and/or the spacer domain (4, 6). Deletion of these domains or grouped mutations therein can eliminate the binding of polyclonal anti-ADAMTS13 IgGs derived from many patients (4, 7–11). However, the exact ADAMTS13 binding sites of these autoantibodies are not known. Current treatment involves daily whole-body plasma exchange to clear anti-ADAMTS13 autoantibodies while supplementing ADAMTS13 metalloprotease. Understanding of the autoantibody inhibitory mechanism might lead to a more targeted, less invasive, and less expensive intervention.
To define antibody–antigen interactions, a number of approaches have been developed. Each has its advantages and disadvantages. X-ray diffraction provides direct structural information that can define the binding interface at atomic resolution (12, 13) but this requires the difficult crystallization of the unperturbed complex. NMR spectroscopy offers a similar level of structural detail but is limited to relatively small proteins that can be expressed with 15N- and 13C-labeled residues and prepared in large amounts at high concentration. Peptide arrays can provide information on linear epitopes but with limited structural resolution and will often miss the more common discontinuous epitopes (14). A different approach, the measurement of protein hydrogen exchange together with a peptide separation approach incorporating mass spectrometry analysis [hydrogen–deuterium exchange mass spectrometry (HX MS)] can specify protein–protein interaction regions even for large protein complexes using only a tiny amount of experimental material (15–19). However, HX MS has so far been limited to fairly low structural resolution not sufficient for pinpointing binding sites. We exploited recent advances in HX MS methodology and data analysis to determine antibody-binding sites at near-amino acid resolution (20–23).
We isolated dozens of human monoclonal autoantibodies against ADAMTS13 from unrelated patients with autoimmune TTP and selected, by phage display, single-chain fragments of their tethered variable regions (scFvs). Three inhibitory scFvs, called 4–20, 4–16, and 3–1, were selected based on their high affinity and inhibitory potency and used in the mapping study. All three bind the same conformationally discontinuous epitope in the spacer domain of ADAMTS13, comprising surface residues in five small flexible loops. All five loops are necessary for high-affinity antibody binding. The same ADAMTS13 epitope is bound by most polyclonal IgGs in TTP patients. These findings define the anti-ADAMTS13 autoantibody epitope.
Additional results show that the same loop system is responsible for binding ADAMTS13 to VWF to organize correct VWF proteolysis. Evidently then, the binding of TTP autoantibodies to the epitope of ADAMTS13 must abrogate the proteolysis step by physically blocking it from binding to its VWF substrate. This information together with detailed knowledge of the epitopic site may help to guide the design of altered ADAMTS13 modules that would resist antibody binding but still allow VWF binding and proteolysis.
Results
Expression and Characterization of Anti-ADAMTS13 scFvs.
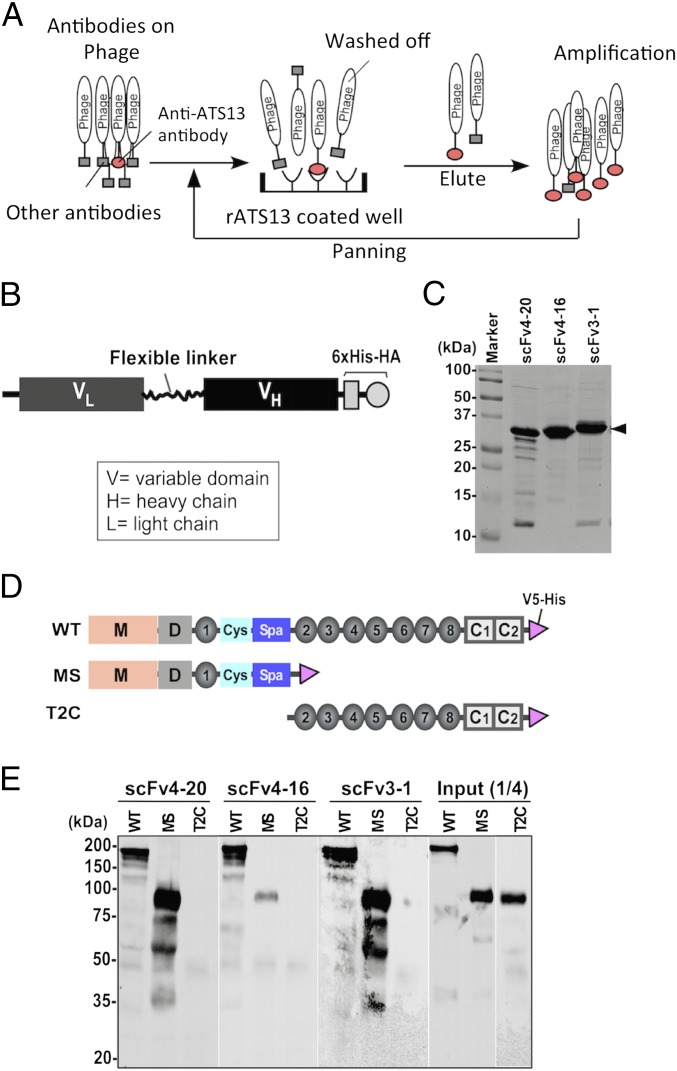
Dozens of monoclonal tethered scFvs of anti-ADAMTS13 antibodies derived from TTP patients were isolated by phage display (Fig. 1A), expressed in Top10′ Escherichia coli, and purified to near homogeneity by affinity chromatography. The scFv constructs comprise the variable regions of a light chain and a heavy chain connected by a flexible linker, and in addition a C-terminal 6×His and hemagglutinin (HA) epitope to facilitate scFv detection and purification (Fig. 1B). Purified scFvs exhibit molecular mass of ∼30 kDa under denaturing and reducing conditions (Fig. 1C). All scFvs were screened for their ability to inhibit the cleavage of the fluorogenic substrate rF-VWF73, a 73-aa fluorescein-labeled peptide that contains the A2 cleavage site of VWF and mimics its ADAMTS13 binding sites (24–26).
Fig. 1.
Isolation, expression, and characterization of anti-ADAMTS13 scFvs. (A) The protocol used for isolation of human anti-ADAMTS13 monoclonal antibodies (scFvs) from TTP patients by phage display (rATS is recombinant ADAMTS13). (B) The scFv expression construct comprises a variable light-chain region (VL), a flexible linker, a fragment of the heavy-chain variable region (VH), and a 6×His-HA (six histidine–hemagglutinin) tag at the C terminus. (C) SDS/PAGE [10% (wt/vol)] with Coomassie blue staining demonstrates the quality of purified scFvs 4–20, 4–16, and 3–1 (∼30 kDa; arrowhead). (D) Schematic representation of constructs used, namely wild-type ADAMTS13 (WT), MDTCS (MS), and TSP1 2–8 repeats plus two CUB domains (T2C), all with a V5-His epitope tag. (E) All three scFvs when bound to anti-HA resin pull down WT and MS but not T2C fragments (Western blotting with anti-V5). Input1/4 means that one-fourth of starting materials was loaded for the Western-blotting comparison.
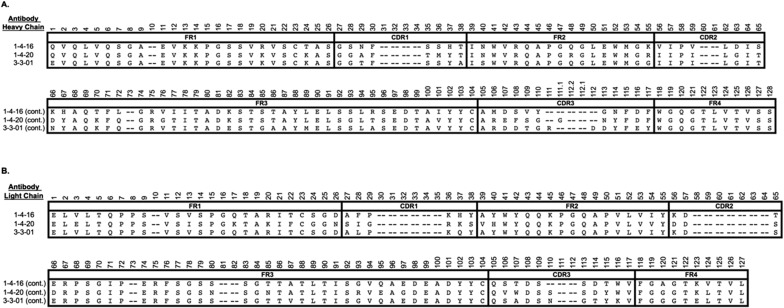
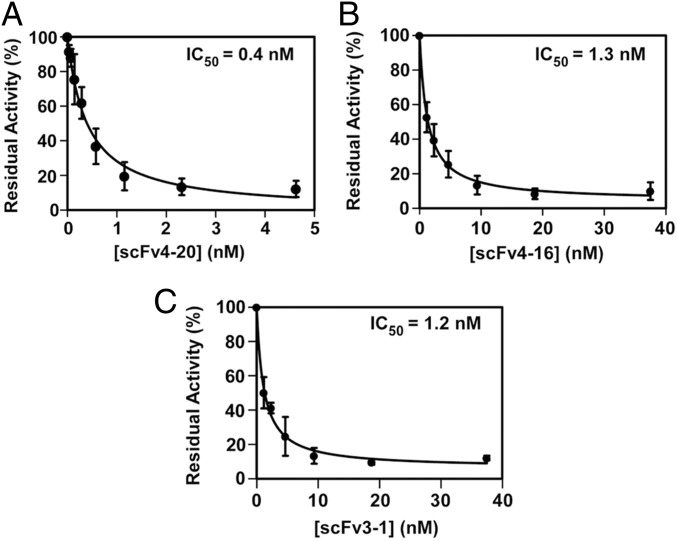
Three scFvs with excellent functional characteristics, 4–20, 4–16, and 3–1, were chosen for epitope characterization. They bind with high affinity to wild-type ADAMTS13 (WT) and to the MDTCS variant truncated after the spacer domain but not to the C-terminal fragment containing TSP1 repeats and CUB domains (T2C) (Fig. 1 D and E). They show high overall amino acid sequence identity (∼80%), but each one has distinct complementarity-determining regions (CDRs) in both heavy and light chains, particularly CDR3 (Fig. S1). They display potent autoantibody protease inhibition (half-maximal inhibitory concentrations of 0.4, 1.3, and 1.2 nM, respectively; Fig. 2 A–C). Injection of scFv4-20 into wild-type mice (CAST/Ei) causes ADAMTS13 deficiency and TTP-like syndrome after Shigatoxin-2 (Stx-2) or recombinant murine VWF (rmVWF) challenge.
Fig. S1.
Alignment of anti-ADAMTS13 heavy (A) and light chains (B) of the scFvs used here. Nucleotide and amino acid sequences of these and dozens of other patient-derived monoclonal TTP antibodies are being deposited in GenBank.
Fig. 2.
Inhibition of plasma ADAMTS13 activity by scFvs. Residual activity of plasma-derived ADAMTS13 (∼0.25 nM) after incubation with increasing concentrations of scFvs 4–20 (A), 4–16 (B), and 3–1 (C) was determined by cleavage of the fluorogenic substrate (rF-VWF73) (2 μM) as described in SI Materials and Methods. Data are the mean ± SD of three independent experiments.
High-Resolution HX MS Mapping of an Antigenic Epitope.
Our preliminary immunoassay results suggested that amino-terminal MDTCS domains but not the carboxyl-terminal TSP1 2–8 plus two CUB (T2C) fragment contains the epitopic binding site for all three scFvs. The MDTCS construct comprises five domains (Metalloprotease, Disintegrin, TSP1, Cys-rich, and Spacer). It is fully functional in catalyzing proteolysis of VWF under various conditions (27–32). Therefore, to reduce the complexity of data analysis, the MDTCS fragment, expressed in Drosophila Schneider 2 (S2) cells (SI Materials and Methods), was used for the epitope-mapping study.
In HX MS experiments, purified recombinant MDTCS was subjected to amide H-to-D exchange either when free in solution or when bound to one of the anti-ADAMTS13 scFvs immobilized on an Affi-gel 15 resin column. To determine HX rates, after timed exposures to D2O, MDTCS was washed off the Affi-gel/scFv column under conditions where HX is largely stopped (low pH and temperature), and then injected into an online analysis system (22). Passage through immobilized acid protease columns (pepsin or fungal protease XIII) rapidly proteolyzed MDTCS into many peptide fragments. The fragments were separated by HPLC followed by online mass spectrometry.
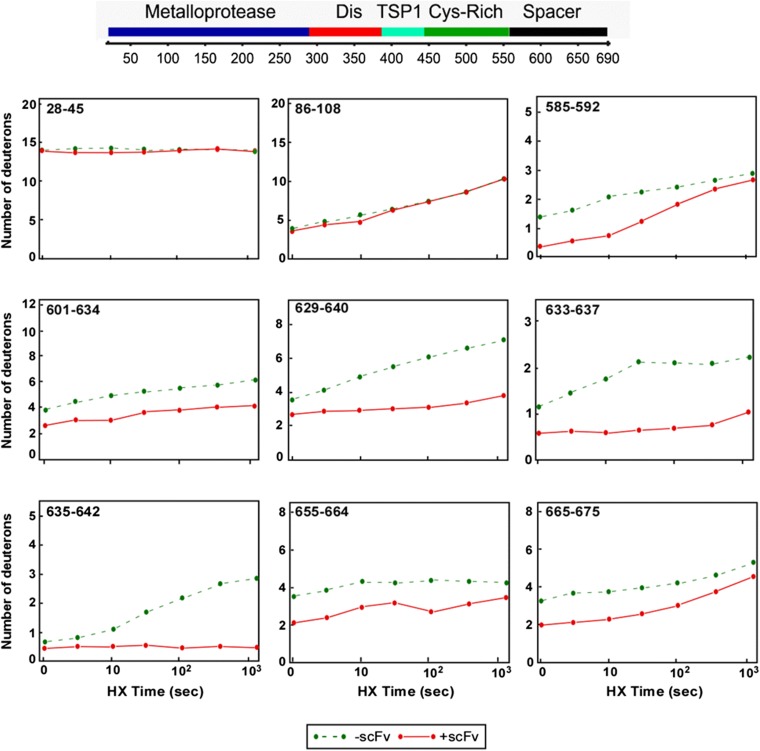
We obtained high-amplitude peptides, high sequence coverage, and low back exchange adequate for monitoring the metalloprotease domain (113 peptides, 73% coverage), the Cys-rich domain (22 peptides, 80% coverage), and the spacer domain (70 peptides, 84% coverage). Few useful peptides were obtained in the disintegrin domain and first TSP1 repeat due to heterogeneous glycosylation (33, 34). The isotopic envelope of each peptide was identified in the MS spectra and analyzed for carried D-label using the ExMS program (21). Typical kinetic HX curves for MDTCS peptides with and without antibody bound are shown in SI Materials and Methods (Fig. S2).
Fig. S2.
HX kinetic profiles of selected peptides of MDTCS fragment (out of 209) in the absence (green) and presence (red) of scFv4-20. HX protection was observed in peptides derived from the spacer domain of ADAMTS13 (e.g., 585–592, 601–634, 629–640, 633–637, and others), but not in other regions (e.g., 28–45, 86–108).
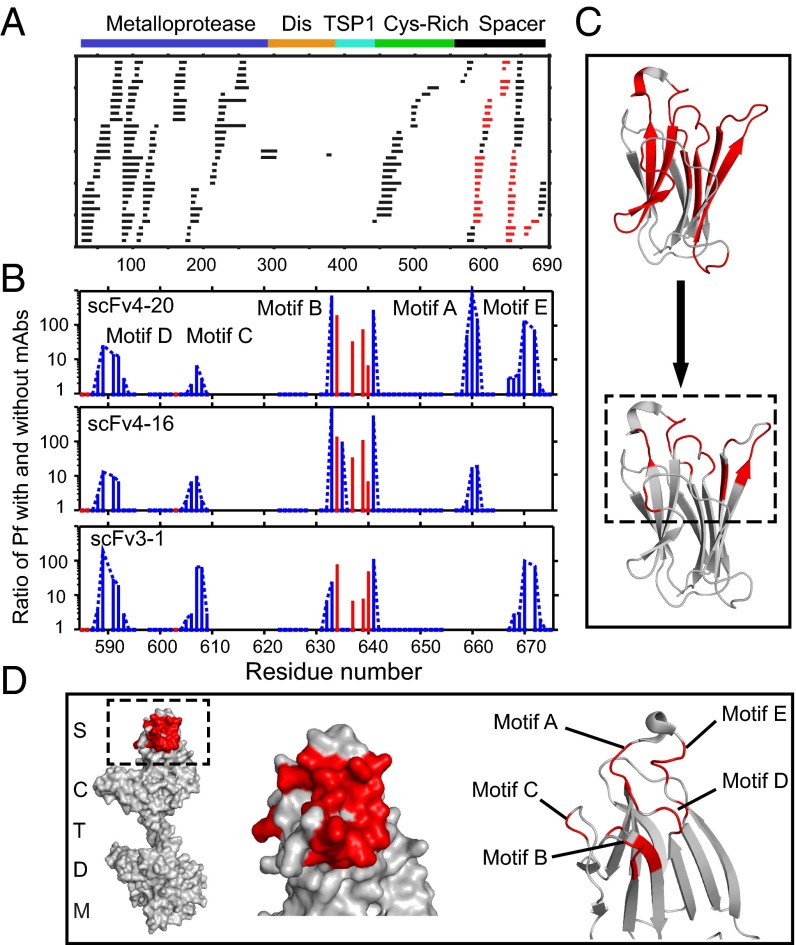
Most MDTCS peptides exhibit no difference in HX rate with or without antibody binding (Fig. 3A, black), but multiple overlapping peptides in the spacer domain were significantly slowed (Fig. 3A, red), indicating the binding of antibody to the corresponding protein segments. We used the HDsite program (20) to more closely define the amino acid positions of HX slowing. HDsite exploits measured D content and the shape of isotopic peptide envelopes, corrects for D-to-H back exchange that occurs during sample analysis, and compares sequentially overlapping peptides. The analysis identifies the affected residues and quantifies their HX rates.
Fig. 3.
Antibody-binding sites in ADAMTS13 revealed by HX MS. (A) Acid protease digestion of human MDTCS fragment produced 208 unique overlapping peptides with 70 derived from the spacer domain and 113 from M. Black peptides show no difference in HX rate with and without scFv bound; red peptides show slowed HX rate upon scFv binding. (B) The ratio of additional protection factor (Pf) with scFv bound. Resolved single residues are in red; switchable residues in blue are connected by dashed lines. Some otherwise-protected regions missing in motifs A and E are ambiguous due to fewer peptides recovered and not necessarily to the absence of antigen–antibody interaction. (C) Representation of the binding epitope (red) at course peptide resolution (Top) and at refined residue resolution (Bottom). (D) Representations of the binding epitope (red) in the spacer domain of ADAMTS13. These results specify the binding epitope in terms of HX slowing in the five-loop region but do not necessarily indicate that the particular residues that show slowed exchange engage in specific interprotein interactions, for reasons described in Discussion.
The relative HX slowing (protection factor) imposed by antibody binding ranges from 2- to 1,000-fold (Fig. 3B and Table S1). Some slowed sites could be specified at single-residue resolution (red in Fig. 3B) and others only at the lesser resolution of two to six sequential “switchable” residues (blue in Fig. 3B). Residues are termed switchable when they cannot be individually resolved by the pattern of overlapping peptides. In this case, HDsite can compute individual site D-occupancies at each time point but cannot specify which occupancy corresponds to which switchable residue.
Table S1.
H-D exchange protection factor (Pf) and Pf increase when bound to scFvs
| Residue | AA | Switch | NoAb Pf | scFv4-20 Pf | scFv4_20 ratio | scFv4_16 Pf | scFv4_16 ratio | scFv3-1 Pf | scFv3-1 ratio |
| 585 | I | 0 | >7E+5 | >7E+5 | — | >7E+5 | — | >7E+5 | — |
| 586 | A | 0 | >7E+5 | >7E+5 | — | >7E+5 | — | >7E+5 | — |
| 587 | N | 1 | >7E+5 | >7E+5 | — | >7E+5 | — | >7E+5 | — |
| 588 | H | 1 | 2.E+04 | 3.E+05 | 15 | 2.E+05 | 10 | 8.E+04 | 4 |
| 589 | R | 1 | 2.E+04 | 1.E+05 | 5 | 5.E+04 | 3 | 4.E+05 | 20 |
| 591 | L | 1 | 3.E+03 | 8.E+04 | 27 | 2.E+04 | 7 | 9.E+04 | 30 |
| 592 | F | 1 | <300 | 4.E+03 | >13 | 4.E+03 | >13 | 6.E+04 | >200 |
| 593 | T | 2 | >7E+5 | >7E+5 | — | >7E+5 | — | >7E+5 | — |
| 594 | H | 2 | 5.E+05 | >7E+5 | >1 | >7E+5 | >1 | >7E+5 | >1 |
| 595 | L | 2 | <300 | 1.E+03 | >3 | <300 | — | 1.E+03 | >3 |
| 598 | R | 3 | 7.E+05 | 8.E+05 | 1 | 9.E+05 | 1 | 1.E+06 | 1 |
| 599 | I | 3 | 2.E+06 | 2.E+06 | 1 | 2.E+06 | 1 | 1.E+06 | 1 |
| 600 | G | 3 | 4.E+03 | 5.E+03 | 1 | 4.E+03 | 1 | 3.E+03 | 1 |
| 601 | G | 3 | <300 | <300 | — | 3.E+02 | — | <300 | — |
| 602 | R | 3 | <300 | <300 | — | <300 | — | <300 | — |
| 603 | Y | 0 | <300 | <300 | — | <300 | — | <300 | — |
| 604 | V | 4 | 3.E+05 | >7E+5 | >2 | >7E+5 | >2 | >7E+5 | >2 |
| 605 | V | 4 | 5.E+05 | >7E+5 | >1 | >7E+5 | >1 | >7E+5 | >1 |
| 606 | A | 4 | <300 | <300 | — | 2.E+04 | >7 | 1.E+04 | >3 |
| 607 | G | 5 | 3.E+03 | 2.E+04 | 7 | 3.E+04 | 10 | 2.E+05 | 67 |
| 608 | K | 5 | <300 | 9.E+02 | >3 | 5.E+02 | >2 | 2.E+04 | >67 |
| 609 | M | 5 | <300 | <300 | — | <300 | — | 1.E+03 | >3 |
| 631 | A | 6 | 4.E+02 | 3.E+05 | 750 | 4.E+05 | 1,000 | 1.E+04 | 25 |
| 632 | L | 6 | <300 | <300 | — | 1.E+02 | — | 2.E+03 | >7 |
| 633 | T | 6 | <300 | <300 | — | <300 | — | <300 | — |
| 634 | E | 0 | 5.E+03 | 1.E+06 | 200 | >7E+5 | 140 | 4.E+05 | 80 |
| 635 | D | 7 | <300 | 3.E+02 | >1 | 3.E+04 | 100 | 3.E+02 | >1 |
| 636 | R | 7 | <300 | < 300 | — | 2.E+02 | — | <300 | — |
| 637 | L | 0 | 2.E+04 | >7E+5 | >35 | >7E+5 | >35 | 1.E+06 | 50 |
| 639 | R | 0 | 9.E+03 | >7E+5 | >78 | 1.E+06 | 111 | 7.E+04 | 8 |
| 640 | L | 0 | 1.E+05 | >7E+5 | >7 | >7E+5 | >7 | >7E+5 | 7 |
| 641 | E | 8 | >7E+5 | >7E+5 | — | >7E+5 | — | >7E+5 | — |
| 642 | E | 8 | 7.E+02 | 2.E+05 | 286 | 4.E+05 | 571 | 8.E+04 | 114 |
| 657 | V | 9 | >7E+5 | >7E+5 | — | >7E+5 | — | — | — |
| 658 | Y | 9 | >7E+5 | >7E+5 | — | >7E+5 | — | — | — |
| 659 | R | 9 | 1.3E+04 | >7E+5 | 54 | 2.4E+05 | 18 | — | — |
| 660 | R | 9 | 8.3E+02 | >7E+5 | 843 | 1.6E+04 | 19 | — | — |
| 661 | Y | 9 | 6.8E+02 | 1.1E+05 | 162 | 4.8E+02 | 1 | — | — |
| 662 | G | 9 | <300 | <300 | — | 5.1E+02 | >2 | — | — |
| 663 | E | 9 | <300 | <300 | — | <300 | — | — | — |
| 664 | E | 9 | <300 | <300 | — | <300 | — | — | — |
| 667 | N | 10 | 2.4E+05 | >7E+5 | >3 | — | — | >7E+5 | >3 |
| 668 | L | 10 | 2.8E+05 | >7E+5 | >3 | — | — | 7.E+05 | 3 |
| 669 | T | 10 | 4.9E+04 | 2.1E+05 | 4 | — | — | 7.4E+04 | 1 |
| 670 | R | 10 | 1.0E+03 | 1.4E+05 | 140 | — | — | 7.0E+04 | 70 |
| 672 | D | 10 | <300 | 2.3E+04 | >77 | — | — | 3.0E+04 | >100 |
| 673 | I | 10 | <300 | 6.4E+02 | >2 | — | — | 4.8E+02 | >2 |
| 674 | T | 10 | <300 | <300 | — | — | — | <300 | — |
| 675 | F | 10 | <300 | <300 | — | — | — | <300 | — |
Fig. 3C compares the resolution available when analysis is limited to the whole peptide level (Top) and the improved resolution (Bottom) provided by the HDsite analysis. HDsite places the protected residues in five flexible loops well defined by multiple sequentially overlapping peptides. All three antiADAMTS13 scFvs appear to protect essentially the same sites (Fig. 3D). The loop motifs not seen to be affected escape analysis due to low peptide recovery and should be considered ambiguous in respect to antibody binding rather than unaffected (Fig. 3B). These results specify the binding epitope in terms of the defined spacer loop region, but they do not necessarily indicate that the particular residues that show slowed exchange engage in specific antibody interactions, for reasons described in Discussion.
Polyclonal IgGs from Acquired TTP Patients Target the Same Antigenic Epitope.
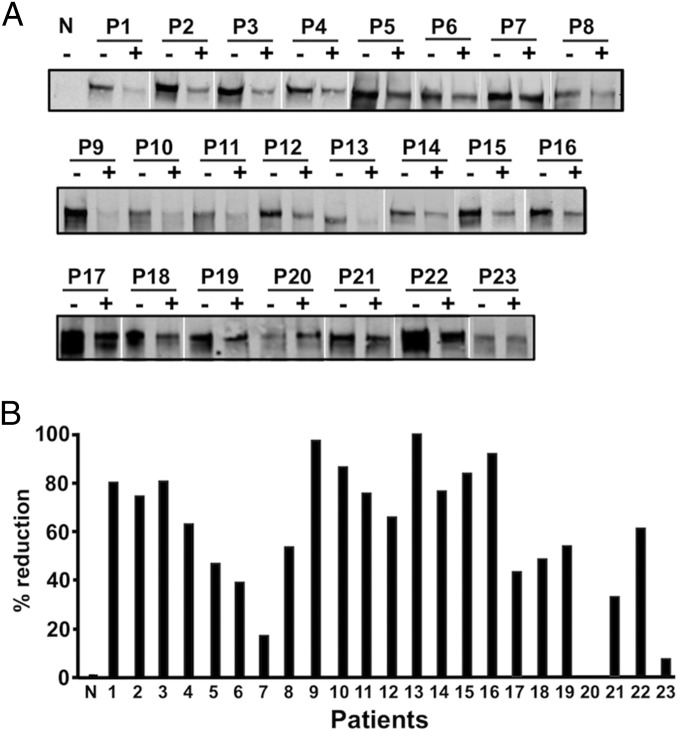
How broadly does the spacer domain epitope represent the target of pathogenic TTP autoantibodies? We addressed this question in competition experiments between polyclonal IgGs in plasma samples from 23 separate TTP patients (Table S2) and scFv4-20, which, as we see, is highly specific for the spacer domain epitope. When scFv4-20 occupied the epitope, polyclonal IgG binding was significantly reduced or even largely eliminated in all but three cases (Fig. 4 A and B). Quantitative differences in these tests reflect the degree of competition between scFv4-20 and patient polyclonal IgGs including their relative concentrations and binding affinities, as well as possible alternative binding sites. It seems clear that the same ADAMTS13 antigenic epitope identified by HX MS is a common binding site for many polyclonal autoantibodies produced by TTP patients. These results are consistent with those reported previously (8–10, 35). The role of autoantibodies reported to bind elsewhere (4, 6, 14) will require further investigation.
Table S2.
Demographic, clinical, and laboratory information of TTP patients included in the study
| Case | Sex | Age, y | Platelet count, ×109/L | TTP diagnosis | ADAMTS13 activity, % | Anti-ADAMTS13 inhibitor | % Suppressed by scFv4-20 |
| Pt1 | M | 27 | 11 | Relapsed | <5 | + | 79.4 |
| Pt2 | F | 56 | 8 | Initial | <5 | + | 75.3 |
| Pt3 | F | 23 | 10 | Exacerbated | <5 | + | 80.8 |
| Pt4 | M | 27 | 106 | Treated | <5 | + | 64.3 |
| Pt5 | M | 20 | 16 | Relapsed | <5 | + | 47.1 |
| Pt6 | F | 55 | 35 | Relapsed | <5 | + | 39.0 |
| Pt7 | M | 68 | 58 | Initial | <5 | + | 17.8 |
| Pt8 | M | 34 | 0 | Initial | <5 | + | 54.2 |
| Pt9 | F | 23 | 11 | Initial | <5 | + | 97.4 |
| Pt10 | M | 34 | 15 | Initial | <5 | + | 86.5 |
| Pt11 | F | 42 | 23 | Initial | <5 | + | 75.5 |
| Pt12 | F | 21 | 9 | Treated | <5 | + | 65.7 |
| Pt13 | F | 45 | 88 | Initial | <5 | + | 100.0 |
| Pt14 | F | 56 | 19 | Initial | <5 | + | 76.1 |
| Pt15 | F | 10 | 31 | Initial | <5 | + | 83.8 |
| Pt16 | F | 48 | 8 | Relapsed | <5 | + | 91.6 |
| Pt17 | F | 55 | 15 | Initial | <5 | + | 12.2 |
| Pt18 | F | 45 | 18 | Initial | <5 | + | 43.3 |
| Pt19 | F | 41 | 23 | Initial | 7.4 | + | 48.9 |
| Pt20 | M | 50 | 190 | Remission | <5 | + | 53.9 |
| Pt21 | F | 77 | 17 | Initial | <5 | + | 0.0 |
| Pt22 | F | 39 | 11 | Initial | <5 | + | 33.0 |
| Pt23 | F | 42 | 15 | Relapsed | <5 | + | 59.9 |
Fig. 4.
Competition for binding to ADAMTS13 between ScFv4-20 and polyclonal anti-ADAMTS13 IgGs from TTP patients. (A) Binding of recombinant ADAMTS13 (arrowhead) to plasma polyclonal IgGs immobilized on protein-G Sepharose-4B from normal (N) and 23 TTP patients (P1–P23) in the absence (−) or presence (+) of scFv4-20 (2 μg) (by immunoprecipitation with anti-V5 IgG and Western blotting). (B) Fractional reduction (in percentage) of recombinant ADAMTS13 binding to polyclonal anti-ADAMTS13 IgGs in TTP patients (P1–P23) by scFv4-20 (by densitometry with NIH ImageJ 1.47).
Deletions in the Epitopic Site of ADAMTS13 Eliminate scFv Binding.
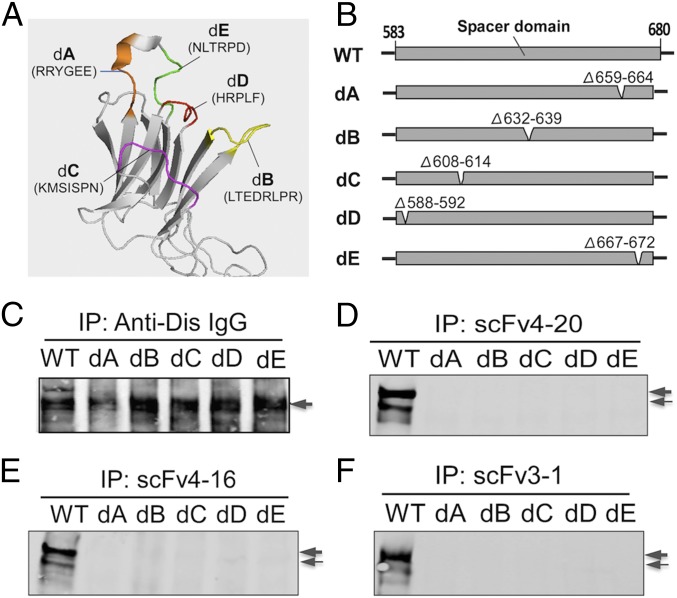
To test the HX MS results, we determined the effect of modifying each of the five epitopic loops in full-length ADAMTS13 on the binding activity of the three scFvs. Each of the loops in the spacer domain was deleted (five to eight residues), one loop at a time. The five deletion variants were designated dA (Δ659–664), dB (Δ632–639), dC (Δ608–614), dD (Δ588–592), and dE (Δ667–672) (Fig. 5 A and B). All were secreted normally from transfected COS7 cells and exhibited a predominant molecular weight of ∼180 kDa, similar to WT (SDS/PAGE and Western blotting analysis). A monoclonal antibody directed against the anti-disintegrin domain was found to bind to WT and all of the deletion mutants (Fig. 5C) as expected because the disintegrin domain was not altered. However, the loss of any one of the ADAMTS13 epitopic loops abolished the binding of all three scFvs (Fig. 5 D–F; by immunoprecipitation and Western blotting).
Fig. 5.
Deletion of any epitopic loop in full-length ADAMTS13 abolishes its binding by scFvs. (A) The position of the five epitopic binding loops in the spacer domain. (B) The partial spacer domain of ADAMTS13 deletion mutants with deletion sites indicated. (C–F) The binding to WT and mutants (arrows) of anti-Dis IgG, scFv4-20, scFv4-16, and scFv3-1 (by Western blotting with anti-V5 IgG after immunoprecipitation). The double band reflects two differently glycosylated ADAMTS13 forms.
These observations support the detailed conformational epitope determined HX MS. It is limited to the loop region of the spacer domain, and the integrity of all five loops is required for high-affinity binding. The same approach opens an avenue for dissecting the autoantibody–ADAMTS13–VWF interaction (below).
Deletions in the Epitopic Site of ADAMTS13 Abolish Its Proteolytic Activity.
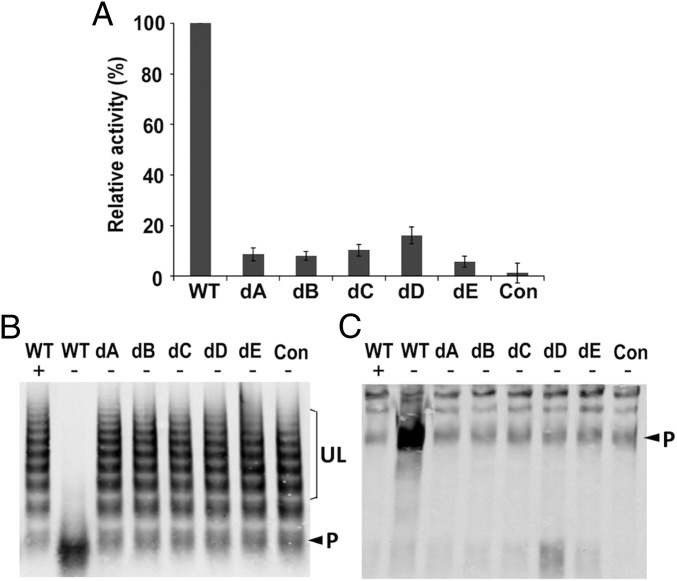
To further explore how the epitopic site might relate to the inhibition of proteolytic activity, we determined the contribution to VWF proteolysis of each of the five epitopic loops in full-length ADAMTS13. WT ADAMTS13 efficiently cleaves both fluorogenic rF-VWF73 substrate peptide (Fig. 6A) and plasma-derived multimeric VWF (Fig. 6 B and C). All five ADAMTS13 deletion mutants (dA, dB, dC, dD, and dE) exhibit greatly reduced activity toward these substrates. Even some point mutations in motif A (36) and alanine substitution of clustered residues in motif B can result in significant impairment of ADAMTS13 proteolytic activity.
Fig. 6.
Deletion of each epitopic site in full-length ADAMTS13 impairs its proteolytic activity. (A) The relative proteolytic activity (in percentage) of ADAMTS13 deletion mutants (dA-dE) [detected by cleavage of the fluorogenic substrate rF-VWF73 in conditioned or mock-transfected (Con) medium; (mean ± SD, n = 3)]. (B and C) The proteolytic cleavage of plasma-derived VWF multimers by purified WT and deletion mutants (dA-dE) in conditioned medium or mock-transfected (Con) medium in the absence (−) or presence (+) of 20 mM EDTA (1.5 M urea, 4 h), detected in 1% SDS-agarose gel (B) and 5% (wt/vol) SDS-polyacrylamide gel (C) (Western blotting with anti-VWF IgG). In B, UL indicates uncleaved large VWF oligomers. P indicates the ADAMTS13 cleavage product mixed with dimeric VWF that is present in normal human plasma. In C, the 350-kDa band (arrowhead) represents either naturally occurring (weak) or ADAMTS13-mediated (strong) cleavage product (P). The large uncleaved VWF oligomers (above 350 kDa) tend not to enter the polyacrylamide gel.
SI Materials and Methods
Preparation of MDTCS Variant.
A plasmid (pcDNA3.1) encoding a human ADAMTS13 variant, i.e., MDTCS (residues 1–685) with a 3×flag tag at the C-terminal end was stably transfected into a Drosophila S2 cell line (Invitrogen) using a modified protocol. Cells were grown in suspension with Schneider’s Drosophila medium supplemented with 10% (wt/vol) FBS (Invitrogen). After Cu2+ induction of stably transfected S2 cells, the MDTCS variant was purified from conditioned medium using anti-flag IgG affinity resin (Sigma). Purity of MDTCS was assessed by 10% (wt/vol) SDS/polyacrylamide gel electrophoresis (SDS/PAGE). Concentration was determined by absorbance at 280 nm using a NanoDrop spectrophotometer (Thermo Fisher Scientific).
Isolation of Monoclonal Anti-ADAMTS13 scFvs by Phage Display.
Phage display library construction and screening were carried out as previously described (46). Briefly, PCR was used to amplify cDNAs encoding the fragments of both the heavy- and light-chain variable regions from spleen and peripheral blood B cells of patients with acquired TTP, which was then cloned into a phage vector pComb3X (a gift from Dr. Carlos Barbas, The Scripps Research Institute, La Jolla, CA). The resulting expression constructs contain a signal peptide, a fragment of light-chain variable region, a flexible linker, and a fragment of a heavy chain variable region, followed by 6×His and a hemagglutinin tag (HA) and the pIII coat protein of M13 filamentous phage. Each individual phage displays scFv protein on its coat and contains the DNA encoding the scFv. The libraries of phages from acquired TTP patients were panned on immobilized rADAMTS13 to isolate those phages that specifically bind rADAMTS13; the bound phages were isolated, and their DNA sequenced. scFvs from two patients, chosen for reasons specified in Results, were studied in this work.
Preparation of Recombinant Anti-ADAMTS13 scFvs.
The construct encoding a specific anti-ADAMTS13 monoclonal scFv (4–20, 4–16, or 3–1) was expressed in Top10′ E. coli cells (Invitrogen). After induction with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (Thermo Fisher Scientific) at 30 °C overnight, the cells were pelleted and lysed with 10 mg of lysozyme, followed by sonication. Recombinant scFvs were purified by Ni2+-chelating affinity chromatography (GE Healthcare), followed by an anti-HA affinity column (Roche) if necessary. Purity was determined by SDS/PAGE with Coomassie blue staining, and concentration was determined by absorbance at 280 nm using a NanoDrop spectrophotometer (Thermo Fisher Scientific).
HX MS.
H-to-D exchange in MDTCS was measured by the fragment separation (HX MS) method in the absence or presence of binding to each antibody scFvs. For antibody-bound MDCTS, one scFv was immobilized on Affi-gel 15 resin (Bio-Rad; following the manufacturer’s protocol), and packed into a 62-µL steel column (2 mm × 2 cm). MDTCS was flushed into the column and equilibrated (50 µL, 0.5 µM MDTCS, 10 mM Hepes, 150 mM NaCl, 5 mM CaCl2, pH 7.4). Excess unbound protein was washed away. To expose the bound MDTCS to H-to-D exchange, 90% (vol/vol) D2O buffer was flushed into the column, and incubated for HX times from 1 to 1,260 min (200 µL, pDr 7.0, 10 mM sodium phosphate, 25 °C). D-labeled MDTCS sample was eluted from the antibody column [100 µL, 0 °C acid buffer, pH 2.5, 50 mM glycine, 2 M guanidinium chloride (GdmCl), 90 mM tris(2-carboxyethyl)phosphine (TCEP)] and injected into an online analysis system (22) where it was carried by flow at 160 μL/min through an immobilized pepsin or fungal XIII protease column for proteolysis. Peptides were caught on a small C8 trap column (1 × 5 mm, 5-μm beads) and washed (3 min), then gradient eluted [9 μL/min, 10–50% (vol/vol) acetonitrile over 12 min], roughly separated in an analytical C18 column (0.3 × 50 mm, 3-μm beads), and then by electrospray into a mass spectrometer (LTQ Orbitrap XL) for a second dimension of separation. Peptides were identified and analyzed for carried D-label by the ExMS program (21) at peptide resolution and by the HDsite program at near–single-residue resolution as previously described (20). Comparative experiments were done with MDTCS in free solution (5 µL, 5.5 µM MDTCS protein mixed with 45 µL of 100% D2O buffer, pDr 7.0, 10 mM sodium phosphate). After different exchange times, 35 µL of cold acid buffer (5.4 M GdmCl, 240 mM TCEP, 134 mM acidified glycine) was added to drop the pH to 2.5 and stop the D-labeling reaction. The sample was injected into the online system and treated as described above. The HX protection factor at near-residue resolution was determined for unbound and antibody-bound peptides by the HDsite analysis. The expectation is that sites in the MDTCS epitopic surface will display slowed HX.
Site-Directed Mutagenesis and Expression of Recombinant ADAMTS13 Variants.
Recombinant ADAMTS13 (rADAMTS13) variants were constructed by QuikChange mutagenesis (Strategene) using pcDNA 3.1-hADAMTS13-V5-His that contains a full-length human ADAMTS13 as a template. The resulting constructs were sequenced at the Nucleic Acid Core Facility at The Children’s Hospital of Philadelphia to confirm accuracy. WT and its variants were expressed in COS7 cells using polyethylenimine (Sigma-Aldrich) according to the manufacturer’s instructions with some modifications. OPTI-MEM serum-free medium (Invitrogen) was added, and cells were cultured for 3–5 d before collection. Conditioned medium was concentrated 250–300 times by filtration with a 100,000 Mr cutoff (Millipore). The 1% protease inhibitor mixture (Sigma-Aldrich) was added to the concentrated conditioned medium, which was stored at −80 °C until assayed. The rADAMTS13 and its variants in the conditioned medium were quantified by a home-designed ELISA as described previously (9). Quantification was double-checked by SDS/PAGE and Western blotting, and compared with a purified V5-tagged rADAMTS13 at known concentration. The rADAMTS13 variants were detected with rabbit anti-V5 IgG (1:5,000) (Novus Biologicals) and goat anti-rabbit IgG conjugated with IRDye 800 (1:10,000) (LI-COR). An Odyssey Infrared Imaging System (LI-COR) was used to quantify the fluorescent signal.
Cleavage of rF-VWF73 by rADAMTS13 and Variants.
Proteolytic activity of rADAMTS13 and variants in conditioned medium was determined by the cleavage of a fluorogenic substrate as previously described (24, 25). A 5ʹ-maleimide fluorescein-labeled VWF73 fragment (rF-VWF73) (0.5∼2 µM) was incubated with either pooled normal human plasma (NHP) (2.5 µL; George King Biotech), or rADAMTS13, or variant (0.5 nM) in the concentrated conditioned medium in 5 mM Bis-Tris, pH 6.0, 25 mM CaCl2, 0.005% Tween 20, and 1 mM Pefabloc in a 96-well opaque white Nunc plate (Thermo Scientific). For inhibition assays, varying concentrations of anti-ADAMTS13 scFvs were incubated with 2.5 µL of NHP for 30 min before addition of rF-VWF73 substrate. A standard curve was prepared by varying the concentration of NHP (0–5 µL) as 1 mL of NHP has the equivalent of 1 unit of active ADAMTS13 activity. The cleavage of rF-VWF73 was measured on a SpectraMax Gemini XPS plate reader (Molecular Devices).
Cleavage of VWF Multimers by rADAMTS13 and Variants.
Purified plasma VWF (∼50 nM) was incubated with purified rADAMTS13 (5 nM) or rADAMTS13 or variants in conditioned medium (1 and 5 nM) in 20 mM Hepes, pH 7.4, containing 150 mM NaCl, 5 mM CaCl2, and 1 µM ZnCl2 on a mixed cellulose filter (Millipore), and then dialyzed against 20 mM Tris⋅HCl, 1.5 M urea (50 mL) at 37 °C for 4 h (10). For antibody inhibition assays, scFvs at various concentrations were incubated with rADAMTS13 or NHP for 30 min before the addition of plasma-derived VWF. Reaction was quenched by mixing with the sample buffer [50 mM Tris, pH 6.8, 10 mM EDTA, 1% SDS, 40% (vol/vol) glycerol, 0.02% bromophenol blue] and heated at 60 °C for 20 min. Ten microliters of each quenched reaction were run on a 1% agarose gel (Lonza) for 3 h at 7 mA to ensure clean multimer separation. The VWF protein was transferred onto a nitrocellulose membrane (Bio-Rad) and detected by rabbit anti-VWF antibody (Dako; 1:5,000) and goat anti-rabbit IgG conjugated with IRDye800 (1:10,000; LI-COR). An Odyssey Infrared Imaging System (LI-COR) was used to measure the fluorescence signal (36).
Patients and Preparation of Plasma Samples.
Twenty-three patients with acquired autoimmune TTP were enrolled into the study, which was approved by the internal review board of The Children’s Hospital of Philadelphia. Each patient was informed and consented to the study participation. Selection criteria were as follows: all patients had thrombocytopenia (<150 × 109/L) and microangiopathic hemolytic anemia and various degrees of organ failure (Table S2). All patients enrolled in the study had received plasma exchange therapy. Whole blood was mostly collected before first plasma exchange and occasionally during treatment or in remission as indicated in Table S2. Blood was anticoagulated with 0.38% sodium citrate, and plasma was separated from cells by centrifugation at 1,000 × g and stored at −80 °C until assay. NHP was obtained from George King Biotech as the positive control.
Immunoprecipitation and Western Blotting.
Anti-HA resin (Roche) was incubated on a rotator with HA-tagged scFvs for 2 h at room temperature in a binding buffer (50 mM Tris⋅HCl, pH 7.5, 300 mM NaCl, 0.25% Triton X-100, 0.25% Tween 20, 1% BSA, and 0.1% casein) (9). Unbound scFv was removed by centrifugation, and ∼400 ng of rADAMTS13 and variants (either purified or in conditioned medium) was added to the scFv-bound resin and incubated overnight at 4 °C with gentle rocking. The anti-HA resin was washed twice with the binding buffer and once with 50 mM Tris⋅HCl (pH 7.5). Bound rADAMTS13 and variants were eluted from the anti-HA resin with 40 μL of sample buffer and heated at 100 °C for 10 min. For competition assay, patient plasma polyclonal IgGs (5–20 µL) were first bound to protein-G Sepharose-4B (20 µL) and then incubated with rADAMTS13 (400 ng) and scFv4-20 (5 µg) at 4 °C overnight. The bound rADAMTS13 protein in the eluate was fractionated by 10% (wt/vol) SDS/PAGE, transferred to a PVDF membrane, and detected by monoclonal anti-V5 antibody (1:5,000; Life Sciences), followed by IRDye800 goat anti-mouse IgG (1:10,000; LI-COR). Fluorescent signals were obtained by scanning with Odyssey Infrared Imaging System (LI-COR) and quantified by densitometry using ImageJ software.
Discussion
HX experiments take advantage of the fact that the backbone amide hydrogens of proteins, one on every amino acid (except proline) in every protein molecule, can undergo facile exchange with solvent hydrogens. HX rates are sensitive to protein structure, structure change, dynamics, and interactions (37, 38), and thus provide a universal probe for protein biophysical behavior resolved in principle to single-amino acid resolution. Under the present conditions [pD(read) 7.0, 25 °C], the average HX time constant of freely exposed amides is ∼0.2 s (39, 40). Amide NH hydrogens protected by H-bonded structure can exchange as much as 10 orders of magnitude more slowly, and thus provide a very large dynamic range within which to distinguish experimental effects.
HX Interpretation.
This work used HX MS to define the epitopic binding site of pathogenic autoantibodies in the ADAMTS13 protease. We sought to find just those ADAMTS13 amide sites that are additionally protected from H-to-D exchange when TTP autoantibodies bind. Contrary to the intuitive conception, the exchange rate of structurally protected hydrogens is poorly correlated with degree of burial, or distance to the aqueous surface, or static solvent accessibility. Instead, structurally protected hydrogens are those that are H-bonded, either to main-chain or side-chain acceptor atoms or to a structurally bound water (41, 42). The HX process for structurally protected hydrogens requires that the protecting H-bond must be transiently broken by some structural fluctuation, and the amide NH must then be brought into solvent contact where normal HX chemistry (reaction with solvent acceptor or donor) can proceed. In this view, antibody binding increases HX protection factors by suppressing dynamic fluctuations that break protecting H-bonds. The bound antibody can be viewed as an external cross-link that tends to suppress dynamic excursions, large and small, of the interacting residues and/or the segments that carry them. Accordingly, binding can slow exchange of immediately involved amides but also others that are not directly involved in specific binding interactions, including amides somewhat outside of the precise binding surface (43, 44).
Residues in the ADAMTS13 epitopic region exhibit a wide range of HX rates and protection factors (Pf) (Fig. 3B and Table S1), both with and without antibody binding. The sizeable Pf values in the unbound protein indicate that these sites are well protected, probably by H-bonding, despite the supposed flexible loop conformation. The protected hydrogens can be further slowed in the complex, whether or not they interact directly with the antibody, because antibody binding may inhibit the structural fluctuations that underlie their exchange with solvent. Residues in the different epitopic loops and even different residues within each loop can experience different degrees of HX slowing, both before and after complex formation (Table S1). This behavior reflects the dynamic reversible breaking and remaking of individual interactions even while the complex remains intact. Similarly disparate HX rates are commonly seen for neighboring residue amides in many proteins because their exchange is determined by different structural fluctuations. The largest additional slowing factor found upon complex formation, ∼1,000-fold, provides an estimate of the maximum fraction of time (10−3 of the time) that the entire complex might be transiently separated (43).
High Resolution HX MS.
The key to high structural resolution in HX MS experiments is to obtain very many sequentially overlapping peptide fragments as in Fig. 3A and then bring to bear adequate computational resources. We used previously described HX MS methods (22, 23) together with the ExMS analysis program (21) to identify and characterize many peptide fragments of ADAMTS13. Given an extensive overlapping peptide dataset, the HDsite analysis program (20) is then able to reduce HX MS data to near–single-residue resolution (Fig. 3B, red). D-occupancy can be attributed to individually specified residues, in principle, when overlapping peptides in the dataset differ by just that residue. Otherwise, HDsite reports two or more adjacent residues as one “switchable” set, as in Fig. 3B (blue).
The application of these methods shows that the binding of different monoclonal scFvs slows HX in five contiguous solvent-exposed loops in the ADAMTS13 spacer domain. This binding region was identified roughly at the peptide level by the HX MS method (Fig. 3 A and C, Top), then at near-residue resolution by the HDsite analysis (Fig. 3 B and C, Bottom), and was validated by the study of small mutational deletions (Fig. 5). The deletion of any one loop (five to eight residues) abrogates scFv binding (Fig. 5), and also the ability to cleave VWF (Fig. 6). Competition experiments show that this same epitope is a common binding site for the polyclonal anti-ADAMTS13 IgGs in almost all of the 23 TTP patients that we tested (Fig. 4). Among the identified epitopic loops, exosite 3 (motif A) is known to play an important role in antibody binding (8, 14, 35, 36). The role of the other loop motifs has not previously been defined nor the fact that all five loops are required for high-affinity ADAMTS13 binding to its inhibiting autoantibodies (Fig. 5) and for functional proteolytic competence (Fig. 6).
Previous efforts to reduce HX MS data to residue resolution have used a subtractive mass centroid-based approach, comparing the altered D-label–dependent peptide mass of peptides that are affected and not affected by the experimental variable. Fig. 3D (Right) shows that almost all peptides in the 5-loop epitope are affected by antibody binding, which invalidates any subtractive method. Nevertheless, the HDsite analysis can discriminate and quantify the D-occupancy of different sites because it exploits more insightful information contained in the isotopic shape of each peptide envelope. The HDsite analysis is able to compute the different residue D-occupancies within each peptide, but it is not possible to assign the different occupancies to specific residues using only single-peptide information (20, 45). HDsite identifies the occupied sites by comparing envelope shape information from overlapping peptides that contain the same amino acids.
Mechanism of Autoimmune TTP.
Autoantibody binding, loop deletions, and some individual mutations (35, 36) in the ADAMTS13 spacer domain epitope disrupt ADAMTS13-mediated proteolysis of VWF (Fig. 6). This must occur either by interfering with the ADAMTS13 to VWF substrate binding step or the proteolysis step itself. It seems unlikely that these various modifications within the same limited spacer domain region could all induce structure changes that move through three intervening domains and disrupt the protease active site in the remote metalloprotease domain (Fig. 3D). In fact, autoantibody binding in the spacer domain produces no detectable HX changes elsewhere in MDTCS, either in the well-covered metalloprotease domain itself or in the intervening Cys-rich domain. These considerations strongly suggest that the TTP antibodies and the spacer loop modifications all interfere with VWF proteolysis in a more straightforward way, by disrupting spacer to VWF binding.
The blocking hypothesis together with the detailed identification of the epitopic site suggests the molecular basis for a possible rationally designed autoantibody-resistant ADAMTS13 variant. One wants to search for mutations in the five-loop ADAMTS13 epitope that would significantly reduce autoantibody binding while retaining sufficient affinity for VWF. A previous study (9) suggests that this approach may well be feasible. Simple i.v. infusion of the altered ADAMTS13 construct might then provide a useful intervention strategy and preempt the currently used rather extreme and expensive daily whole-body plasma replacement therapy. In pursuing this effort, it should be appreciated as noted above that the specific residues measured to be slowed in the complex, although very similar for the different monoclonal antibodies tested, need not be the ones that contribute most to direct protein–protein interaction, whereas unprotected (non–H-bonded) residues may be important.
Materials and Methods
For details, see SI Materials and Methods.
The human ADAMTS13 variant, MDTCS (residues 1–685), was expressed in Drosophila S2 cells (Invitrogen). Phage display library construction and screening of patient-derived scFvs against ADAMTS13 were carried out as previously described (46) using the phage vector pComb3X (a gift from Dr. Carlos Barbas, The Scripps Research Institute, La Jolla, CA) (Fig. 1A). The resulting constructs contain a signal peptide, a fragment of light-chain variable region, a flexible linker, and a fragment of a heavy-chain variable region, followed by 6×His and a HA tag and the pIII coat protein of M13 filamentous phage (Fig. 1B). Phage libraries were panned on immobilized rADAMTS13 (Fig. 1A). Selected monoclonal scFvs were expressed in Top10′ E. coli and purified. The purity of recombinant scFvs was determined by 10% (wt/vol) SDS/PAGE with Coomassie blue staining (Fig. 1C).
Recombinant ADAMTS13 variants were constructed by QuikChange mutagenesis. Proteolytic activity was determined by the cleavage of MDTCS and a fluorogenic substrate derived from the VWF A2 domain, as previously described (24). Polyclonal antibody preparations were from 23 patients with acquired TTP, mostly collected before their first plasma exchange and occasionally during treatment or in remission, as detailed in Table S2. This study was approved by the internal review board of The Children's Hospital of Philadelphia. Each patient was informed and consented to the study participation.
H-to-D exchange was measured for MDTCS both in free solution and when bound to an immobilized column-bound scFv, by the HX MS methods previously described (20–23).
Acknowledgments
This study was supported in part by research grants from the American Society of Hematology Bridge Fund and HL115187-01A1 (to X.L.Z.), HL007971 (to V.C.C.), P50 HL81012 (to D.L.S.), RIBS-PNU-2012-08790001 (to W.K.L.), and GM031847, MCB 1020649, and The Mathers Foundation (to S.W.E.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512561112/-/DCSupplemental.
References
- 1.Moake JL. Thrombotic thrombocytopenic purpura: The systemic clumping “plague.”. Annu Rev Med. 2002;53:75–88. doi: 10.1146/annurev.med.53.082901.103948. [DOI] [PubMed] [Google Scholar]
- 2.Bell WR, Braine HG, Ness PM, Kickler TS. Improved survival in thrombotic thrombocytopenic purpura-hemolytic uremic syndrome. Clinical experience in 108 patients. N Engl J Med. 1991;325(6):398–403. doi: 10.1056/NEJM199108083250605. [DOI] [PubMed] [Google Scholar]
- 3.Tsai HM, Lian EC. Antibodies to von Willebrand factor-cleaving protease in acute thrombotic thrombocytopenic purpura. N Engl J Med. 1998;339(22):1585–1594. doi: 10.1056/NEJM199811263392203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng XL, et al. Multiple domains of ADAMTS13 are targeted by autoantibodies against ADAMTS13 in patients with acquired idiopathic thrombotic thrombocytopenic purpura. Haematologica. 2010;95(9):1555–1562. doi: 10.3324/haematol.2009.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furlan M, et al. von Willebrand factor-cleaving protease in thrombotic thrombocytopenic purpura and the hemolytic-uremic syndrome. N Engl J Med. 1998;339(22):1578–1584. doi: 10.1056/NEJM199811263392202. [DOI] [PubMed] [Google Scholar]
- 6.Klaus C, et al. Epitope mapping of ADAMTS13 autoantibodies in acquired thrombotic thrombocytopenic purpura. Blood. 2004;103(12):4514–4519. doi: 10.1182/blood-2003-12-4165. [DOI] [PubMed] [Google Scholar]
- 7.Pos W, et al. VH1-69 germline encoded antibodies directed towards ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. J Thromb Haemost. 2009;7(3):421–428. doi: 10.1111/j.1538-7836.2008.03250.x. [DOI] [PubMed] [Google Scholar]
- 8.Pos W, et al. Residues Arg568 and Phe592 contribute to an antigenic surface for anti-ADAMTS13 antibodies in the spacer domain. Haematologica. 2011;96(11):1670–1677. doi: 10.3324/haematol.2010.036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jian C, et al. Gain-of-function ADAMTS13 variants that are resistant to autoantibodies against ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. Blood. 2012;119(16):3836–3843. doi: 10.1182/blood-2011-12-399501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luken BM, et al. Amino acid regions 572–579 and 657–666 of the spacer domain of ADAMTS13 provide a common antigenic core required for binding of antibodies in patients with acquired TTP. Thromb Haemost. 2006;96(3):295–301. doi: 10.1160/TH06-03-0135. [DOI] [PubMed] [Google Scholar]
- 11.Luken BM, et al. The spacer domain of ADAMTS13 contains a major binding site for antibodies in patients with thrombotic thrombocytopenic purpura. Thromb Haemost. 2005;93(2):267–274. doi: 10.1160/TH04-05-0301. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, et al. Selection and analysis of an optimized anti-VEGF antibody: Crystal structure of an affinity-matured Fab in complex with antigen. J Mol Biol. 1999;293(4):865–881. doi: 10.1006/jmbi.1999.3192. [DOI] [PubMed] [Google Scholar]
- 13.Beyer BM, et al. Crystal structures of the pro-inflammatory cytokine interleukin-23 and its complex with a high-affinity neutralizing antibody. J Mol Biol. 2008;382(4):942–955. doi: 10.1016/j.jmb.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Schaller M, Vogel M, Kentouche K, Lämmle B, Kremer Hovinga JA. The splenic autoimmune response to ADAMTS13 in thrombotic thrombocytopenic purpura contains recurrent antigen-binding CDR3 motifs. Blood. 2014;124(23):3469–3479. doi: 10.1182/blood-2014-04-561142. [DOI] [PubMed] [Google Scholar]
- 15.Brock A. Fragmentation hydrogen exchange mass spectrometry: A review of methodology and applications. Protein Expr Purif. 2012;84(1):19–37. doi: 10.1016/j.pep.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Coales SJ, Tuske SJ, Tomasso JC, Hamuro Y. Epitope mapping by amide hydrogen/deuterium exchange coupled with immobilization of antibody, on-line proteolysis, liquid chromatography and mass spectrometry. Rapid Commun Mass Spectrom. 2009;23(5):639–647. doi: 10.1002/rcm.3921. [DOI] [PubMed] [Google Scholar]
- 17.Pandit D, et al. Mapping of discontinuous conformational epitopes by amide hydrogen/deuterium exchange mass spectrometry and computational docking. J Mol Recognit. 2012;25(3):114–124. doi: 10.1002/jmr.1169. [DOI] [PubMed] [Google Scholar]
- 18.Houde D, Engen JR. Conformational analysis of recombinant monoclonal antibodies with hydrogen/deuterium exchange mass spectrometry. Methods Mol Biol. 2013;988:269–289. doi: 10.1007/978-1-62703-327-5_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, et al. Epitope mapping of a 95 kDa antigen in complex with antibody by solution-phase amide backbone hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2011;83(18):7129–7136. doi: 10.1021/ac201501z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kan ZY, Walters BT, Mayne L, Englander SW. Protein hydrogen exchange at residue resolution by proteolytic fragmentation mass spectrometry analysis. Proc Natl Acad Sci USA. 2013;110(41):16438–16443. doi: 10.1073/pnas.1315532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kan ZY, Mayne L, Chetty PS, Englander SW. ExMS: Data analysis for HX-MS experiments. J Am Soc Mass Spectrom. 2011;22(11):1906–1915. doi: 10.1007/s13361-011-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayne L, et al. Many overlapping peptides for protein hydrogen exchange experiments by the fragment separation-mass spectrometry method. J Am Soc Mass Spectrom. 2011;22(11):1898–1905. doi: 10.1007/s13361-011-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walters BT, Ricciuti A, Mayne L, Englander SW. Minimizing back exchange in the hydrogen exchange-mass spectrometry experiment. J Am Soc Mass Spectrom. 2012;23(12):2132–2139. doi: 10.1007/s13361-012-0476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, et al. Creation of a recombinant peptide substrate for fluorescence resonance energy transfer-based protease assays. Anal Biochem. 2006;358(2):298–300. doi: 10.1016/j.ab.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 25.Raife TJ, et al. Leukocyte proteases cleave von Willebrand factor at or near the ADAMTS13 cleavage site. Blood. 2009;114(8):1666–1674. doi: 10.1182/blood-2009-01-195461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furlan M, Robles R, Lämmle B. Partial purification and characterization of a protease from human plasma cleaving von Willebrand factor to fragments produced by in vivo proteolysis. Blood. 1996;87(10):4223–4234. [PubMed] [Google Scholar]
- 27.Ai J, Smith P, Wang S, Zhang P, Zheng XL. The proximal carboxyl-terminal domains of ADAMTS13 determine substrate specificity and are all required for cleavage of von Willebrand factor. J Biol Chem. 2005;280(33):29428–29434. doi: 10.1074/jbc.M505513200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bao J, Xiao J, Mao Y, Zheng XL. Carboxyl terminus of ADAMTS13 directly inhibits platelet aggregation and ultra large von Willebrand factor string formation under flow in a free-thiol-dependent manner. Arterioscler Thromb Vasc Biol. 2014;34(2):397–407. doi: 10.1161/ATVBAHA.113.302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Y, Xiao J, Falls E, Zheng XL. A shear-based assay for assessing plasma ADAMTS13 activity and inhibitors in patients with thrombotic thrombocytopenic purpura. Transfusion. 2011;51(7):1580–1591. doi: 10.1111/j.1537-2995.2010.03020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin SY, et al. AAV-mediated expression of an ADAMTS13 variant prevents shigatoxin-induced thrombotic thrombocytopenic purpura. Blood. 2013;121(19):3825–3829, S3821–S3823. doi: 10.1182/blood-2013-02-486779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao W, Anderson PJ, Majerus EM, Tuley EA, Sadler JE. Exosite interactions contribute to tension-induced cleavage of von Willebrand factor by the antithrombotic ADAMTS13 metalloprotease. Proc Natl Acad Sci USA. 2006;103(50):19099–19104. doi: 10.1073/pnas.0607264104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao W, Anderson PJ, Sadler JE. Extensive contacts between ADAMTS13 exosites and von Willebrand factor domain A2 contribute to substrate specificity. Blood. 2008;112(5):1713–1719. doi: 10.1182/blood-2008-04-148759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng X, et al. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. J Biol Chem. 2001;276(44):41059–41063. doi: 10.1074/jbc.C100515200. [DOI] [PubMed] [Google Scholar]
- 34.Sorvillo N, et al. Identification of N-linked glycosylation and putative O-fucosylation, C-mannosylation sites in plasma derived ADAMTS13. J Thromb Haemost. 2014;12(5):670–679. doi: 10.1111/jth.12535. [DOI] [PubMed] [Google Scholar]
- 35.Pos W, et al. An autoantibody epitope comprising residues R660, Y661, and Y665 in the ADAMTS13 spacer domain identifies a binding site for the A2 domain of VWF. Blood. 2010;115(8):1640–1649. doi: 10.1182/blood-2009-06-229203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin SY, Skipwith CG, Zheng XL. Amino acid residues Arg(659), Arg(660), and Tyr(661) in the spacer domain of ADAMTS13 are critical for cleavage of von Willebrand factor. Blood. 2010;115(11):2300–2310. doi: 10.1182/blood-2009-07-235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Englander SW, Hiller R. Dynamics and thermodynamics of hyperthermophilic proteins by hydrogen exchange. Methods Enzymol. 2001;334:342–350. doi: 10.1016/s0076-6879(01)34481-6. [DOI] [PubMed] [Google Scholar]
- 38.Englander SW, Sosnick TR, Englander JJ, Mayne L. Mechanisms and uses of hydrogen exchange. Curr Opin Struct Biol. 1996;6(1):18–23. doi: 10.1016/s0959-440x(96)80090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connelly GP, Bai Y, Jeng MF, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17(1):87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 41.Skinner JJ, Lim WK, Bedard S, Black BE, Englander SW. Protein hydrogen exchange: Testing current models. Protein Sci. 2012;21(7):987–995. doi: 10.1002/pro.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skinner JJ, Lim WK, Bedard S, Black BE, Englander SW. Protein dynamics viewed by hydrogen exchange. Protein Sci. 2012;21(7):996–1005. doi: 10.1002/pro.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paterson Y, Englander SW, Roder H. An antibody binding site on cytochrome c defined by hydrogen exchange and two-dimensional NMR. Science. 1990;249(4970):755–759. doi: 10.1126/science.1697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams DC, Jr, Rule GS, Poljak RJ, Benjamin DC. Reduction in the amide hydrogen exchange rates of an anti-lysozyme Fv fragment due to formation of the Fv-lysozyme complex. J Mol Biol. 1997;270(5):751–762. doi: 10.1006/jmbi.1997.1122. [DOI] [PubMed] [Google Scholar]
- 45.Hu W, et al. Stepwise protein folding at near amino acid resolution by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci USA. 2013;110(19):7684–7689. doi: 10.1073/pnas.1305887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Payne AS, et al. Genetic and functional characterization of human pemphigus vulgaris monoclonal autoantibodies isolated by phage display. J Clin Invest. 2005;115(4):888–899. doi: 10.1172/JCI24185. [DOI] [PMC free article] [PubMed] [Google Scholar]