Significance
Outcomes for individuals with central nervous system (CNS) malignancies remain abysmal. A major challenge in managing these patients is the lack of reliable biomarkers to monitor tumor dynamics. Consequently, many patients undergo invasive surgical procedures to determine disease status or experience treatment delays when radiographic testing fails to show disease progression. We show here that primary CNS malignancies shed detectable levels of tumor DNA into the surrounding cerebrospinal fluid (CSF), which could serve as a sensitive and exquisitely specific marker for quantifying tumor burden without invasive biopsies. Therefore, assessment of such tumor-derived DNA in the CSF has the potential to improve the management of patients with primary CNS tumors.
Keywords: CSF-tDNA, CNS tumors, biomarker
Abstract
Cell-free DNA shed by cancer cells has been shown to be a rich source of putative tumor-specific biomarkers. Because cell-free DNA from brain and spinal cord tumors cannot usually be detected in the blood, we studied whether the cerebrospinal fluid (CSF) that bathes the CNS is enriched for tumor DNA, here termed CSF-tDNA. We analyzed 35 primary CNS malignancies and found at least one mutation in each tumor using targeted or genome-wide sequencing. Using these patient-specific mutations as biomarkers, we identified detectable levels of CSF-tDNA in 74% [95% confidence interval (95% CI) = 57–88%] of cases. All medulloblastomas, ependymomas, and high-grade gliomas that abutted a CSF space were detectable (100% of 21 cases; 95% CI = 88–100%), whereas no CSF-tDNA was detected in patients whose tumors were not directly adjacent to a CSF reservoir (P < 0.0001, Fisher’s exact test). These results suggest that CSF-tDNA could be useful for the management of patients with primary tumors of the brain or spinal cord.
Approximately 25,000 individuals each year are diagnosed with a malignant brain or spinal cord tumor in the United States, and more than one-half of these patients will die from their disease (1). Although there are a number of different subtypes of primary CNS cancers, nearly all are treated with maximal safe surgical resection followed by radiation and in some cases, chemotherapy. Given the lack of clinically available biomarkers for CNS malignancies, the conventional method for disease monitoring in these patients is radiographic using either computed tomography or MRI (2). Unfortunately, anatomic changes detected by these imaging modalities are often nonspecific and slow to change, even in the face of progressing or regressing disease. Moreover, it can be difficult to discriminate between treatment effect and cancer growth with imaging alone (3). Patients must, therefore, have additional surgeries for definitive tissue diagnosis or inappropriately wait for radiographic findings to change as their disease progresses. As a result, there is a great need for more sensitive and specific tumor biomarkers in neurooncology.
The recent success of detecting circulating tumor cells in the peripheral blood of glioblastoma patients represents an important step toward this goal, with reported sensitivities between 21% and 39% (4–6). Circulating tumor DNA (ctDNA) is found in the plasma of patients with most forms of malignancies (7–11). However, brain tumors, including high-grade gliomas and medulloblastomas, are an exception, with only a minority giving rise to detectable levels of ctDNA, perhaps because of the blood–brain barrier (8).
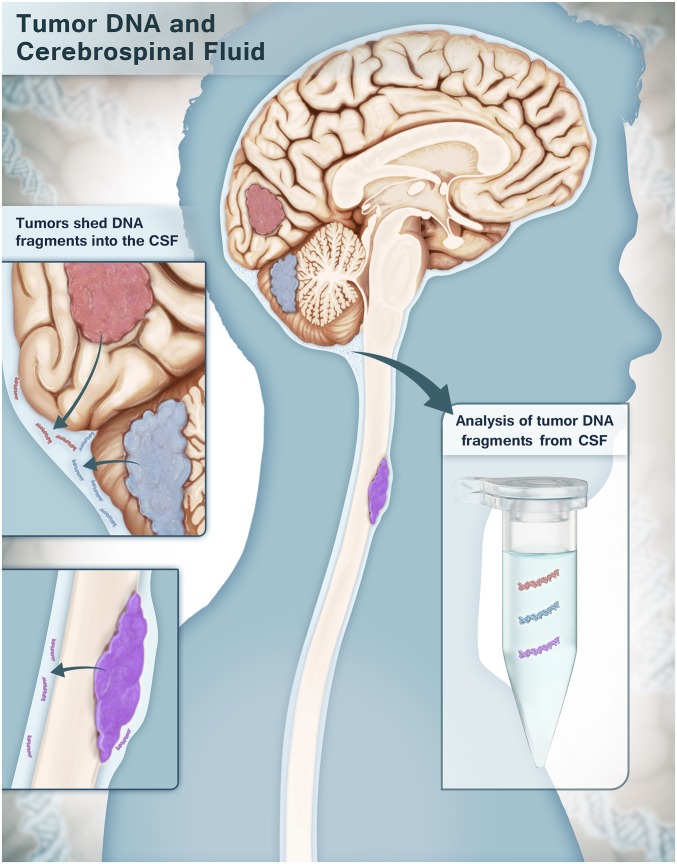
Other studies have shown that tumor-derived DNA can be found in anatomically relevant fluids, such as urine in bladder cancer patients, sputum in lung cancer patients, stool in patients with colorectal carcinomas, and endocervical fluid in patients with gynecological malignancies (12–17). Based on this concept, we wondered whether primary brain and spinal cord tumors might shed appreciable levels of tDNA into the cerebrospinal fluid (CSF) that bathes the CNS (Fig. 1). We coined the term “CSF-tDNA” to describe tumor DNA shed into the CSF. The experiments below were designed to test this hypothesis in an exploratory study of tumors of diverse histology and locations within the CNS.
Fig. 1.
Schematic showing the shedding of CSF-tDNA from CNS malignancies. Tumor cells from primary brain and spinal cord tumors shed DNA into the CSF that bathes the CNS. DNA purified from the CSF is analyzed for tumor-specific mutations.
Results
Patient and Tumor Characteristics.
Thirty-five patients with CNS cancers were enrolled in this study. Their ages, sexes, races, and preoperative symptoms are listed in Table S1. In total, 6 patients had medulloblastomas, and 29 patients had gliomas; 7, 9, 2, and 17 of the tumors were classified as World Health Organization (WHO) grades I–IV, respectively. Twenty-nine (83%) of 35 patients provided CSF during the initial surgery, whereas the remaining 6 (17%) did so during a repeat resection. The tumors were distributed throughout the brain and spinal cord, with 14 arising in the posterior fossa (including six medulloblastomas), 8 arising in the supratentorial compartment of the brain, and 13 arising in the spinal cord (Table 1).
Table S1.
Patient demographics
| Patient | Sex | Recurrence | Age (y) | Race | Preoperative symptoms | Duration of symptoms (mo) | Hydrocephalus | Enhancing on CT or MRI | Tumor size (cm3) |
| CGLI 02 | Female | No | 33 | W | Dysesthesias, urinary retention | 33 | No | No | 2 × 1 × 1 |
| CGLI 03 | Male | No | 13 | W | Incidental | Not applicable | No | Yes | 3 × 3.2 × 3.8 |
| CGLI 06 | Female | No | 13 | AA | Headache, vision loss | 2 | Yes | Yes | 4.9 × 5 × 5.6 |
| CGLI 11 | Female | No | 50 | W | Dysesthesias, numbness | 36 | No | Yes | 5.3 × 1 × 1.2 |
| CGLI 12 | Male | No | 5 | W | Headache, vomiting | 1 | Yes | Yes | 4.1 × 4.2 × 3.8 |
| CGLI 13 | Male | No | 27 | W | Back pain, sciatica | 15 | No | NA | NA |
| CGLI 14 | Female | No | 62 | W | Back pain | 5 | No | Yes | 4.5 × 0.8 × 1.2 |
| CGLI 15 | Male | No | 52 | W | Neck pain | 1 | No | Yes | 2.1 × 0.9 × 1.2 |
| CGLI 20 | Female | No | 14 | W | Headache, vomiting | 3 | Yes | Yes | 3.4 × 3.2 × 2.6 |
| CGLI 22 | Female | No | 18 | A | Headache, vomiting | 3 | No | Yes | 5 × 4.4 × 4.3 |
| CGLI 25 | Female | No | 72 | W | Back pain, arm pain | 3 | No | Yes | 4.3 × 1.2 × 0.7 |
| CGLI 26 | Female | No | 46 | W | Left leg pain | 13 | No | No | 5 × 1.4 × 1.2 |
| CGLI 28 | Female | No | 79 | A | Left arm weakness | 3 | No | Yes | 4.4 × 5.7 × 5.8 |
| CGLI 29 | Male | Yes | 18 | W | Neck pain | 28 | No | Yes | 3.1 × 1.1 |
| CGLI 31 | Male | No | 74 | AA | Seizure | 0.5 | No | Yes | 4.5 × 5.2 × 4.3 |
| CGLI 35 | Male | No | 70 | AA | Memory loss | 2 | No | Yes | 6.1 × 3 × 3.5 |
| CGLI 36 | Male | No | 11 | W | Back pain | 2 | No | Yes | 6.5 × 1.7 × 2.2 |
| CGLI 39 | Male | No | 56 | W | Paresthesias | 3 | No | Yes | 1.7 × 1.1 × 1.6 |
| CGLI 40 | Male | No | 7 | W | Headache, ataxia | 5 | No | Yes | 4.0 × 4.0 × 4.8 |
| CGLI 41 | Male | Yes | 34 | W | Gait imbalance, incoordination | 0.5 | No | Yes | 4.0 × 3.0 × 2.9 |
| CGLI 42 | Female | No | 31 | W | Back pain | 24 | No | NA | NA |
| CGLI 43 | Male | Yes | 66 | W | Dysesthesias | 12 | No | Yes | 3.8 × 1.4 × 1.4 |
| CGLI 44 | Female | No | 6 | A | Headache, vomiting | 2 | Yes | Yes | 3.4 × 3.8 × 3.4 |
| CGLI 47 | Female | Yes | 71 | W | Headache | 2 | No | Yes | 6.0 × 3.7 × 3.3 |
| CGLI 48 | Male | No | 56 | W | Blurry vision, mental status change | 2 | No | Yes | 6.1 × 7.7 × 4.9 |
| CGLI 50 | Female | No | 69 | W | Vision | 1 | No | Yes | 5.1 × 5.0 × 3.8 |
| CGLI 51 | Female | No | 84 | W | Asymptomatic | Not applicable | No | Yes | 4.0 × 3.3 × 2.9 |
| CGLI 55 | Male | No | 7 | ME | Headache, difficulty walking | 1 | No | Yes | 3.1 × 3.8 × 3.4 |
| CGLI 56 | Male | No | 3 | W | Headache, difficulty walking | 1 | Yes | Yes | 4.1 × 4.2 × 4.0 |
| CGLI 58 | Male | No | 32 | ME | Numbness, myelopathy | 24 | No | No | 5.1 × 1.9 × 1.2 |
| CGLI 60 | Female | No | 9 | W | Headache | 3 | No | Yes | 2.4 × 2.2 × 2.0 |
| CGLI 61 | Female | No | 12 | W | Headache, nausea, vomiting | 1 | No | Yes | 5.5 × 5.3 × 5.2 |
| CGLI 63 | Male | No | 22 | ME | Headache, vomiting, ataxia | 1 | No | Yes | 3.0 × 2.2 × 2.0 |
| CGLI 101 | Male | Yes | 49 | W | Headache, nausea, vomiting | NA | Yes | Yes | 2.6 × 2.0 × 1.6 |
| CGLI 254 | Male | Yes | 7 | W | Headache, ataxia, vomiting | NA | Yes | Yes | 1.7 × 1.2 × 1.2 |
A, Asian; AA, African American; CT, computed tomography; ME, Middle Eastern; NA, not available; W, white.
Table 1.
Tumor characteristics and the detection of CSF-tDNA
| Patient | Diagnosis | Tumor grade | Tumor location | Location of CSF sampling | Tumor abutting CSF space | CSF-tDNA |
| CGLI 02 | Glioma | WHO II, diffuse astrocytoma | T11 spinal cord | Spinal subarachnoid space | Yes | Not detected |
| CGLI 03 | Anaplastic astrocytoma | WHO III, anaplastic astrocytoma | Pons | Basal cistern | Yes | Positive |
| CGLI 06 | Pilocytic astrocytoma | WHO I, pilocytic astrocytoma | Cerebellar vermis | Basal cistern | Yes | Positive |
| CGLI 11 | Spinal ependymoma | WHO II, ependymoma | C7-T3 spinal cord | Spinal subarachnoid space | Yes | Positive |
| CGLI 12 | Intracranial ependymoma | WHO II, ependymoma | Fourth ventricle | Basal cistern | Yes | Positive |
| CGLI 13 | Myxopapillary ependymoma | WHO I, myxopapillary ependymoma | L2-3 spinal cord | Spinal subarachnoid space | NA | Positive |
| CGLI 14 | Intramedullary spinal cord lesion | Low-grade neoplasm | T7-9 spinal cord | Spinal subarachnoid space | Yes | Positive |
| CGLI 15 | Spinal ependymoma | WHO II, ependymoma | C3-4 spinal cord | Spinal subarachnoid space | No | Not detected |
| CGLI 20 | Medulloblastoma | WHO IV, medulloblastoma | Fourth ventricle | Basal cistern | Yes | Positive |
| CGLI 22 | Pilocytic astrocytoma | WHO I, pilocytic astrocytoma | Cerebellar hemisphere | Basal cistern | No | Not detected |
| CGLI 25 | Myxopapillary ependymoma | WHO I, myxopapillary ependymoma | L2-3 spinal cord | Spinal subarachnoid space | Yes | Positive |
| CGLI 26 | Intramedullary spinal cord tumor | WHO II, infiltrating astrocytoma with oligodendroglial features | C3-6 spinal cord | Spinal subarachnoid space | Yes | Positive |
| CGLI 28 | Anaplastic astrocytoma | WHO III, anaplastic astrocytoma | Right frontal/butterfly | Ventricle | Yes | Positive |
| CGLI 29 | Glioblastoma | WHO IV, glioblastoma | C4-6 spinal cord | Spinal subarachnoid space | Yes | Positive |
| CGLI 31 | Glioblastoma | WHO IV, glioblastoma | Right frontal/butterfly | Ventricle | Yes | Positive |
| CGLI 35 | Glioblastoma | WHO IV, glioblastoma | Right temporal | Ventricle | Yes | Positive |
| CGLI 36 | Spinal cord glioblastoma | WHO IV, glioblastoma | T10-L1 spinal cord | Spinal subarachnoid space | Yes | Positive |
| CGLI 39 | Intramedullary spinal cord tumor | WHO II, low-grade glioma likely ependymoma | C2-3 spinal cord | Spinal subarachnoid space | No | Not detected |
| CGLI 40 | Medulloblastoma | WHO IV, medulloblastoma | Fourth ventricle | Basal cistern | Yes | Positive |
| CGLI 41 | Glioblastoma | WHO IV, glioblastoma | Cerebellar hemisphere | Basal cistern | Yes | Positive |
| CGLI 42 | Spinal ependymoma | WHO II, ependymoma | T1-7 spinal cord | Spinal subarachnoid space | NA | Positive |
| CGLI 43 | Low-grade glioma | WHO II, low-grade glioma | T10 spinal cord | Spinal subarachnoid space | Yes | Not detected |
| CGLI 44 | Pilocytic astrocytoma | WHO I, pilocytic astrocytoma | Cerebellar vermis | Basal cistern | No | Not detected |
| CGLI 47 | Glioblastoma | WHO IV, glioblastoma | Right temporal | Ventricle | Yes | Positive |
| CGLI 48 | Glioblastoma | WHO IV, glioblastoma | Left temporal | Ventricle | Yes | Positive |
| CGLI 50 | Glioblastoma | WHO IV, glioblastoma | Right temporal | Ventricle | Yes | Positive |
| CGLI 51 | Glioblastoma | WHO IV, glioblastoma | Right frontal | Ventricle | Yes | Positive |
| CGLI 55 | Brainstem glioblastoma | WHO IV, glioblastoma | Midbrain | Basal cistern | Yes | Positive |
| CGLI 56 | Medulloblastoma | WHO IV, medulloblastoma | Fourth ventricle | Basal cistern | Yes | Positive |
| CGLI 58 | Diffuse astrocytoma | WHO II, diffuse astrocytoma | T2-4 spinal cord | Spinal subarachnoid space | Yes | Not detected |
| CGLI 60 | Medulloblastoma | WHO IV, medulloblastoma | Fourth ventricle | Basal cistern | Yes | Positive |
| CGLI 61 | Pilocytic astrocytoma | WHO I, pilocytic astrocytoma | Cerebellar vermis | Basal cistern | Yes | Not detected |
| CGLI 63 | Medulloblastoma | WHO IV, medulloblastoma | Cerebellum | Basal cistern | No | Not detected |
| CGLI 101 | Glioblastoma | WHO IV, glioblastoma | Cerebellar vermis | Basal cistern | Yes | Positive |
| CGLI 254 | Medulloblastoma | WHO IV, medulloblastoma | Fourth ventricle | Basal cistern | Yes | Positive |
NA, not available.
Identification of Somatic Mutations.
At least one mutation was identified in each of 35 tumors analyzed using a tiered approach [targeted sequencing followed by whole-exome sequencing (WES)] described in Materials and Methods.
With the targeted sequencing approach, we identified mutations in 13 tumors. The mutations in these samples occurred in TP53 (tumor protein p53; n = 5), IDH1 (isocitrate dehydrogenase 1; n = 2), or the TERT promoter (telomerase reverse transcriptase; n = 6) (Table S2). In the remaining 22 tumors, WES was used to identify at least one mutation per sample (Dataset S1). Genes mutated in these samples included well-known drivers, such as NF2, PIK3R1, PTCH1, and PTEN (18). The fractions of mutant alleles in tumors were generally high, averaging 46% (with an SD of 18%). This finding is consistent with the expected early development of driver gene mutations during tumor evolution and the presence of nonneoplastic cells in all tumors, even macrodissected ones, such as the samples used here. All mutations identified were confirmed to be absent in DNA from matched noncancerous (normal) cells from each patient.
Table S2.
Mutations detected in the CSF or tumor of each patient
| Patient | Volume of CSF analyzed (mL) | DNA in CSF (ng) | Sequencing method | Mutation | Location | Gene | Transcript | Protein | cDNA | Mutant allele fraction in tumor (%) | Mutant allele fraction in CSF (%) |
| CGLI 02 | 4 | 245 | WES | NM_144631.5 (ZNF513):c.413C > T | Exon | ZNF513 | NM_144631.5 | p.P138L | c.413C > T | 49 | Not detected |
| CGLI 03 | 7 | 314 | WES | NM_181523.2 (PIK3R1):c.1690A > G | Exon | PIK3R1 | NM_181523.2 | p.N564D | c.1690A > G | 39 | 0.1 |
| CGLI 06 | 8 | 148 | WES | NR_024604.1 (CLCA3P):c.446G > A | Exon | CLCA3P | NR_024604.1 | p.R149Q | c.446G > A | 36 | 0.2 |
| CGLI 11 | 5 | 290 | WES | NM_133450.3 (ANKS3):c.1738G > A | Exon | ANKS3 | NM_133450.3 | p.E580K | c.1738G > A | 47 | 0.3 |
| CGLI 12 | 5 | 71 | WES | NM_003531.2 (HIST1H3C):c.83T > A | Exon | HIST1H3C | NM_003531.2 | p.K28M | c.83T > A | 44 | 0.2 |
| CGLI 13 | 1.5 | 6 | WES | NM_144965.1 (TTC16):c.1016T > A | Exon | TTC16 | NM_144965.1 | p.V339E | c.1016T > A | 48 | 1.0 |
| CGLI 14 | 7.5 | 187 | Panel | NM_000546.5 (TP53):c.638G > A | Exon | TP53 | NM_000546.5 | p.R213Q | c.638G > A | 59 | 19.6 |
| CGLI 15 | 5 | 482 | WES | NM_004990.3 (MARS):c.1487C > T | Exon | MARS | NM_004990.3 | p.T496I | c.1487C > T | 13 | Not detected |
| CGLI 20 | 3 | 127 | WES | NM_198229.2 (RGS12):c.2980C > T | Exon | RGS12 | NM_198229.2 | p.R994W | c.2980C > T | 52 | 52.6 |
| CGLI 22 | 5 | 8 | WES | NM_001080522.2 (CC2D2A):c.1938G > A | Exon | CC2D2A | NM_001080522.2 | p.W646X | c.1938G > A | 54 | Not detected |
| CGLI 25 | 10 | 253 | WES | NM_001795.3 (CDH5):c.1211G > A | Exon | CDH5 | NM_001795.3 | p.S404N | c.1211G > A | 50 | 0.3 |
| CGLI 26 | 7 | 20 | Panel | NM_005896.2 (IDH1):c.394C > T | Exon | IDH1 | NM_005896.2 | p.R132C | c.394C > T | 43 | 0.3 |
| CGLI 28 | 1 | 2,361 | Panel | NM_005896.2 (IDH1):c.395G > A | Exon | IDH1 | NM_005896.2 | p.R132H | c.395G > A | 33 | 33.0 |
| CGLI 29 | 3.5 | 206 | Panel | NM_000546.5 (TP53):c.742C > T | Exon | TP53 | NM_000546.5 | p.R248W | c.742C > T | 69 | 0.2 |
| CGLI 31 | 4 | 1,416 | Panel | NM_198253.2 (TERT):c.1–124C > T | Promoter | TERT | NM_198253.2 | NA | c.1–124C > T | 37 | 8.2 |
| CGLI 35 | 2.5 | 432 | Panel | NM_198253.2 (TERT):c.1–124C > T | Promoter | TERT | NM_198253.2 | NA | c.1–124C > T | 46 | 1.4 |
| CGLI 36 | 5.5 | 448 | Panel | NM_000546.5 (TP53):c.742C > T | Exon | TP53 | NM_000546.5 | p.R248W | c.742C > T | 65 | 14.3 |
| CGLI 39 | 8.5 | 154 | WES | NM_000268.3 (NF2):c.592C > T | Exon | NF2 | NM_000268.3 | p.R198X | c.592C > T | 80 | Not detected |
| CGLI 40 | 1.5 | 1,169 | WES | NM_000388.3 (CASR):c.2549C > T | Exon | CASR | NM_000388.3 | p.A850V | c.2549C > T | 45 | 2.8 |
| CGLI 41 | 4.25 | 87 | Panel | NM_000546.5 (TP53):c.396G > C | Exon | TP53 | NM_000546.5 | p.K132N | c.396G > C | 43 | 1.4 |
| CGLI 42 | 0.75 | 20 | WES | NM_001848.2 (COL6A1):c.1417G > A | Exon | COL6A1 | NM_001848.2 | p.G473R | c.1417G > A | 39 | 0.5 |
| CGLI 43 | 4.5 | 100 | WES | NM_000268.3 (NF2):c.1009C > T | Exon | NF2 | NM_000268.3 | p.Q337* | c.1009C > T | 61 | Not detected |
| CGLI 44 | 1.5 | 64 | WES | NM_001004439.1 (ITGA11):c.2140C > T | Exon | ITGA11 | NM_001004439.1 | p.R714* | c.2140C > T | 16 | Not detected |
| CGLI 47 | 6.8 | 29 | Panel | NM_198253.2 (TERT):c.1–124C > T | Promoter | TERT | NM_198253.2 | NA | c.1–124C > T | 44 | 22.1 |
| CGLI 48 | 5 | 1,400 | Panel | NM_198253.2 (TERT):c.1–124C > T | Promoter | TERT | NM_198253.2 | NA | c.1–124C > T | 52 | 1.0 |
| CGLI 50 | 5.6 | 820 | Panel | NM_198253.2 (TERT):c.1–124C > T | Promoter | TERT | NM_198253.2 | NA | c.1–124C > T | 47 | 18.8 |
| CGLI 51 | 0.75 | 64 | Panel | NM_198253.2 (TERT):c.1–124C > T | Promoter | TERT | NM_198253.2 | NA | c.1–124C > T | 10 | 5.9 |
| CGLI 55 | 9 | 146 | WES | NM_000314.4 (PTEN):c.388C > T | Exon | PTEN | NM_000314.4 | p.R130* | c.388C > T | 66 | 33.2 |
| CGLI 56 | 6.5 | 1,155 | WES | NM_014691.2 (AQR):c.1746delT | Exon | AQR | NM_014691.2 | p.P582fs | c.1746delT | 39 | 1.8 |
| CGLI 58 | 9 | 90 | WES | NM_052853.3 (ADCK2):c.1057T > A | Exon | ADCK2 | NM_052853.3 | p.F353I | c.1057T > A | 23 | Not detected |
| CGLI 60 | 8 | 1,335 | WES | NM_004687.4 (MTMR4):c.2110C > G | Exon | MTMR4 | NM_004687.4 | p.L704V | c.2110C > G | 40 | 18.7 |
| CGLI 61 | 3 | 99 | WES | NM_001122679.1 (TENM2):c.799G > A | Exon | TENM2 | NM_001122679.1 | p.A267T | c.799G > A | 21 | Not detected |
| CGLI 63 | 4 | 411 | WES | NM_000264.3 (PTCH1):c.746+1C > A | Splice site | PTCH1 | NM_000264.3 | NA | c.746+1C > A | 71 | Not detected |
| CGLI 101 | 3.5 | 14 | Panel | NM_000546.5 (TP53):c.731G > A | Exon | TP53 | NM_000546.5 | p.G244D | c.731G > A | 36 | 0.9 |
| CGLI 254 | 2.5 | 168 | WES | NM_021140.2 (KDM6A):c.4153C > T | Exon | KDM6A | NM_021140.2 | p.Q1385X | c.4153C > T | 91 | 77.3 |
NA, not available; panel, directed sequencing of codons 130–139 of IDH1; codons 126–155, 144–178, and 250–262 of IDH2; all coding exons of TP53; and the TERT promoter (Materials and Methods).
The presence of one of the mutations detected in each patient’s tumor was then assessed in the CSF of the same patient using a sensitive sequencing-based method. This method reliably detects mutations with allele fractions as low as 0.01% (8, 19). An average of 4.8 mL CSF (SD of 2.6) was collected from 35 patients (Table S2). DNA could be purified from all CSF samples, although the amounts varied considerably (average of 417 ng; SD of 553 ng) (Table S2). Primers were designed to amplify each of 35 mutations as previously described (8, 19). Using this technology, we found that 74% of 35 CSF samples contained detectable levels of tumor DNA. The detectability of tumor DNA present in the CSF was not correlated with demographic characteristics, symptom duration, presence of hydrocephalus, contrast enhancement on imaging, or mutation type (Table S3). The fraction of mutant alleles in the CSF was, as expected, usually lower than the fraction in the primary tumors, and it was also much more variable than in the primary tumors. The average detectable mutant allele fraction in CSF was 12.2% (range = 0.1–77%).
Table S3.
Associations between clinical characteristics and detection of CSF-tDNA
| Characteristic assessed | All patients (n = 35) | CSF undetected (n = 9) | CSF detected (n = 26) | P value |
| Age (y) median (range) | 32 (3–84) | 32 (6–66) | 32.5 (3–84) | 0.65 |
| Sex, no. (%) | 1 | |||
| Female | 16 (46) | 4 (44) | 12 (46) | |
| Male | 19 (54) | 5 (55) | 14 (54) | |
| Recurrent, no. (%) | 1 | |||
| No | 29 (83) | 8 (89) | 21 (81) | |
| Yes | 6 (17) | 1 (11) | 5 (19) | |
| Race, no. (%) | 0.08 | |||
| A | 3 (9) | 2 (22) | 1 (4) | |
| AA | 3 (9) | 0 (0) | 3 (12) | |
| ME | 3 (9) | 2 (22) | 1 (4) | |
| W | 26 (74) | 5 (56) | 21 (81) | |
| Hydrocephalus, no. (%) | 0.65 | |||
| No | 28 (80) | 8 (89) | 20 (77) | |
| Yes | 7 (20) | 1 (11) | 6 (23) | |
| Tumor grade, no. (%) | 0.01 | |||
| 1 | 6 (17) | 3 (33) | 3 (12) | |
| 2 | 9 (26) | 5 (56) | 4 (15) | |
| 3 | 2 (6) | 0 (0) | 2 (8) | |
| 4 | 17 (49) | 1 (11) | 16 (62) | |
| NA | 1 (3) | 0 (0) | 1 (4) | |
| Tumor grade, no. (%) | 0.004 | |||
| 1 and 2 | 15 (43) | 8 (89) | 7 (27) | |
| 3 and 4 | 19 (54) | 1 (11) | 18 (69) | |
| NA | 1 (3) | 0 (0) | 1 (4) | |
| Enhancing CT or MRI, no. (%) | 0.17 | |||
| No | 3 (9) | 2 (22) | 1 (4) | |
| Yes | 30 (86) | 7 (78) | 23 (88) | |
| NA | 2 (6) | 0 (0) | 2 (8) | |
| Tumor location, no. (%) | 0.16 | |||
| Infratentorial | 14 (40) | 4 (44) | 10 (38) | |
| Spinal | 13 (37) | 5 (56) | 8 (31) | |
| Supratentorial | 8 (23) | 0 (0) | 8 (31) | |
| Abutting CSF space, no. (%) | <0.001 | |||
| No | 5 (14) | 5 (56) | 0 (0) | |
| Yes | 28 (80) | 4 (44) | 24 (92) | |
| NA | 2 (6) | 0 (0) | 2 (8) | |
| Sequencing method, no. (%) | 0.01 | |||
| Panel | 13 (37) | 0 (0) | 13 (50) | |
| WES | 22 (63) | 9 (100) | 13 (50) | |
| Location, no. (%) | 0.1 | |||
| Exon | 28 (80) | 8 (89) | 20 (77) | |
| Promoter | 6 (17) | 0 (0) | 6 (23) | |
| Splice site | 1 (3) | 1 (11) | 0 (0) | |
| Location, no. (%) | 0.3 | |||
| Promoter | 6 (17) | 0 (0) | 6 (23) | |
| Exon and splice site | 29 (83) | 9 (100) | 20 (77) | |
| Symptom duration | ||||
| Median (range) | 3 (0.5–36) | 3 (1–33) | 2.5 (0.5–36) | 0.65 |
| Tumor size | ||||
| Median (range) | 34.8 (2–230) | 11.6 (2–152) | 37.4 (2.45–230) | 0.41 |
| CSF volume (mL) | ||||
| Median (range) | 5 (0.75–10) | 4.5 (1.5–9) | 5 (0.75–10) | 0.88 |
| Quantity of DNA in CSF (ng) | ||||
| Median (range) | 168 (6–2,361) | 100 (8–482) | 196 (6–2,361) | 0.03 |
| Mutant allele fraction in tumor | ||||
| Median (range) | 0.45 (0.1–0.91) | 0.49 (0.13–0.80) | 0.44 (0.10–0.91) | 0.68 |
A, Asian; AA, African American; CT, computed tomography; ME, Middle Eastern; NA, not available; panel, directed sequencing of codons 130–139 of IDH1; codons 126–155, 144–178, and 250–262 of IDH2; all coding exons of TP53; and the TERT promoter (Materials and Methods); W, white.
Relationship Between Mutations and Clinical Features.
The great variation in mutant allele fraction among the CSF samples suggested that there might be some anatomical or biological factor underlying the differences. The tumors were distributed among the brain and spinal cord (Table 1), and malignancies arising in both organs were detected at similar frequencies (P = 0.16; t test). High-grade (WHO grades III and IV) tumors were more likely to have detectable CSF-tDNA than low-grade lesions (P = 0.004) (Table S3), which was evidenced by the fact that all but one high-grade tumor (18 of 19) was detected. The levels of CSF-tDNA were also higher in high-grade lesions than in low-grade lesions (mutant allele fractions of 16.3 ± 21.2% vs. 2.8 ± 6.8%). Eighteen of 19 (∼95%) high-grade (WHO grade III or IV) tumors had detectable levels of CSF-tDNA. However, tumor size was not a statistically significant factor in predicting CSF-tDNA detectability or level (P = 0.41) (Table S3).
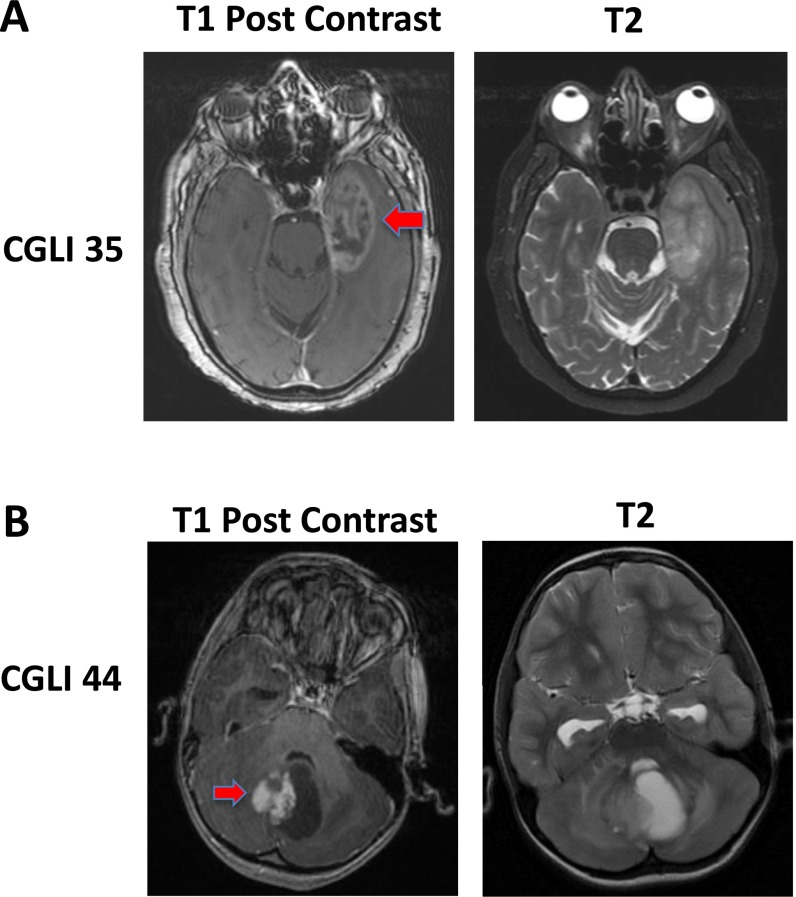
Another important factor associated with CSF-tDNA levels was anatomic location. MRI scans were examined for the presence of contrast enhancement adjacent to a large CSF space (Table 1). Representative examples are provided in Fig. S1. Patients with lesions adjacent to a CSF reservoir in the brain or spinal cord were much more likely to have detectable levels of CSF-tDNA than those with the remaining lesions. Such reservoirs included the cortical surfaces and ventricles as well as the basal and other cisterns. Accordingly, 86% of 28 cases in which tumors were adjacent to a CSF reservoir had detectable levels of CSF-tDNA. These cases included all 13 high-grade gliomas, all 3 ependymomas, and all 5 medulloblastomas that were in contact with the CSF. The four tumors in CSF contact that were not detectable were all low-grade gliomas. Moreover, zero of five patients whose tumors were entirely encapsulated by the brain or spinal cord parenchyma had detectable levels of CSF-tDNA (P < 0.001) (Table S2). On multivariate logistic regression, only the location of tumors with respect to CSF and the tumor grade were statistically significant (Table S4).
Fig. S1.
Representative magnetic resonance images. (A) An example of a tumor (red arrow) abutting a CSF space is shown. (B) An example of tumor (red arrow) not in contact with a CSF space is shown. Corresponding T2 images are provided for easier visualization of CSF.
Table S4.
Univariate and multivariate analyses to identify clinical characteristics associated with the detection of CSF-tDNA
| Characteristic assessed in univariate or multivariate analysis | Odds ratio (95% confidence interval) | P value |
| Univariate logistic regression | ||
| Sex, female vs. male | 1.07 (0.23–4.92) | 0.93 |
| Recurrent, yes vs. no | 1.91 (0.19–18.93) | 0.58 |
| Race | ||
| A vs. W | 0.15 (0.01–1.92) | 0.15 |
| AA vs. W | 1.79 (0.05–62.48) | 0.75 |
| ME vs. W | 0.15 (0.01–1.92) | 0.15 |
| AA vs. A | 11.67 (0.19–736) | 0.25 |
| ME vs. A | 1.00 (0.04–27.27) | 1 |
| ME vs. AA | 0.09 (0.001–5.40) | 0.25 |
| Hydrocephalus, yes vs. no | 2.40 (0.25–23.24) | 0.45 |
| Tumor grade, III and IV vs I and II | 20.57 (2.16–196) | 0.01 |
| Enhancing imaging, yes vs. no | 6.57 (0.52–83.75) | 0.15 |
| Tumor location | ||
| Spinal vs. infratentorial | 0.66 (0.13–3.27) | 0.61 |
| Supratentorial vs. infratentorial | 7.29 (0.29–185) | 0.23 |
| Supratentorial vs. spinal | 11.0 (0.44–276) | 0.14 |
| Abutting CSF space, yes vs. no | 59.9 (2.14–1,669) | 0.02 |
| Sequencing method, panel vs. WES | 19.0 (0.90–401) | 0.06 |
| Location, promoter vs. exon and splice site | 6.03 (0.25–148) | 0.27 |
| Age | 1.01 (0.98–1.04) | 0.64 |
| Symptom duration | 0.98 (0.91–1.06) | 0.63 |
| Tumor size | 1.01 (0.99–1.02) | 0.41 |
| CSF volume | 0.98 (0.72–1.32) | 0.88 |
| Quantity of DNA in CSF | 1.00 (1.00–1.01) | 0.2 |
| Mutant allele fraction in tumor | 3.55 (0.04–298) | 0.58 |
| Multivariate logistic regression | ||
| Tumor grade, III and IV vs I and II | 26.51 (1.45–485) | 0.03 |
| CSF space, yes vs. no | 90.59 (1.07–7,670) | 0.05 |
A, Asian; AA, African American; ME, Middle Eastern; panel, directed sequencing of codons 130–139 of IDH1; codons 126–155, 144–178, and 250–262 of IDH2; all coding exons of TP53; and the TERT promoter (Materials and Methods); W, white.
Genome-Wide Sequencing of DNA from the CSF.
The results described above were found after identifying at least one mutation in the primary tumor of each patient. In four patients with either brainstem or intramedullary spinal cord tumors, we also tested whether CSF-tDNA could be detected directly in their CSF by WES without prior knowledge of the tumor genotype. These four samples were selected based on the critical and highly sensitive location of the malignancies, making surgery treacherous. We found that two of four cases analyzed had levels of CSF-tDNA that were comparable with the levels identified through single-amplicon sequencing (Safe-SeqS) when the same mutation was assessed (Table 2). Both detectable cases had greater than 10% mutant allele fractions in the CSF as measured by single-amplicon sequencing. In contrast, the two cases in which WES was unable to identify CSF-tDNA had mutant allele fractions <1% as assessed by single-amplicon sequencing. As controls, we also performed WES on matched normal tissues and tumor tissues. The mutations were found in the tumors at a high frequency, but they were absent in normal tissues.
Table 2.
Detection of CSF-tDNA using WES
| Patient and sample type | Mutation | Genomic coordinate | Distinct coverage (SafeSeqS) | Mutant (%; SafeSeqS) | Distinct coverage (WES) | Mutant (%; WES) |
| CGLI 03 | ||||||
| CSF | PIK3R1 p.N564D, c.A1690G | Chr5:67591097 | 57,921 | 0.1 | 76 | 0.0 |
| Normal | PIK3R1 p.N564D, c.A1690G | Chr5:67591097 | 284 | 0.0 | 61 | 0.0 |
| Primary tumor | PIK3R1 p.N564D, c.A1690G | Chr5:67591097 | 377 | 38.7 | 147 | 12.9 |
| CGLI 29 | ||||||
| CSF | TP53 p.R248W, c.C742T | Chr17:7577539 | 13,964 | 0.2 | 64 | 0.0 |
| Normal | TP53 p.R248W, c.C742T | Chr17:7577539 | NA | 0.0 | 55 | 0.0 |
| Primary tumor | TP53 p.R248W, c.C742T | Chr17:7577539 | 6,418 | 69.0 | 58 | 70.7 |
| CGLI 36 | ||||||
| CSF | TP53 p.R248W, c.C742T | Chr17:7577539 | 376,434 | 14.3 | 74 | 9.5 |
| Normal | TP53 p.R248W, c.C742T | Chr17:7577539 | 57,818 | 0.0 | 75 | 0.0 |
| Primary tumor | TP53 p.R248W, c.C742T | Chr17:7577539 | 44,981 | 65.0 | 25 | 72.0 |
| CGLI 55 | ||||||
| CSF | PTEN p.R130*, c.C388T | Chr10:89692904 | 251,609 | 33.2 | 63 | 42.9 |
| Primary tumor | PTEN p.R130*, c.C388T | Chr10:89692904 | 91,515 | 65.9 | 56 | 66.1 |
Genomic coordinates refer to the human reference genome hg19 release (Genome Reference Consortium GRCh37, February of 2009). NA, not available.
Discussion
Minimally invasive techniques to monitor disease burden have been a challenge for many diseases of the CNS, including cancer. This challenge is highlighted by the high risks associated with neurosurgical procedures and the widely recognized limitations of current imaging modalities. In cancer patients, there is no reliable way of parsing out treatment effects from tumor recurrence, causing many patients to undergo unnecessary repeat surgeries. For example, in ∼30% of patients with glioblastoma who undergo a repeat resection for presumptive recurrence, pathologic examination of the resected specimen reveals necrosis, scarring, or other treatment-related effects rather than recurrent disease (20). Conversely, while patients are waiting for or recovering from surgery for suspicious lesions, chemotherapy or radiation therapy cannot be administered, providing time for unabated tumor growth. Finally, patients are often kept on ineffective medication regimens until definitive signs of tumor progression appear on imaging. This delay in detection precludes potential opportunities to undergo new targeted therapies that might be effective for their disease (21). The health costs of these missed opportunities will increase with the expected advances in therapeutic modalities.
Given the need for sensitive and specific markers to monitor tumor dynamics, we asked whether tumor-derived DNA could be found in the CSF of patients with primary CNS tumors. This study was stimulated by our inability to consistently detect ctDNA in the plasma of these patients (8) and inspired by previous demonstrations that tumor-derived DNA could be found in fluids located in the proximity of neoplastic lesions. For example, a recent pilot study by Pan et al. (22) suggests that tumor-derived DNA can be detected in the spinal fluid of individuals whose primary tumors have metastasized to the brain. Although lumbar puncture to obtain CSF is not a noninvasive procedure, it qualifies as minimally invasive and is currently routinely performed to follow some brain tumor patients, particularly those with medulloblastomas (23, 24). Unfortunately, the examination of these CSF samples by cytology is usually of limited use, with relatively low sensitivities achieved even using large volumes of CSF (25, 26). Only 1 of 35 patients evaluated in our study had concomitant cytologic studies of CSF, precluding direct comparison. The results of this study suggest that the rates of tumor-derived DNA found in the CSF (74%) closely approximate the levels found in body fluids adjacent to other tumor types. For example, urine in bladder cancer was found to have tumor-derived DNA in 70% of cases, whereas sputum in lung cancer was positive in 79% of cases (27, 28). Although the rate of detection observed in this study was not 100%, its sensitivity was comparable with or superior to other noninvasive tests for malignancies in general. Moreover and as noted below, it was particularly sensitive for tumors that abutted a CSF reservoir or cortical surface. Finally, from a technological standpoint, the average fraction of mutant DNA (12.2%) far exceeded the limit of detection of the sequencing assay used (0.01%). This assay could be performed with any commercially available next generation sequencing instrument at relatively small cost.
Our study revealed a significant association between the location and type of the tumor and the presence of CSF-tDNA. In particular, we were able to detect all 13 WHO grade III or IV gliomas (also known as anaplastic astrocytoma and glioblastoma, respectively), all 5 medulloblastomas, and all 3 ependymomas that abutted a CSF reservoir or cortical surface. It is in these aggressive tumors where the need for a robust biomarker is most desperate. There are also emerging data that some brain tumors, particularly those with genotypes susceptible to targeted therapies, may be able to be treated primarily with medical therapies, thereby obviating the need for surgery if appropriate noninvasive diagnostic tools were available (29–32). It is also worth noting that surgical resection nearly always creates an opening extending from the surface to the deep-seated tumor. This passageway typically persists and may enable tDNA from any residual or recurrent tumor to enter the CSF. Even without such surgically induced openings, the vast majority of medulloblastomas and ependymomas arise within or communicate with a ventricular reservoir, making them well-suited for CSF monitoring (24–26, 33, 34). Future studies will be required to directly compare CSF-tDNA with CSF cytology. Rather than replacing cytology, we envision that CSF-tDNA will be used in combination with it and other biomarkers under development as well as with radiographic and clinical parameters (35–38). This could substantially increase the accuracy of the estimates of tumor burden at various points during the management of patients.
Given the invasive and risky nature of surgical interventions on the brain and spinal cord, it would be useful to be able to identify a neoplastic process without performing surgery. Our results provide a glimpse of the potential for this form of diagnosis in the future. We evaluated four patients: one patient with a tumor in the midbrain, one patient with a tumor in the pons, and two patients with a tumor in the spinal cord. Using WES, we were able to detect CSF-tDNA in two of four cases by comparing the data with those obtained by targeted sequencing with SafeSeqS. The results were consistent with expectation, in that the mutant fractions revealed by genome-wide sequencing were in accord with those identified by targeted sequencing (Table 2). Additional cases will need to be tested to elucidate the potential of this approach in patients in whom biopsies are challenging, but our results show that genome-wide analysis of the DNA from CSF is feasible in at least some cases.
Although the results described above are promising, we caution that this is an exploratory study designed primarily to determine whether it was possible to detect CSF-tDNA in patients with primary CNS tumors. A secondary goal was to document the anatomical and pathologic characteristics of the tumors that shed DNA into the CSF. The most important technical limitation of our study is that CSF samples were obtained at the time of surgery, and they were often from the ventricles rather than from a lumbar puncture. CSF has been shown to quickly circulate throughout the ventricles and spinal reservoirs (39, 40). It is, therefore, very likely that the DNA in the spinal fluid obtained through lumbar puncture will be similar to that of the ventricles, although the fluid obtained from lumbar puncture is farther away from the site of malignancy. An additional consideration is that, in individuals with a bulky mass that obstructs spinal fluid flow or elevates intracranial pressure, a lumbar puncture might be unsafe. However, these patients will almost always require surgical decompression to reduce the mass effect generated by the tumor, and CSF could be safely obtained after opening the dura. The exact method and location of CSF sampling in patients with CNS neoplasms will need to be individualized, and they will be based on a number of factors, including tumor location, ease of CSF sampling, and clinical characteristics. For example, patients may initially undergo CSF sampling from an intracranial space at the time of surgery to determine baseline levels of CSF-tDNA, but lumbar punctures could be used to longitudinally monitor CSF-tDNA levels.
Now that it has been documented that most primary brain tumors release tDNA into the CSF, the stage is set for a longitudinal study of the clinical use of this biomarker. Our results suggest specific guidelines for such a follow-up study. The optimal patients to follow would be those with medulloblastomas, ependymomas, or high-grade gliomas that abut a CSF space, because the CSF-tDNA assay is particularly sensitive in such cases and these tumor types are relatively common. CSF-tDNA should be evaluated intraoperatively to establish a baseline, and a concomitant lumbar puncture should be performed when possible to ensure concordance between the two fluid samples. Subsequent evaluations of CSF obtained through lumbar puncture or an implanted reservoir should be compared with other clinical and laboratory features, with the goal of determining the use of CSF-tDNA to detect minimal residual disease. For example, patients whose mass persists on MRI but CSF-tDNA is undetectable might be spared a second biopsy. Alternatively, patients in whom residual disease is evident on CSF-tDNA analysis but equivocal on imaging analysis might be well-served by additional therapy. In the future, it is likely that most brain tumors will be routinely assessed for mutations in various genes of interest for both prognostic and therapeutic purposes (41–43). The availability of such sequencing data should make the approach described here more cost-effective and easier to implement.
Materials and Methods
Patient Samples.
All samples were collected after approval was obtained from the Johns Hopkins Institutional Review Board and informed consent was provided. Whole blood and CSF were collected at the time of surgery before surgical manipulation of the tumor. A WBC pellet was prepared from the blood sample after hypotonic lysis of RBCs by centrifugation at 200 × g. CSF was frozen in its entirety at −80 °C until DNA purification, and the entire volume of CSF (cells plus fluid) was used for DNA purification. The amount of CSF used averaged 4.8 mL (range = 0.75–10 mL). When fresh tumor tissue from surgical specimens was available, it was immediately frozen at −80 °C. When frozen tissue was not available, formalin-fixed, paraffin-embedded tissues were used for DNA purification. In either case (fresh frozen or formalin-fixed, paraffin-embedded), tumors were macrodissected to ensure neoplastic cellularity exceeding 50%. DNA was purified from the white cell pellet, CSF, and tumor using an ALLPrep Kit (Qiagen).
Statistical Analysis.
Clinical characteristics were compared between the CSF samples with and without detectable CSF-tDNA with Fisher’s exact test or t test. Correlation coefficients among outcomes were estimated using Pearson correlation statistics. A logistic regression model was used to estimate the odds of detecting CSF-tDNA under different conditions. All P values are two-sided, and all analyses were conducted using SAS software (version 9.2; SAS Institute).
Tumor Mutational Profiling.
A tiered approach was used to determine a somatic mutation within each tumor. Initially, a PCR-based approach testing for mutations in codons 130–139 of IDH1; codons 126–155, 144–178, and 250–262 of IDH2; all coding exons of TP53; and the TERT promoter was used (44–48). If no mutations were present within these genes, paired-end libraries of DNA from the tumors and WBC pellets were prepared and captured (SureSelect; Agilent) as previously described (47). Massively parallel sequencing was carried out on an Illumina HiSeq Instrument at either the Goldman Sequencing Facility at Johns Hopkins Medical Institutions or Personal Genome Diagnostics. Mutations were identified as previously described (47, 49–52).
Mutation Detection in CSF.
DNA from tumor, WBCs, and CSF was used to validate the somatic mutations identified by targeted sequencing and determine whether these mutations could be found in the CSF; 3–5 ng tumor and WBC DNA was used for each assay, whereas all DNA from the CSF (for cases with <20 ng CSF DNA available) or 20 ng CSF DNA was used for each assay (Table S2). For this purpose, primers were designed to amplify an ∼100-bp region surrounding each mutation. The two primers had universal sequences at their 5′ ends, allowing a second round of PCR to be performed using a second set of primers containing these sequences (19, 47). The sequences of the primers used to assess each mutation are listed in Table S5. Oligonucleotides used in this study were synthesized by TriLink Biotechnologies. The final PCR products (after two rounds of PCR) were purified with AMPure (Beckman) and sequenced using an Illumina MiSeq Instrument. The data were analyzed with the SafeSeqS Pipeline, allowing mutations occurring as infrequently as 0.01% to be detected and quantified with confidence using the experimental conditions applied (19). In every case, DNA from the normal cells served as a control to ensure that the mutations were not the result of errors generated during the DNA purification, amplification, or sequencing processes. Four paired-end libraries for CSF samples were also generated and exome-captured (Table 2). Preparation of the genomic library was performed using the TruSeq DNA Sample Prep Kit (Illumina) according to the manufacturer’s recommendations. Exomic capture (SureSelect; Agilent) and massively parallel sequencing were carried out as described above.
Table S5.
Primers used for mutation detection
| Patient | Gene | Mutation | Location | Forward primer sequence | Reverse primer sequence |
| CGLI 02 | ZNF513 | NM_144631.5 (ZNF513):c.413C > T | Exon | CACACAGGAAACAGCTATGACCATGCTTCAGGTGGCTCGAGTAGTG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNGTGAGGGGCCGTGTTGT |
| CGLI 03 | PIK3R1 | NM_181523.2 (PIK3R1):c.1690A > G | Exon | CACACAGGAAACAGCTATGACCATGTTTCTTTTGCCTGCAGGATT | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNCCTGAATTGTAGCAATCACCAA |
| CGLI 06 | CLCA3P | NR_024604.1 (CLCA3P):c.446G > A | Exon | CACACAGGAAACAGCTATGACCATGTGGACAATGTGGAGATAAAGGAC | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNCCACCAGTGGAAATAAGAACAGA |
| CGLI 11 | ANKS3 | NM_133450.3 (ANKS3):c.1738G > A | Exon | CACACAGGAAACAGCTATGACCATGCTGGTCCTGCCTCACTCG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNTCCCTCCCCTAGAAGGTCTG |
| CGLI 12 | HIST1H3C | NM_003531.2 (HIST1H3C):c.83T > A | Exon | CACACAGGAAACAGCTATGACCATGGAAGCAAACAGCTCGCAAGT | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNGCGGTAGCGATGAGGTTTC |
| CGLI 13 | TTC16 | NM_144965.1 (TTC16):c.1016T > A | Exon | CACACAGGAAACAGCTATGACCATGGCCAGCTGTTGCTGACCTA | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNAGAGTCCTTTCTCCTGCTGCT |
| CGLI 14 | TP53 | NM_000546.5 (TP53):c.638G > A | Exon | CACACAGGAAACAGCTATGACCATGAGACCTCAGGCGGCTCATAG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNTTGCGTGTGGAGTATTTGGA |
| CGLI 15 | MARS | NM_004990.3 (MARS):c.1487C > T | Exon | CACACAGGAAACAGCTATGACCATGGAGTGGTTGGGGAGGACATT | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNGTTCCCCATTTGAGGTCTCG |
| CGLI 20 | RGS12 | NM_198229.2 (RGS12):c.2980C > T | Exon | CACACAGGAAACAGCTATGACCATGCTGTCAAGGCGGGCTTCT | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNGCAGCACGAGGGTCAGG |
| CGLI 22 | CC2D2A | NM_001080522.2 (CC2D2A):c.1938G > A | Exon | CACACAGGAAACAGCTATGACCATGATCGGGCAGTGATAGAGCAG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNTCATTGGGTGTTACGCTTCC |
| CGLI 25 | CDH5 | NM_001795.3 (CDH5):c.1211G > A | Exon | CACACAGGAAACAGCTATGACCATGGAAGCCTCTGATTGGCACAG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNAGCAGGCATAGGAACATCCA |
| CGLI 26 | IDH1 | NM_005896.2 (IDH1):c.394C > T | Exon | CACACAGGAAACAGCTATGACCATGGCTTGTGAGTGGATGGGTAAAACCTATCAT | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNATGCAAAATCACATTATTGCCAACATGACT |
| CGLI 28 | IDH1 | NM_005896.2 (IDH1):c.395G > A | Exon | CACACAGGAAACAGCTATGACCATGGCTTGTGAGTGGATGGGTAAAACCTATCAT | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNATGCAAAATCACATTATTGCCAACATGACT |
| CGLI 29 | TP53 | NM_000546.5 (TP53):c.742C > T | Exon | CACACAGGAAACAGCTATGACCATGTCCACTACAACTACATGTGTAACAGTTC | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNTGTGATGATGGTGAGGATGG |
| CGLI 31 | TERT | NM_198253.2 (TERT):c.1–124C > T | Promoter | CACACAGGAAACAGCTATGACCATGGCGGAAAGGAAGGGGAG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNCCCGTCCCGACCCCT |
| CGLI 35 | TERT | NM_198253.2 (TERT):c.1–124C > T | Promoter | CACACAGGAAACAGCTATGACCATGGCGGAAAGGAAGGGGAG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNCCCGTCCCGACCCCT |
| CGLI 36 | TP53 | NM_000546.5 (TP53):c.742C > T | Exon | CACACAGGAAACAGCTATGACCATGTGGCTCTGACTGTACCACCATC | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNGTGGCAAGTGGCTCCTGA |
| CGLI 39 | NF2 | NM_000268.3 (NF2):c.592C > T | Exon | CACACAGGAAACAGCTATGACCATGCGGAAATGTGGGAGGAGAG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNAAAGCCCATAAAGGAATGTAAACC |
| CGLI 40 | CASR | NM_000388.3 (CASR):c.2549C > T | Exon | CACACAGGAAACAGCTATGACCATGCCAGCACCTATGGCAAGTTT | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNCGGGATGGCTTGAAGAGA |
| CGLI 41 | TP53 | NM_000546.5 (TP53):c.396G > C | Exon | CACACAGGAAACAGCTATGACCATGGCCCTGACTTTCAACTCTGTCT | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNGGGGGTGTGGAATCAACC |
| CGLI 42 | COL6A1 | NM_001848.2 (COL6A1):c.1417G > A | Exon | CACACAGGAAACAGCTATGACCATGGTCCAACGTGCCATATCCAT | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNGGTGGGGAGTGGGACTCA |
| CGLI 43 | NF2 | NM_000268.3 (NF2):c.1009C > T | Exon | CACACAGGAAACAGCTATGACCATGCCTTGTGGCACCCTAGGTC | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNGTTCAGCCTCCTCCCTCATC |
| CGLI 44 | ITGA11 | NM_001004439.1 (ITGA11):c.2140C > T | Exon | CACACAGGAAACAGCTATGACCATGGGAAGTTGATCCGCTCACA | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNCATCAGATACAACGCCACCAT |
| CGLI 47 | TERT | NM_198253.2 (TERT):c.1–124C > T | Promoter | CACACAGGAAACAGCTATGACCATGGCGGAAAGGAAGGGGAG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNCCCGTCCCGACCCCT |
| CGLI 48 | TERT | NM_198253.2 (TERT):c.1–124C > T | Promoter | CACACAGGAAACAGCTATGACCATGGCGGAAAGGAAGGGGAG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNCCCGTCCCGACCCCT |
| CGLI 50 | TERT | NM_198253.2 (TERT):c.1–124C > T | Promoter | CACACAGGAAACAGCTATGACCATGGCGGAAAGGAAGGGGAG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNCCCGTCCCGACCCCT |
| CGLI 51 | TERT | NM_198253.2 (TERT):c.1–124C > T | Promoter | CACACAGGAAACAGCTATGACCATGGCGGAAAGGAAGGGGAG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNCCCGTCCCGACCCCT |
| CGLI 55 | PTEN | NM_000314.4 (PTEN):c.388C > T | Exon | CACACAGGAAACAGCTATGACCATGGCAATTCACTGTAAAGCTGGAAA | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNTGGTCCTTACTTCCCCATAGAA |
| CGLI 56 | AQR | NM_014691.2 (AQR):c.1746delT | Exon | CACACAGGAAACAGCTATGACCATGTCTGACATAAACCAGGCCAAC | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNACCAGGTCTTCGTAAGCATGA |
| CGLI 58 | ADCK2 | NM_052853.3 (ADCK2):c.1057T > A | Exon | CACACAGGAAACAGCTATGACCATGCCGAGTCCTGGGAGTTTTG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNTACAGCTGAGGGGAGGAGAA |
| CGLI 60 | MTMR4 | NM_004687.4 (MTMR4):c.2110C > G | Exon | CACACAGGAAACAGCTATGACCATGCGAGGCACTGCGGTATTAAG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNTGGGTCTTCCTCCTCCTCTG |
| CGLI 61 | TENM2 | NM_001122679.1 (TENM2):c.799G > A | Exon | CACACAGGAAACAGCTATGACCATGACAGCCAGTCGACTCTGAGG | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNATCTGACTCCGCCGATTG |
| CGLI 63 | PTCH1 | NM_000264.3 (PTCH1):c.746+1C > A | Splice site | CACACAGGAAACAGCTATGACCATGAATGGAACAAACAATGATAAGCAA | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNGGAAGGGGCGAAATTACAGT |
| CGLI 101 | TP53 | NM_000546.5 (TP53):c.731G > A | Exon | CACACAGGAAACAGCTATGACCATGTCCACTACAACTACATGTGTAACAGTTC | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNTGTGATGATGGTGAGGATGG |
| CGLI 254 | KDM6A | NM_021140.2 (KDM6A):c.4153C > T | Exon | CACACAGGAAACAGCTATGACCATGACTCACATGTTGATTTGACTTACTAATGTA | CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNACAGTACAAAATGGAGGACCTGA |
Ns in primer sequences represent bases with an equal probability of A, C, T, and G.
Supplementary Material
Acknowledgments
We thank our patients for their courage and generosity. We also thank N. Silliman, J. Schaefer, C. Blair, K. Judge, and M. Popoli for technical assistance. This work was supported by a Burroughs Wellcome Career Award for Medical Scientists, Alex’s Lemonade Stand/Malia’s Cord Foundation, the Pediatric Brain Tumor Foundation, a Johns Hopkins Clinician Scientist Award, Doris Duke Charitable Foundation Grant 2014107, The Virginia and D. K. Ludwig Fund for Cancer Research, The Banyan Gate Foundation, Swim Across America, The Sol Goldman Sequencing Facility at Johns Hopkins, and NIH Grants CA43460 and NS70024.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1511694112/-/DCSupplemental.
References
- 1.Ostrom QT, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro-oncol. 2013;15(Suppl 2):ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kros JM, et al. Circulating glioma biomarkers. Neuro-oncol. 2014 doi: 10.1093/neuonc/nou207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Mieghem E, et al. Defining pseudoprogression in glioblastoma multiforme. Eur J Neurol. 2013;20(10):1335–1341. doi: 10.1111/ene.12192. [DOI] [PubMed] [Google Scholar]
- 4.Müller C, et al. Hematogenous dissemination of glioblastoma multiforme. Sci Transl Med. 2014;6(247):247ra101. doi: 10.1126/scitranslmed.3009095. [DOI] [PubMed] [Google Scholar]
- 5.Macarthur KM, et al. Detection of brain tumor cells in the peripheral blood by a telomerase promoter-based assay. Cancer Res. 2014;74(8):2152–2159. doi: 10.1158/0008-5472.CAN-13-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan JP, et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014;4(11):1299–1309. doi: 10.1158/2159-8290.CD-14-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diehl F, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson SJ, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 10.Newman AM, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martignetti JA, et al. Personalized ovarian cancer disease surveillance and detection of candidate therapeutic drug target in circulating tumor DNA. Neoplasia. 2014;16(1):97–103. doi: 10.1593/neo.131900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sidransky D, et al. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256(5053):102–105. doi: 10.1126/science.1566048. [DOI] [PubMed] [Google Scholar]
- 13.Sidransky D, et al. Identification of p53 gene mutations in bladder cancers and urine samples. Science. 1991;252(5006):706–709. doi: 10.1126/science.2024123. [DOI] [PubMed] [Google Scholar]
- 14.Ralla B, et al. Nucleic acid-based biomarkers in body fluids of patients with urologic malignancies. Crit Rev Clin Lab Sci. 2014;51(4):200–231. doi: 10.3109/10408363.2014.914888. [DOI] [PubMed] [Google Scholar]
- 15.Hubers AJ, Prinsen CF, Sozzi G, Witte BI, Thunnissen E. Molecular sputum analysis for the diagnosis of lung cancer. Br J Cancer. 2013;109(3):530–537. doi: 10.1038/bjc.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diehl F, et al. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology. 2008;135(2):489–498. doi: 10.1053/j.gastro.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinde I, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med. 2013;5(167):167ra4. doi: 10.1126/scitranslmed.3004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108(23):9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodworth GF, et al. Histopathological correlates with survival in reoperated glioblastomas. J Neurooncol. 2013;113(3):485–493. doi: 10.1007/s11060-013-1141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krueger DA, et al. Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis. N Engl J Med. 2010;363(19):1801–1811. doi: 10.1056/NEJMoa1001671. [DOI] [PubMed] [Google Scholar]
- 22.Pan W, Gu W, Nagpal S, Gephart MH, Quake SR. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61(3):514–522. doi: 10.1373/clinchem.2014.235457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Hoff K, Rutkowski S. Medulloblastoma. Curr Treat Options Neurol. 2012;14(4):416–426. doi: 10.1007/s11940-012-0183-8. [DOI] [PubMed] [Google Scholar]
- 24.Bartlett F, Kortmann R, Saran F. Medulloblastoma. Clin Oncol (R Coll Radiol) 2013;25(1):36–45. doi: 10.1016/j.clon.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Glass JP, Melamed M, Chernik NL, Posner JB. Malignant cells in cerebrospinal fluid (CSF): The meaning of a positive CSF cytology. Neurology. 1979;29(10):1369–1375. doi: 10.1212/wnl.29.10.1369. [DOI] [PubMed] [Google Scholar]
- 26.Preusser M, Hainfellner JA. CSF and laboratory analysis (tumor markers) Handb Clin Neurol. 2012;104:143–148. doi: 10.1016/B978-0-444-52138-5.00011-6. [DOI] [PubMed] [Google Scholar]
- 27.Allory Y, et al. Telomerase reverse transcriptase promoter mutations in bladder cancer: High frequency across stages, detection in urine, and lack of association with outcome. Eur Urol. 2014;65(2):360–366. doi: 10.1016/j.eururo.2013.08.052. [DOI] [PubMed] [Google Scholar]
- 28.Destro A, et al. K-ras and p16(INK4A)alterations in sputum of NSCLC patients and in heavy asymptomatic chronic smokers. Lung Cancer. 2004;44(1):23–32. doi: 10.1016/j.lungcan.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Mack SC, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506(7489):445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gajjar A, Pfister SM, Taylor MD, Gilbertson RJ. Molecular insights into pediatric brain tumors have the potential to transform therapy. Clin Cancer Res. 2014;20(22):5630–5640. doi: 10.1158/1078-0432.CCR-14-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudin CM, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361(12):1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson MC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24(12):1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 33.Moreno L, et al. Utility of cerebrospinal fluid cytology in newly diagnosed childhood ependymoma. J Pediatr Hematol Oncol. 2010;32(6):515–518. doi: 10.1097/MPH.0b013e3181d7adf5. [DOI] [PubMed] [Google Scholar]
- 34.Weston CL, Glantz MJ, Connor JR. Detection of cancer cells in the cerebrospinal fluid: Current methods and future directions. Fluids Barriers CNS. 2011;8(1):14. doi: 10.1186/2045-8118-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khwaja FW, et al. Proteomic identification of biomarkers in the cerebrospinal fluid (CSF) of astrocytoma patients. J Proteome Res. 2007;6(2):559–570. doi: 10.1021/pr060240z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy S, et al. Protein biomarker identification in the CSF of patients with CNS lymphoma. J Clin Oncol. 2008;26(1):96–105. doi: 10.1200/JCO.2007.12.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bougel S, et al. Methylation of the hTERT promoter: A novel cancer biomarker for leptomeningeal metastasis detection in cerebrospinal fluids. Clin Cancer Res. 2013;19(8):2216–2223. doi: 10.1158/1078-0432.CCR-12-1246. [DOI] [PubMed] [Google Scholar]
- 38.Samuel N, Remke M, Rutka JT, Raught B, Malkin D. Proteomic analyses of CSF aimed at biomarker development for pediatric brain tumors. J Neurooncol. 2014;118(2):225–238. doi: 10.1007/s11060-014-1432-3. [DOI] [PubMed] [Google Scholar]
- 39.Chamberlain MC, Kormanik PA, Glantz MJ. A comparison between ventricular and lumbar cerebrospinal fluid cytology in adult patients with leptomeningeal metastases. Neuro-oncol. 2001;3(1):42–45. doi: 10.1093/neuonc/3.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gajjar A, et al. Comparison of lumbar and shunt cerebrospinal fluid specimens for cytologic detection of leptomeningeal disease in pediatric patients with brain tumors. J Clin Oncol. 1999;17(6):1825–1828. doi: 10.1200/JCO.1999.17.6.1825. [DOI] [PubMed] [Google Scholar]
- 41.Thomas L, Di Stefano AL, Ducray F. Predictive biomarkers in adult gliomas: The present and the future. Curr Opin Oncol. 2013;25(6):689–694. doi: 10.1097/CCO.0000000000000002. [DOI] [PubMed] [Google Scholar]
- 42.Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol. 2014;232(2):165–177. doi: 10.1002/path.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gajjar AJ, Robinson GW. Medulloblastoma-translating discoveries from the bench to the bedside. Nat Rev Clin Oncol. 2014;11(12):714–722. doi: 10.1038/nrclinonc.2014.181. [DOI] [PubMed] [Google Scholar]
- 44.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 45.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Killela PJ, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110(15):6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bettegowda C, et al. Exomic sequencing of four rare central nervous system tumor types. Oncotarget. 2013;4(4):572–583. doi: 10.18632/oncotarget.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinde I, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013;73(24):7162–7167. doi: 10.1158/0008-5472.CAN-13-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang M, et al. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46(11):1170–1172. doi: 10.1038/ng.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agrawal N, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2(10):899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bettegowda C, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.