Abstract
The parasitic flagellate Trypanosoma vivax is a cause of animal trypanosomiasis across Africa and South America. The parasite has a digenetic life cycle, passing between mammalian hosts and insect vectors, and a series of developmental forms adapted to each life cycle stage. Each point in the life cycle presents radically different challenges to parasite metabolism and physiology and distinct host interactions requiring remodeling of the parasite cell surface. Transcriptomic and proteomic studies of the related parasites T. brucei and T. congolense have shown how gene expression is regulated during their development. New methods for in vitro culture of the T. vivax insect stages have allowed us to describe global gene expression throughout the complete T. vivax life cycle for the first time. We combined transcriptomic and proteomic analysis of each life stage using RNA-seq and mass spectrometry respectively, to identify genes with patterns of preferential transcription or expression. While T. vivax conforms to a pattern of highly conserved gene expression found in other African trypanosomes, (e.g. developmental regulation of energy metabolism, restricted expression of a dominant variant antigen, and expression of ‘Fam50’ proteins in the insect mouthparts), we identified significant differences in gene expression affecting metabolism in the fly and a suite of T. vivax-specific genes with predicted cell-surface expression that are preferentially expressed in the mammal (‘Fam29, 30, 42’) or the vector (‘Fam34, 35, 43’). T. vivax differs significantly from other African trypanosomes in the developmentally-regulated proteins likely to be expressed on its cell surface and thus, in the structure of the host-parasite interface. These unique features may yet explain the species differences in life cycle and could, in the form of bloodstream-stage proteins that do not undergo antigenic variation, provide targets for therapy.
Author Summary
Trypanosoma vivax is a single-celled parasite that infects cattle and non-domesticated animals through the bite of the tsetse fly. The parasite causes animal trypanosomiasis, a chronic condition resulting in severe anemia, muscle wastage and ultimately death if untreated. This disease is endemic across sub-Saharan Africa but has also spread to South America and causes considerable losses in animal productivity, impeding economic development in the world’s poorest nations. To develop new ways of preventing and treating animal trypanosomiasis, we need an accurate understanding of how the parasite causes disease. In this study, we present an analysis of gene expression throughout the T. vivax life cycle that compares the abundance of gene transcripts (mRNA) and proteins in the mammalian and insect hosts. We have identified genes that are preferentially expressed in each life stage, including many that are unique to T. vivax and probably expressed on its cell surface. Our findings provide a comprehensive understanding of how gene expression is regulated in T. vivax and further refine a pool of T. vivax-specific genes that could be exploited to prevent and treat animal trypanosomiasis.
Introduction
African trypanosomes are unicellular vector-borne hemoparasites of humans, domestic livestock and wild animals. They cause African trypanosomiasis, an endemic disease of sub-Saharan Africa otherwise known as sleeping sickness in humans and nagana in animals, and are transmitted between vertebrate hosts by the bite of tsetse flies (Glossina spp.). This endemic disease causes considerable morbidity in livestock herds and associated losses in animal productivity. The threat of Animal African trypanosomiasis in tsetse-infested areas also prevents effective exploitation of available pasture, thereby impeding economic development in the world’s poorest nations.
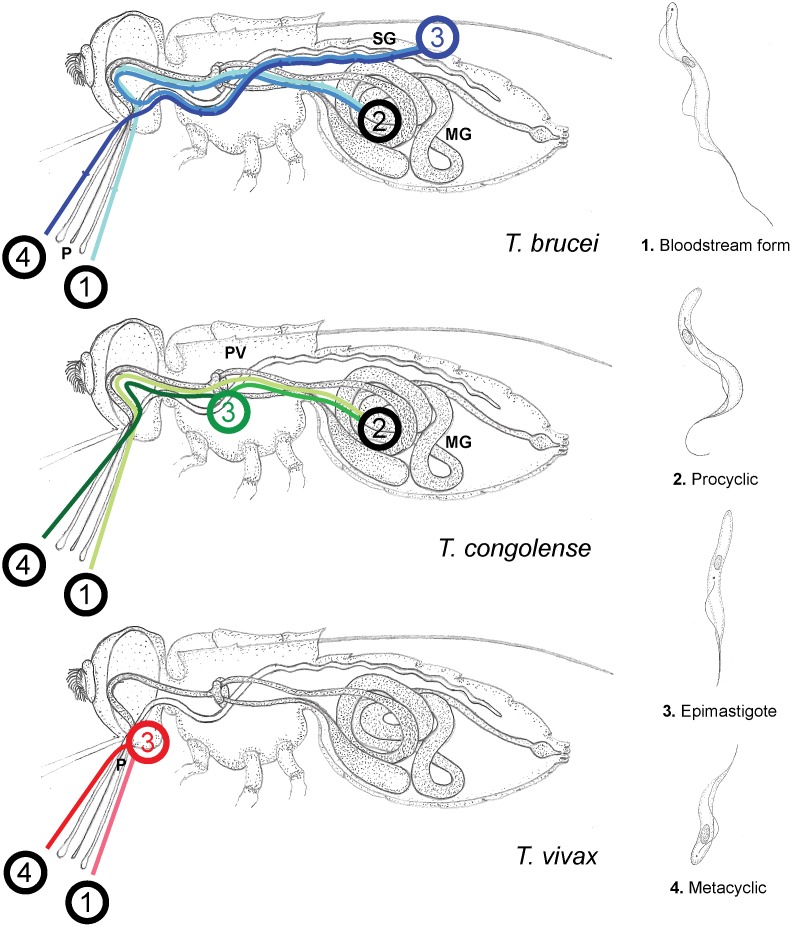
There are several species of African trypanosome that vary in life cycle, host range and pathology. Trypanosoma brucei is predominantly an animal pathogen that has evolved the ability to infect humans on multiple occasions [1], while T. congolense and T. vivax are exclusively animal pathogens. During their life cycles, T. brucei and T. congolense exist as procyclic forms in the mid-gut of the tsetse fly before migrating into the salivary glands and proventriculus respectively, where they develop into epimastigotes and then metacyclic trypomastigotes that are able to infect vertebrates (see Fig 1). In contrast, T. vivax lacks a procyclic stage in the insect mid-gut and has no complex migration within the insect; rather, T. vivax develops directly into epimastigote forms within the insect proboscis [2] (Fig 1). This difference might explain why T. vivax can be transmitted by other kinds of biting insect [3–4] and has therefore spread beyond the sub-Saharan distribution of the tsetse fly into northern Africa and South America [5–6].
Fig 1. Life cycle variation among African trypanosomes.
The passage and development of African trypanosomes through the fly vector varies by species. The figure shows the route taken by each species through the vector and position of each developmental stage. Insect anatomy is abbreviated thus: proboscis (P), mid-gut (MG), salivary gland (SG) and proventriculus (PV). The life cycle within the fly is defined here by four stages: (1) Ingestion of bloodstream-form parasites; (2) migration of parasites to the insect mid-gut with differentiation into procyclic forms (T. vivax lacks this stage); (3) migration anteriorly to the proboscis (T. vivax), proventriculus (T. congolense) or salivary gland (T. brucei) and differentiation into epimastigote forms; and (4) differentiation into metacyclic forms and inoculation into the vertebrate host upon insect feeding.
In recent years our understanding of trypanosome biology has progressed substantially through the determination of genome sequences for T. brucei [7–10] and for T. congolense and T. vivax [11], as well as numerous analyses of gene expression, (largely confined to T. brucei, except for three studies [12–14]), using transcriptomic [15–20] and proteomic [21–23] approaches. Consequently, we now know that the complex life cycle of African trypanosomes is facilitated by considerable developmental regulation of gene expression. Developmental regulation in T. brucei is particularly apparent in the expression of major surface glycoproteins belonging to the procyclic, epimastigote and bloodstream forms respectively, i.e. procyclin [24], B rucei Alanine-Rich Protein (BARP; [25]) and the Variant Surface Glycoprotein (VSG).
There are compelling reasons for supposing that gene expression in T. vivax will be different to T. brucei in important ways, not least due to differences in life cycle development (Fig 1), but also because the T. vivax genome contains substantially different repertoires of VSG and BARP-like genes (and no procyclin at all), as well as numerous gene families that appear to be unique [26]. As in vitro cultivation of insect stages has not previously been possible, gene expression in T. vivax has only been analyzed in the bloodstream form, and then only through transcriptomic analysis [14]. Moreover, given that gene regulation is achieved largely through post-transcriptional modifications in trypanosomes (reviewed in [27]), differences between transcript and peptide abundances across the life cycle are expected. We recently established in vitro cultures of the insect stages of T. vivax [28], and so a comparison of gene expression across African trypanosome species is now possible.
Using transcriptome sequencing and proteomics, we have analyzed differences in gene expression between T. vivax epimastigote, metacyclic and bloodstream forms. Our results show that the numerous T. vivax-specific genes predicted to function on the parasite cell surface are transcribed and often developmentally regulated. Genome-wide patterns of developmental regulation are conserved across African trypanosome species, with some notable exceptions concerning pyruvate metabolism in T. vivax, which might indicate an important species difference in energy metabolism. Comparative genomics suggests that T. vivax differs quite considerably from the model T. brucei; by illuminating the expression of distinctive features in the T. vivax genome, this study moves us closer to understanding their phenotypic effects.
Methods
Ethics statement
All mice were housed in the Institut Pasteur animal care facilities in compliance with European animal welfare regulations (European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes CETS No.: 123). Institut Pasteur is a member of Committee #1 of the Comité Régional d’Ethique pour l’Expérimentation Animale (CREEA), Ile de France. Animal housing conditions and the protocols used in the work described herein were approved by the ‘‘Direction des Transports et de la Protection du Public, Sous-Direction de la Protection Sanitaire et de l’Environnement, Police Sanitaire des Animaux” (#B 75-15-28), in accordance with the Ethics Charter of animal experimentation that includes appropriate procedures to minimize pain and animal suffering. Authorization (to PM) to perform experiments on vertebrate animals is granted by license #75–846 issued by the Paris Department of Veterinary Services, DDSV.
Cell culture
Trypanosoma (Duttonella) vivax IL 1392 was originally derived from the Zaria Y486 Nigerian isolate. Bloodstream form parasites were maintained in vivo by continuous passage in mice, as previously described [29]. Once parasitemia reached at least 5x108 parasites per ml blood was collected by cardiac puncture onto heparin (2500 IU/kg), and was then diluted 1: 10 (v/v) with PBS 0.5% glucose to 5x107 parasites per ml. Parasites were separated from red blood cells by differential centrifugation using a swing-out rotor (Jouan GR412, Fisher Bioblock Scientific, Strasbourg, France). Diluted blood was processed by one round of centrifugation (5 min at 200 g) and the supernatant withdrawn with a pipette without disturbing the red blood cell layer and the thin interface containing the white blood cells. Parasite enriched suspension was submitted to a second round of centrifugation (5 min at 200 g) to eliminate all residual cells. The supernatant was then centrifuged for 10 min at 1800 g and bloodstream form-containing pellets devoid of host cells were submitted to two further PBS washes under the same centrifugation conditions. Bloodstream form-containing pellets were further treated for RNA or protein extractions.
T. vivax epimastigote cultures have been previously described [28]. Briefly, bloodstream forms purified as described above from infected mice differentiated into epimastigotes in TV3 media: IMDM 50%, DMEM (without glucose) 10% heat-inactivated fetal calf serum (FBS, MP Biomedicals or Invitrogen) and/or 10% heat-inactivated goat serum (GS, Invitrogen), 0.03 mM bathocuproinedisulfonic acid, 0.45 mM L-cysteine, 0.2 mM hypoxanthine, 0.14 mM ß-mercaptoethanol, 4mM L-proline, 0.05 mM thymidine, and 25 mM HEPES pH7.4. All supplements were obtained from Sigma Aldrich except HEPES (Invitrogen, Cergy Pontoise). Epimastigote growth cultures were maintained in vitro by serial passages. Epimastigotes attached to the surface of the culture flask formed micro-colonies and covered the entire surface after two weeks; the number of cells in the supernatant increased proportionally to the density of the adherent cell layer. Adherent epimastigotes were recovered from the flask by scraping and washed three times with PBS. As previously described, metacyclic forms are produced during in vitro growth and are found in the cell culture supernatant [28]. Metacyclic forms were isolated from the cell culture using an approach derived from “bovine plasma aggregation method” [30]. Supernatant from a dense culture (14 days) was remove from the flask, 30% non-inactivated goat serum was added to the cells and incubated at 27°C for 30 min. During the incubation period, epimastigotes aggregate into cell clumps, while metacyclic forms continue to swim freely. The metacyclic forms were then separated from the epimastigote clumps by passing the trypanosome suspension through a 5 μm pore size filter (Millipore Cat. Bedford, MS, USA). The metacyclic forms were then concentrated and washed by centrifugation at 750g for 15 min in 14 ml conical centrifuge tubes and RNA or protein prepared from the resultant cells pellets.
Sample preparation for RNA-seq
Total RNA was isolated using an RNeasy Mini Kit (Qiagen, Courtaboeuf, France) in accordance with the manufacturer's instructions. RNA purity and concentration were evaluated by spectrophotometry using NanoDrop ND-2000 (ThermoFisher). RNA quality and the relative contributions of total and small RNA were assessed by the Agilent 2100 Bioanalyzer microfluidics-based platform (Agilent Technologies, Santa Clara, USA). Four biological replicates were prepared for bloodstream form and metacyclic cells each. Five replicates were produced for epimastigote cells.
RNA sequencing
For each replicate, poly-adenylated RNA (mRNA) was purified from total RNA using an oligo-dT magnetic bead pull-down, using TruSeq RNA Sample Prep v2 kits (Illumina). The mRNA was then fragmented using metal ion-catalyzed hydrolysis. A random-primed cDNA library was synthesized and double-strand cDNA was used as the input to a standard Illumina library preparation, with a fragment size of 400bp. The libraries were amplified with 10 cycles of PCR using KAPA Hifi Polymerase. Samples were quantified and pooled based on a post-PCR Agilent Bioanalyzer, followed by size-selection using the LabChip XT Caliper. The multiplexed library was sequenced on the Illumina HiSeq 2000 with forward and reverse primers, according to the manufacturers standard protocol, resulting in 100-nucleotide paired-end reads. Sequenced data was analyzed and quality controlled and individual indexed library BAM files created.
Transcriptomic data analysis
Paired-end RNA-seq data were mapped to the T. vivax Y486 reference strain [11] (downloaded from TritrypDB release 6.0) using Bowtie2 [31] under the default parameters and within the Galaxy bioinformatics platform [32]. Transcript abundance for each replicate was estimated across the genome using Cufflinks [33] and measured in Fragments Per Kilobase Mapped (FPKM). The option for quartile normalization within Galaxy was applied to maximize our ability to detect preferential expression of low abundance transcripts against the background of highly abundant species. The option for bias detection and correction was enforced. The option for multi-read correction was applied because some of our genes of interest are multi-copy and may map to multiple locations. Fold change in transcript abundance, and significance of differential expression, was estimated using Cuffdiff [33] for three pairwise comparisons of T. vivax life stages, combining all replicates in each case. Cuffdiff applies the Benjamini-Hochberg correction for multiple testing when assessing the significance of fold changes. To ensure accurate assessment of differential expression, transcript abundance was corroborated using a second method, edgeR [34]. Correlations for fold change in transcript abundance returned by Cufflinks and edgeR displayed high congruence when comparing life stages (r2 = 0.89–0.91). Significant differences in transcript expression were defined as at least 2-fold enrichment between conditions and q < 0.05, where q is the p value corrected for false discovery rate (FDR).
Sample preparation for proteomics
Protein from cell lysates was dispensed into low protein-binding microcentrifuge tubes (Sarstedt, Leicester, UK) and made up to 160 μl by addition of 25 mM ammonium bicarbonate. The proteins were denatured using 10 μl of 1% (w/v) RapiGest (Waters MS Technologies, Manchester, UK) in 25 mM ammonium bicarbonate followed by three cycles of freeze-thaw, and two cycles of 10 min sonication in a water bath. The sample was then incubated at 80°C for 10 min and reduced with 3 mM dithiothreitol (Sigma-Aldrich, Dorset, UK) at 60°C for 10 min then alkylated with 9 mM iodoacetamide (Sigma-Aldrich, Dorset, UK) at room temperature for 30 min in the dark. Proteomic grade trypsin (Sigma-Aldrich, Dorset, UK) was added at a protein:trypsin ratio of 50:1 and samples incubated at 37°C overnight. Three biological replicates were prepared for each cell type.
LC-MS/MS analysis
Peptide mixtures were analyzed by on-line nanoflow liquid chromatography using the nanoACQUITY-nLC system (Waters MS technologies, Manchester, UK) coupled to an LTQ-Orbitrap Velos (ThermoFisher Scientific, Bremen, Germany) mass spectrometer equipped with the manufacturer’s nanospray ion source. The analytical column (nanoACQUITY UPLCT BEH130 C18 15 cm x 75 μm, 1.7 μm capillary column) was maintained at 35°C and a flow-rate of 300nl/min. The gradient consisted of 3–40% acetonitrile in 0.1% formic acid for 90 min then a ramp of 40–85% acetonitrile in 0.1% formic acid for 3 min. Full scan MS spectra (m/z range 300–2000) were acquired by the Orbitrap at a resolution of 30,000. Analysis was performed in data-dependent mode. The top 20 most intense ions from MS1 scan (full MS) were selected for tandem MS by collision induced dissociation (CID) and all product spectra were acquired in the LTQ ion trap. Ion trap and Orbitrap maximal injection times were set to 50 ms and 500 ms, respectively.
Proteomic data analysis
Thermo RAW files were imported into Progenesis LC–MS (version 4.1, Nonlinear Dynamics, UK). Runs were time aligned using default settings and using an auto selected run as reference. Peaks were picked by the software and filtered to include only peaks with a charge state of between +2 and +6. Peptide intensities were normalized against the reference run by Progenesis LC-MS and these intensities are used to highlight differences in protein expression between control and treated samples with supporting statistical analysis (ANOVA and q-values) calculated by the Progenesis LC-MS software. Spectral data were transformed to mgf files with Progenesis LC-MS and exported for peptide identification using the Mascot (version 2.3.02, Matrix Science) search engine. Tandem MS data were searched against a custom database that contained the common contamination and protein sequences predicted for the T. vivax reference genome (downloaded from TriTrypDB v-6.0). Search parameters were as follows; precursor mass tolerance set to 10ppm and fragment mass tolerance set to 0.5 Da. One missed tryptic cleavage was permitted. Carbamidomethylation (cysteine) was set as a fixed modification and oxidation (methionine) set as a variable modification. Mascot search results were further processed using the machine learning algorithm Percolator. The false discovery rates were set at 1% and at least two unique peptides were required for reporting protein identifications. Protein abundance (iBAQ) was calculated as the sum of all the peak intensities (from Progenesis output) divided by the number of theoretically observable tryptic peptides [35]. Protein abundance was normalized by dividing the protein iBAQ value by the summed iBAQ values for that sample. The reported abundance is the mean of the biological replicates.
Data accessibility
All cDNA sequence data are available from the European Nucleotide Archive (http://www.ebi.ac.uk/ena), accession number ERP001753. Details of the transcriptomic experiments are also available from the Array Express website (https://www.ebi.ac.uk/arrayexpress/), accession number E-ERAD-100. The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD001617.
Results
Estimation of transcript and peptide abundance
By exploiting new protocols for the in vitro cultivation of T. vivax epimastigote and metacyclic forms, we have produced comparative transcriptomic and proteomic data for the whole T. vivax life cycle using RNAseq and LC-MS/MS approaches respectively. Transcripts were detected for 10116 T. vivax Y486 genes (85.1% of all genes); 8994 of these transcripts (88.9%) were observed with at least 10 FPKM. The abundance of each transcript, as estimated using Cufflinks [33], is described in S1 Table. The most abundant transcripts in the bloodstream form were derived from tubulins, diverse ribosomal proteins and VSG-like sequences TvY486_0009580 (17331 FPKM) and TvY486_0018880 (9669 FPKM), which are assumed to have been the active VSG at the time of sequencing. Besides these, abundant transcripts encoding named proteins concern glyceraldehyde 3-phosphate dehydrogenase (TvY486_0603710; 1512 FPKM), a receptor-type adenylate cyclase (TvY486_0029610; 1210 FPKM), cathepsin B-like cysteine peptidase (TvY486_0600060; 794 FPKM), and an uncharacterized gene specific to T. vivax (TvY486_0900440; 1646 FPKM). The most abundant transcripts in the epimastigote and metacyclic cells encoded the same set of highly abundant tubulins and ribosomal proteins, but not the putative VSG, displaying instead an abundance of BARP-like proteins TvY486_0012620 (975 FPKM) and TvY486_1114940 (847 FPKM). Abundance estimates across our independent replicates were consistent, with strong positive correlations of replicates (ranging from 0.94 to 0.99) across all life stages (S1 Fig); and when fold change in transcript abundance is compared between life stages using edgeR, replicates cluster by stage illustrating their consistency (S2 Fig).
Peptide abundance, as defined by quantitative analysis with MASCOT, is described in S2 Table. 11099 peptides were counted corresponding to 1952 proteins (16.3% of predicted proteome). Of these, 1245 were sufficiently abundant to be quantified by iBAQ (i.e. two unique peptides were observed with a FDR of 0.01). Of these, 798 had a q value < 0.05, meaning that differential expression can be reliably inferred. The most abundant peptides in the bloodstream form were alpha and beta tubulin, various histones, putative VSG (TvY486_0009580/TvY486_0018880; i.e. coinciding with the most abundant VSG-like transcripts), and metabolic enzymes such as fructose-bisphosphate aldolase, enolase, glutamate dehydrogenase, arginine kinase, phosphoglycerate mutase, succinyl-coA:3-ketoacid-coenzyme A transferase and glycerol-3-phosphate dehydrogenase. The most abundant peptides in the epimastigote and metacyclic stages largely belonged to the same set of proteins, except that VSG were not observed and glycolytic enzymes were less abundant. As with transcript abundance, the proteome was consistent between independent replicates for each life stage, as illustrated in a principle component analysis in which replicates cluster tightly by stage (S3 Fig).
The degree to which relative abundance of transcripts and peptides concur throughout the life cycle is an important question with implications for regulation of gene expression, especially in trypanosomatids in which regulation is thought to be mostly post-transcriptional [27]. Correlations of transcript and peptide abundance across the genome (a-c) and for differentially expressed genes in each life stage (d-f) are shown in S4 Fig. These graphs show that the correlation is poor for all genes (r2 between 0.22 and 0.36) but improved for genes with evidence of developmental regulation (r2 between 0.38 and 0.65).
Developmental regulation of transcripts
Differential expression is defined as significant where transcript abundance displays at least two-fold enrichment and where q < 0.05. We found that 11.2% (1137) of transcripts showed significant differential expression in one or more stage comparison; we refer to these as ‘developmentally regulated’ and they are listed in S3 Table. In bloodstream forms, 518 transcripts were significantly more abundant relative to epimastigotes, and 382 transcripts were significantly more abundant in bloodstream forms relative to metacyclics. The greatest enrichment in favor of bloodstream forms concerned the putative active VSG (TvY486_0018880; fold-change (FC) = 110.9); other large fold-changes that implicated named sequences concerned three receptor-type adenylate cyclases (TvY486_0026190, TvY486_0003180 and TvY486_0029610; FC = 49.8, 18.9 and 10.8 respectively), a glycerol-3-phosphate dehydrogenase (TvY486_0802930; FC = 9.4) and a phospholipase A1 (TvY486_0102170; FC = 15.6). Besides these instances, the majority of transcripts (83%) preferentially expressed in bloodstream forms encode hypothetical proteins. Among these are hypothetical proteins belonging to T. vivax-specific families that are included in a Cell-Surface Phylome (CSP) that we published previously [26] for gene families predicted to be expressed on the cell surfaces of the three principal African trypanosome species; i.e. Fam30 (e.g. TvY486_0003670; FC = 29.4), Fam28 (e.g. TvY486_0030920; FC = 27.5), Fam34 (e.g. TvY486_0009950; FC = 26.5) and Fam31 (e.g. TvY486_0000210; FC = 23.4). Yet another family of uncharacterized genes, unique to T. vivax but not included in the CSP presently, show greater differential expression in bloodstream forms than any other family except for VSG. This gene family occurs 25 times among transcripts up-regulated in bloodstream forms relative to epimastigotes (S3 Table) and provides four of the 20 largest fold-changes in favor of bloodstream forms (e.g. TvY486_0033680, FC = 38.1). A BLASTp analysis shows that this family has at least 44 members across the T. vivax genome but none of these paralogs were observed to be preferentially expressed in either epimastigote or metacyclic form.
In epimastigotes, we identified 393 transcripts that were developmentally regulated, 387 of which are significantly more abundant in epimastigotes relative to bloodstream forms, while 8 transcripts are significantly more abundant in epimastigotes relative to metacyclic forms (see S3 Table). The dearth of preferential expression in epimastigotes relative to metacyclics was only slightly relieved by analysis with edgeR, which reported 23 cases. Since it was necessary to grow epimastigote cultures to high density in order to achieve a high proportion of metacyclic cells, it is possible that the lack of significant differences between these cell types is due to the effects of high density on growth. As with bloodstream forms, most developmentally regulated transcripts encode hypothetical proteins (65.1%), including those with the greatest fold changes in expression, i.e. TvY486_1110640 (FC = 73.1). Transcripts preferentially expressed in epimastigotes also concern further T. vivax-specific, CSP gene families, namely Fam35 (11 paralogs; FC = 5.4–43.7) and Fam43 (three paralogs; FC = 35.0–53.8). However, these gene families were more abundant still in metacyclic forms (see below), indicating that their main focus of expression was not the epimastigote. Aside from these uncharacterized gene families, transcripts implicated in cellular respiration were also seen, for example, components of the electron transfer chain such as cytochrome c1 (TvY486_0801280; FC = 13.0), cytochrome c (TvY486_0804690; FC = 13.2) and cytochrome c oxidase subunits (FC between 4.3–20.9). Also transcripts for multiple cation transporters (FC between 3.8–24.7) and a meiotic recombination protein DMC1 (TvY486_0904120; FC = 10.5).
In the final, metacyclic life stage, 357 transcripts were significantly more abundant relative to bloodstream forms, and these gave a very similar picture to the enriched transcripts in epimastigotes. A further 136 transcripts were significantly more abundant relative to epimastigotes (see S3 Table) including the T. vivax-specific, CSP families 34, 35 and 43 (see above), and other transcripts encoding DNA polymerase kappa (TvY486_1109280; FC = 2.8), an adenylate cyclase (TvY486_0029610; FC = 2.5), and various reverse transcriptases derived from SLACS elements (average FC = 6.8).
With respect to these observations, it should be noted that further analysis using edgeR produced very similar results to Cufflinks, with only 8.6% of the gene set displaying significant differential expression. Also, there was substantial overlap in the identities of developmentally regulated transcripts between comparisons; thus, of 518 transcripts significantly enriched in bloodstream forms relative to epimastigotes, 372 of these were also enriched relative to metacyclics; similarly, of 387 transcripts significantly enriched in epimastigotes relative to bloodstream forms, 285 of these were also enriched in metacyclics relative to the latter. From this it should be clear that, of the three life stages, the epimastigotes and metacyclic transcriptomes were most alike.
We examined the developmental regulated transcripts for Gene Ontology (GO) terms that were significantly enriched using a Fishers Exact test in BLAST2GO [36]. This confirmed that transcripts preferentially expressed in bloodstream forms are enriched for terms associated with glycolysis (GO:0006096; q = 3.38e-04) and glycosome (GO:0020015; p = 1.83e-03), while those preferentially expressed in epimastigotes are enriched for cytochrome-c oxidase activity (GO:0004129; p = 2.01e-13) and ATP synthesis coupled proton transport (GO:0015986; p = 1.43e-03). While suggestive of consistent differences in energy metabolism between life stages, differences in transcript abundance do not guarantee disparity in protein expression; thus, we sought to corroborate these observations with our proteomic data.
Developmental regulation of proteins
A protein shows significant differential expression if a constitutive peptide displays at least 2-fold enrichment and q < 0.05. Under these criteria, 595 or 30.5% of observed proteins (74.6% of quantifiable peptides) were developmentally regulated and these are listed in S4 Table. In the bloodstream form, 131 proteins were significantly more abundant relative to epimastigotes, while 63 proteins were significantly more abundant relative to metacyclics. Compared to differentially expressed transcripts, there is a smaller proportion of hypothetical proteins (29%), suggesting that our proteomic analysis has captured the most abundant components of cellular physiology that are relatively well understood. Accordingly, various structural proteins and diverse enzymes are listed, most notably components of the glycolytic pathway such as phosphoglycerate mutase (TvY486_0302920; FC = 6.6), fructose-bisphosphate aldolase (TvY486_1005670; FC = 3.5) and glycerol-3-phosphate dehydrogenase (TvY486_0802930; FC = 5.5). Analysis of functional terms associated with peptides preferentially expressed in bloodstream forms shows that glucose metabolism is significantly enriched (GO:0006006; FDR = 5.6e-05) relative to epimastigotes, while the citric acid cycle is significantly enriched (KEGG; FDR = 4.6e-04) relative to metacyclics.
The multi-copy, T. vivax-specific transcripts described above as being the most highly abundant family in bloodstream forms were also identified among our peptides. Four members of this family are preferentially expressed in bloodstream forms, one relative to epimastigotes (TvY486_0008730; FC = 15.9) and three more relative to metacyclics. None were expressed in either insect stage, further indicating that this is a novel and very prominent feature of bloodstream forms.
In the epimastigote form, 147 and 104 proteins were significantly more abundant relative to bloodstream forms and metacyclics respectively. GO terms associated with oxidation-reduction processes (GO:0055114; FDR = 1.1e-03) and amino acid metabolism (GO:0006520; FDR = 2.3e-02) were found to be significantly enriched. Peptides suggestive of oxidative phosphorylation, although no other elements of mitochondrial energy metabolism, were also significantly more abundant relative to metacyclics.
In the metacyclic stage, 62 peptides were significantly more abundant relative to bloodstream forms, and 89 peptides were significantly more abundant relative to epimastigotes. In both cases, the greatest fold changes pertain to hypothetical proteins encoded by members of CSP gene families (see below). Among the 88 peptides, various proteins with roles in intracellular trafficking were implicated, for example, the vesicle formation protein Sec24C (TvY486_0300580; FC = 11.5; [37]) and signal recognition particle receptor (TvY486_1110560; FC = 4.9), as well as several dynein heavy chains (FC between 2.2 and 7.1). A Fishers Exact test shows that functions associated with intracellular transport (GO:0046907; FDR = 2.0e-02) are significantly enriched. The same test shows that purine ribonucleotide catabolism (GO:0009154; FDR = 2.1e-03), (which refers here to the ATP-binding requirements of the same intracellular transport processes), is also significantly enriched.
It should be noted that, unlike the cohorts of stage-specific transcripts, there was no overlap in the membership of these various stage-specific peptide groups, i.e. there were no peptides significantly enriched in both epimastigotes and metacyclics relative to bloodstream forms.
Interspecies comparison of developmental regulation
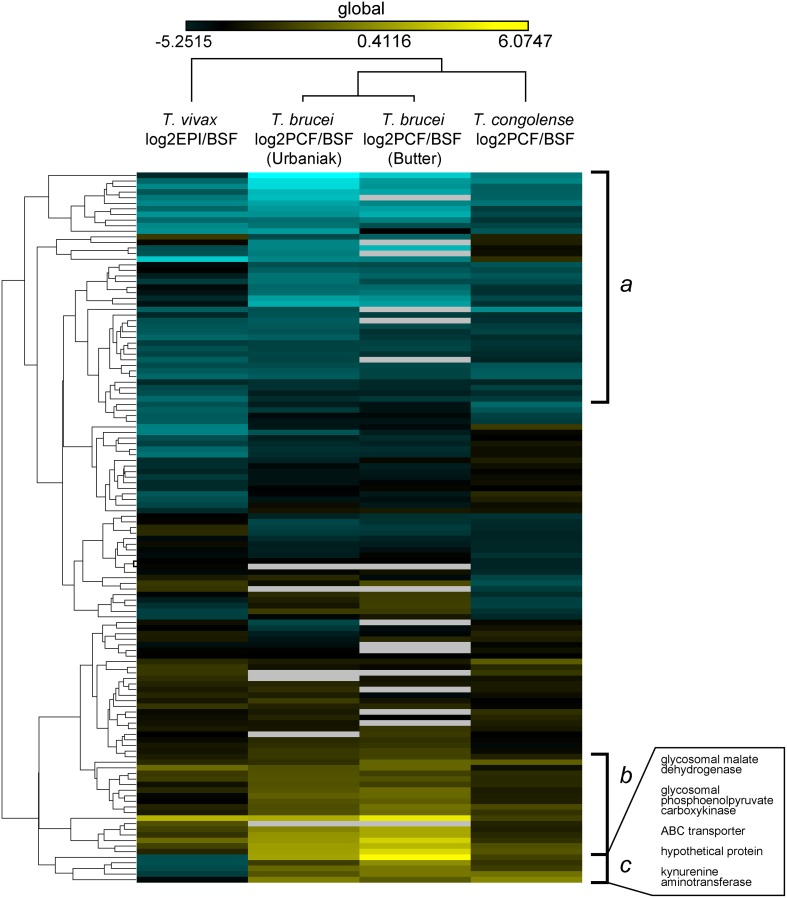
Transcriptomic and proteomic data from diverse African trypanosomes indicate consistently that life stages differ with respect to primary energy metabolism. Previous studies of global gene expression T. brucei and T. congolense have shown developmental regulation of such genes, as well as those encoding the principal cell surface glycoproteins [12–13, 16–19, 21–22]. We would like to know how conserved such developmental regulation is through evolutionary time, and thus, how regulatory evolution might have contributed to phenotypic differences between species. Comparative genomics predicts that T. vivax lacks certain enigmatic components of T. brucei and T. congolense cell surfaces, such as the VSG-related transferrin receptor of bloodstream forms and procyclin, as well as some elements of Fam50 (see below) [26]. To relate global gene expression across the African trypanosomes, we compared our proteome with existing data sets for T. brucei [21–22] and T. congolense [13], calculating the fold change in abundance from the non-infective insect stage (i.e. epimastigote for T. vivax and procyclic form for T. brucei and T. congolense) to the bloodstream form, for each protein observed in all species (N = 128). Fig 2 compares relative peptide abundance across four proteomic datasets from three species.
Fig 2. Fold changes in peptide abundance for 128 proteins when comparing insect stage (i.e. epimastigote (EPI) or procyclic form (PCF) and bloodstream forms (BSF), across three species.
The phylogram shown at the top describes the overall similarity of the four datasets. The dendrogram at the left describes how the proteins cluster by expression profile. Blue shades indicate insect-stage expression; yellow shades indicate vertebrate-stage expression. Subset ‘a’ are preferentially expressed in insect stages of all species. Subset ‘b’ are enriched during the vertebrate stage of all species. Subset ‘c’ is preferentially expressed in insect stages of T. vivax only.
Subset a contains proteins that are preferentially expressed in the bloodstream form in all species; the GO term for glycolysis (GO: 0006096; FDR = 6.4e-06) is enriched among these proteins. This indicates that the use of substrate-level phosphorylation as the dominant process for ATP generation in the bloodstream form is a consistent feature of African trypanosomes. Subset b contains proteins with the opposite expression profile, i.e. preferentially expressed in the non-infective insect stage in all species. Analysis of GO terms associated with these proteins shows that proton-transporting ATP synthase activity (GO:0046933; FDR = 1.8e-03), ATP synthesis coupled proton transport (GO:0015986; FDR = 1.8e-03) and oxidation-reduction process (GO:0055114; FDR = 3.1e-03) are enriched. This is consistent with the widespread use oxidative phosphorylation in the low-glucose environment of the insect vector to generate ATP via a proton-motive force across the mitochondrial membrane. Hence, at the broadest level, developmental regulation is conserved across all species, as their shared insect host will have predicted. However, there are obvious differences also.
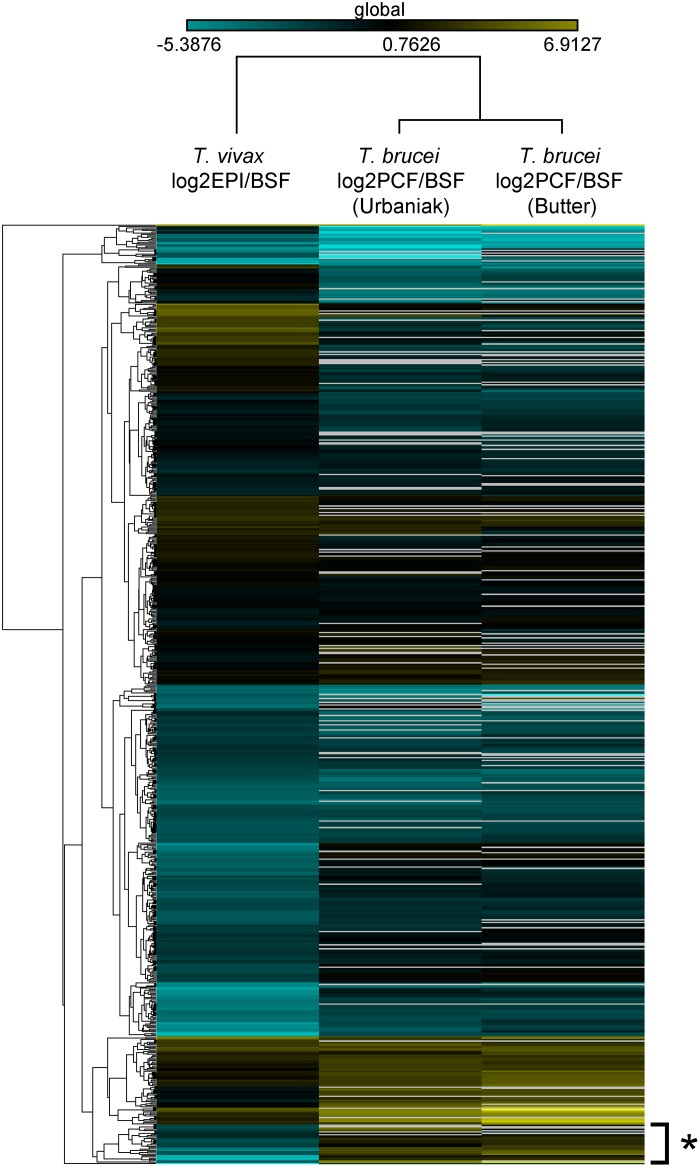
Against this background of conserved developmental regulation, we are interested in genes that are regulated differently in T. vivax, and which might contribute to its unique phenotypes. Subset c contains proteins that are significantly more abundant in the insect stages of T. brucei and T. congolense than the vertebrate stage, but preferentially expressed in T. vivax bloodstream forms. In the larger dataset of Fig 3, this cohort is expanded to 714 proteins by excluding T. congolense (for which proteome coverage is lowest); the number of proteins falling into subset c is increased to 27 and these are listed in Table 1. The expression profile of these proteins in T. vivax is not simply a lack of regulation or low expression generally, since many are highly abundant. Ten of the proteins in Table 1 appear in the top 10% of our proteome when ranked by abundance. Analysis of the GO terms associated with these proteins shows enrichment for succinate-CoA ligase (GDP-forming) activity (GO:0004776; FDR = 1.8e-02), pyruvate dehydrogenase (acetyl-transferring) activity (GO:0004739; FDR = 1.8e-02) and glucose metabolic process (GO:0006006; FDR = 2.8e-02). Hence, comparison of differentially expressed genes across African trypanosomes shows that much is conserved at the regulatory level but that important differences exist, even in the most essential physiology.
Fig 3. Fold changes in peptide abundance for 714 proteins when comparing insect stage (i.e. epimastigote (EPI) or procyclic form (PCF) and bloodstream forms (BSF), in T. brucei and T. vivax.
The asterisk denotes an expanded cohort of insect-specific proteins in T. vivax, corresponding to subset c in Fig 2.
Table 1. Fold change in peptide abundance for 27 loci that show preferential expression in the bloodstream form of T. vivax and in the insect (procyclic) stage of T. brucei.
| Gene ID | Product | T. vivax | T. brucei 1* | T. brucei 2** |
|---|---|---|---|---|
| TvY486_0902340 | ABC transporter, putative | -2.44 | 2.66 | 2.85 |
| TvY486_1007540 | ABC transporter, putative | -1.20 | 2.75 | 3.11 |
| TvY486_0904230 | BRCT domain-containing hypothetical protein, conserved | -3.44 | 1.64 | 2.02 |
| TvY486_0907550 | CHAT domain-containing hypothetical protein, conserved | -0.68 | 3.15 | 0.81 |
| TvY486_1007360 | dihydrolipoamide acetyltransferase precursor, putative | -0.20 | 1.16 | 1.81 |
| TvY486_0806870 | dihydrolipoamide dehydrogenase, putative | -0.17 | ND | 1.37 |
| TvY486_0604120 | dynein heavy chain-containing protein, putative | -2.06 | 2.49 | 2.07 |
| TvY486_1002790 | eukaryotic translation initiation factor 5, putative | -3.66 | 1.56 | 1.06 |
| TvY486_1014790 | glycosomal malate dehydrogenase, putative | -1.28 | 4.09 | 6.07 |
| TvY486_0039220 | glycosomal phosphoenolpyruvate carboxykinase, putative | -1.31 | 1.84 | 3.63 |
| TvY486_0101860 | hypothetical protein, conserved | -0.57 | 2.63 | ND |
| TvY486_0300460 | hypothetical protein, conserved | -1.04 | ND | 1.31 |
| TvY486_0303060 | hypothetical protein, conserved | -0.68 | 2.06 | 2.02 |
| TvY486_1011060 | hypothetical protein, conserved | -1.74 | 2.67 | 2.15 |
| TvY486_1111980 | hypothetical protein, conserved | -0.96 | 1.51 | 1.80 |
| TvY486_1114040 | hypothetical protein, conserved (fragment) | -0.07 | 1.10 | 1.24 |
| TvY486_0907650 | kinesin, putative | -3.61 | 1.88 | 1.89 |
| TvY486_1111670 | kinesin-like protein, putative | -0.20 | 0.95 | ND |
| TvY486_0901260 | mitochondrial pyruvate carrier protein, putative | -0.74 | 1.02 | 2.43 |
| TvY486_0702860 | nitroreductase 4-like protein, putative | -0.93 | 2.32 | 3.11 |
| TvY486_0100140 | phosphoglycerate kinase, putative | -1.01 | 3.76 | ND |
| TvY486_0604100 | phosphoglycerate kinase, putative | -1.28 | 3.76 | ND |
| TvY486_0301070 | pyruvate dehydrogenase E1 beta subunit, putative | -0.77 | 1.00 | 1.70 |
| TvY486_1012350 | pyruvate dehydrogenase E1 component alpha subunit, putative | -0.65 | 0.95 | 1.89 |
| TvY486_0902590 | RNA binding protein, putative | -3.39 | 1.09 | 1.52 |
| TvY486_1007240 | succinyl-CoA ligase [GDP-forming] beta-chain, putative | -0.96 | 1.80 | 1.76 |
| TvY486_0301600 | succinyl-CoA synthetase alpha subunit, putative | -1.01 | 1.49 | 1.74 |
Note: Fold change in peptide abundance is calculated as log2(epimastigote/bloodstream form) for T. vivax and log2(procyclic/bloodstream form) for T. brucei.
* Urbaniak et al. 2012
** Butter et al. 2013.
ND: no data.
Expression of T. vivax-specific, cell surface-expressed gene families
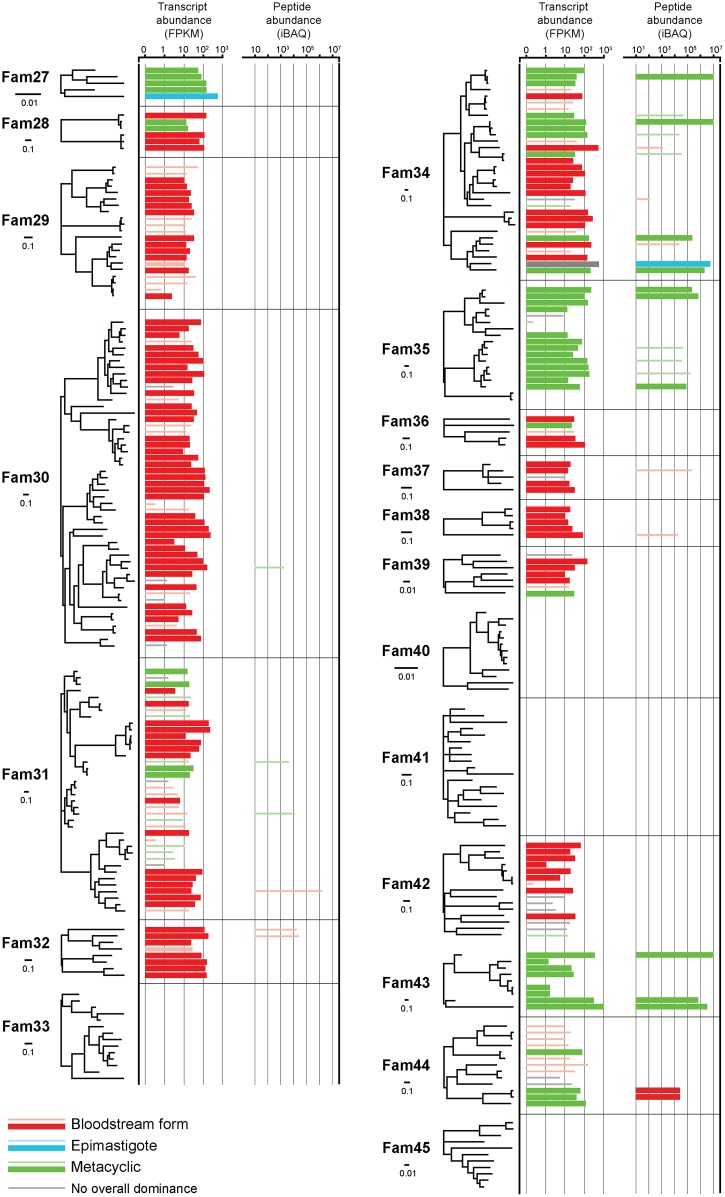
We have previously identified several gene families, known as Fam27-45, that are predicted to encode cell surface proteins and which, being unique to T. vivax, might distinguish the parasite from T. brucei and T. congolense [26]. Fam27-45 are among the most highly expressed genes and these data are extracted in Table 2. Many of the CSP families unique to T. vivax also appear to be developmentally regulated at the transcript level. For example, Fam27 (five paralogs), Fam35 (11 paralogs) and Fam 43 (five paralogs) are preferentially transcribed in insect stages. Conversely, Fam29 (13 paralogs), Fam30 (38 paralogs) and Fam32 (seven paralogs) are preferentially transcribed in bloodstream forms. Indeed, rarely are transcripts belonging to one of the families found throughout the lifecycle; Fam34 (25 paralogs) being one such case. Fig 4 summarizes the evidence for differential expression of Fam27-45 at the transcript and peptide level. In four cases, (Fams 33, 40, 41 and 45), both transcriptomic or proteomic evidence for expression is lacking and we conclude that these sequence families do not encode protein-coding genes (and so should be removed from the CSP). As expected, proteomic evidence for gene expression is not as prevalent as transcriptomic data, although it generally corroborates the latter when it is available.
Table 2. Preferential expression of T. vivax-specific Cell Surface Phylome (CSP) gene families based on fold change (FC) in transcript (a) and peptide (b) abundance across three life cycle stages.
| a) | Gene ID | CSP ID | Value 1 | Stage | Value 2 | Stage | FC | Max. stage |
| TvY486_0019430 | Fam27 | 97.6 | BSF | 2488.1 | EPI | 21.83 | EPI | |
| TvY486_0038280 | Fam27 | 74.4 | BSF | 325.6 | MET | 4.54 | MET | |
| TvY486_0038300 | Fam27 | 137.6 | BSF | 606.6 | MET | 4.58 | MET | |
| TvY486_0043530 | Fam27 | 71.0 | BSF | 317.2 | MET | 4.66 | MET | |
| TvY486_0044810 | Fam27 | 126.3 | BSF | 701.7 | MET | 6.12 | MET | |
| TvY486_0001150 | Fam28 | 43.3 | MET | 10.5 | EPI | 4.19 | MET | |
| TvY486_0005050 | Fam28 | 310.0 | BSF | 17.7 | EPI | 17.09 | BSF | |
| TvY486_0017990 | Fam28 | 274.6 | BSF | 12.5 | EPI | 19.89 | BSF | |
| TvY486_0019710 | Fam28 | 60.5 | MET | 14.2 | EPI | 4.38 | MET | |
| TvY486_0030920 | Fam28 | 203.3 | BSF | 4.7 | MET | 29.40 | BSF | |
| TvY486_0040160 | Fam28 | 488.7 | BSF | 28.5 | EPI | 16.82 | BSF | |
| TvY486_0010970 | Fam29 | 3.4 | BSF | 0.7 | EPI | 4.88 | BSF | |
| TvY486_0011750 | Fam29 | 72.4 | BSF | 13.5 | MET | 5.88 | BSF | |
| TvY486_0013730 | Fam29 | 65.3 | BSF | 17.0 | MET | 3.76 | BSF | |
| TvY486_0014410 | Fam29 | 37.6 | BSF | 9.2 | EPI | 4.12 | BSF | |
| TvY486_0014700 | Fam29 | 85.2 | BSF | 23.3 | EPI | 3.50 | BSF | |
| TvY486_0015070 | Fam29 | 122.2 | BSF | 41.5 | MET | 2.42 | BSF | |
| TvY486_0015280 | Fam29 | 70.0 | BSF | 16.5 | MET | 4.33 | BSF | |
| TvY486_0030720 | Fam29 | 126.5 | BSF | 26.5 | MET | 5.09 | BSF | |
| TvY486_0033210 | Fam29 | 33.0 | BSF | 9.7 | MET | 3.10 | BSF | |
| TvY486_0036530 | Fam29 | 42.2 | BSF | 11.4 | MET | 3.57 | BSF | |
| TvY486_0040000 | Fam29 | 124.6 | BSF | 29.8 | MET | 4.26 | BSF | |
| TvY486_0040440 | Fam29 | 45.6 | BSF | 15.1 | MET | 2.53 | BSF | |
| TvY486_0000090 | Fam30 | 63.0 | BSF | 8.8 | EPI | 8.08 | BSF | |
| TvY486_0001730 | Fam30 | 62.8 | BSF | 13.1 | EPI | 5.13 | BSF | |
| TvY486_0001740 | Fam30 | 191.8 | BSF | 33.7 | EPI | 6.29 | BSF | |
| TvY486_0003600 | Fam30 | 301.9 | BSF | 9.1 | EPI | 25.58 | BSF | |
| TvY486_0003670 | Fam30 | 233.4 | BSF | 5.5 | EPI | 29.35 | BSF | |
| TvY486_0004540 | Fam30 | 25.7 | BSF | 8.2 | EPI | 2.71 | BSF | |
| TvY486_0005120 | Fam30 | 20.6 | BSF | 5.1 | EPI | 4.07 | BSF | |
| TvY486_0005240 | Fam30 | 165.2 | BSF | 7.5 | EPI | 19.95 | BSF | |
| TvY486_0005910 | Fam30 | 266.6 | BSF | 6.6 | EPI | 28.49 | BSF | |
| TvY486_0006060 | Fam30 | 127.5 | BSF | 11.4 | EPI | 12.14 | BSF | |
| TvY486_0006100 | Fam30 | 101.4 | BSF | 14.9 | EPI | 7.67 | BSF | |
| TvY486_0007350 | Fam30 | 33.6 | BSF | 11.0 | MET | 2.58 | BSF | |
| TvY486_0007870 | Fam30 | 354.5 | BSF | 10.2 | EPI | 26.25 | BSF | |
| TvY486_0008460 | Fam30 | 156.8 | BSF | 11.3 | MET | 14.43 | BSF | |
| TvY486_0011830 | Fam30 | 129.1 | BSF | 30.0 | EPI | 4.44 | BSF | |
| TvY486_0014950 | Fam30 | 372.6 | BSF | 34.0 | EPI | 11.94 | BSF | |
| TvY486_0016140 | Fam30 | 531.7 | BSF | 40.6 | MET | 13.77 | BSF | |
| TvY486_0016150 | Fam30 | 474.4 | BSF | 43.0 | EPI | 12.00 | BSF | |
| TvY486_0020240 | Fam30 | 682.5 | BSF | 29.7 | EPI | 20.43 | BSF | |
| TvY486_0020730 | Fam30 | 95.4 | BSF | 9.1 | EPI | 11.48 | BSF | |
| TvY486_0021030 | Fam30 | 162.7 | BSF | 14.1 | EPI | 12.45 | BSF | |
| TvY486_0022710 | Fam30 | 195.4 | BSF | 18.6 | EPI | 11.54 | BSF | |
| TvY486_0023840 | Fam30 | 42.9 | BSF | 11.5 | MET | 3.60 | BSF | |
| TvY486_0027870 | Fam30 | 107.7 | BSF | 21.5 | EPI | 5.39 | BSF | |
| TvY486_0029560 | Fam30 | 248.4 | BSF | 23.5 | EPI | 11.56 | BSF | |
| TvY486_0029680 | Fam30 | 36.8 | BSF | 3.6 | EPI | 11.32 | BSF | |
| TvY486_0029750 | Fam30 | 156.6 | BSF | 21.5 | EPI | 8.19 | BSF | |
| TvY486_0031200 | Fam30 | 22.8 | BSF | 5.6 | EPI | 4.13 | BSF | |
| TvY486_0031450 | Fam30 | 211.0 | BSF | 51.4 | EPI | 4.15 | BSF | |
| TvY486_0032170 | Fam30 | 169.2 | BSF | 19.3 | EPI | 9.80 | BSF | |
| TvY486_0032760 | Fam30 | 234.1 | BSF | 44.5 | EPI | 5.74 | BSF | |
| TvY486_0033670 | Fam30 | 200.1 | BSF | 16.0 | EPI | 13.27 | BSF | |
| TvY486_0036500 | Fam30 | 71.8 | BSF | 9.2 | EPI | 8.83 | BSF | |
| TvY486_0037590 | Fam30 | 347.9 | BSF | 19.3 | EPI | 17.39 | BSF | |
| TvY486_0037600 | Fam30 | 120.6 | BSF | 18.9 | EPI | 7.16 | BSF | |
| TvY486_0038800 | Fam30 | 254.9 | BSF | 14.6 | EPI | 16.98 | BSF | |
| TvY486_0041770 | Fam30 | 71.6 | BSF | 11.9 | EPI | 6.72 | BSF | |
| TvY486_0043800 | Fam30 | 167.5 | BSF | 7.0 | EPI | 21.02 | BSF | |
| TvY486_0044700 | Fam30 | 91.2 | BSF | 11.0 | EPI | 9.32 | BSF | |
| TvY486_0000210 | Fam31 | 510.7 | BSF | 16.8 | MET | 24.29 | BSF | |
| TvY486_0007400 | Fam31 | 186.1 | BSF | 15.1 | EPI | 13.11 | BSF | |
| TvY486_0008000 | Fam31 | 168.5 | BSF | 9.2 | EPI | 17.61 | BSF | |
| TvY486_0010510 | Fam31 | 62.2 | BSF | 23.2 | EPI | 2.03 | BSF | |
| TvY486_0011600 | Fam31 | 80.1 | MET | 30.1 | EPI | 1.99 | MET | |
| TvY486_0014260 | Fam31 | 202.0 | BSF | 24.6 | MET | 9.22 | BSF | |
| TvY486_0017860 | Fam31 | 148.6 | MET | 34.8 | EPI | 4.39 | MET | |
| TvY486_0020480 | Fam31 | 174.0 | BSF | 9.8 | EPI | 17.24 | BSF | |
| TvY486_0023090 | Fam31 | 949.0 | BSF | 34.7 | MET | 22.78 | BSF | |
| TvY486_0023590 | Fam31 | 227.5 | BSF | 24.3 | MET | 10.42 | BSF | |
| TvY486_0026820 | Fam31 | 25.4 | BSF | 8.7 | EPI | 2.40 | BSF | |
| TvY486_0027490 | Fam31 | 168.7 | BSF | 12.0 | EPI | 14.58 | BSF | |
| TvY486_0029930 | Fam31 | 54.4 | MET | 14.5 | EPI | 3.64 | MET | |
| TvY486_0034320 | Fam31 | 27.2 | BSF | 8.9 | MET | 2.59 | BSF | |
| TvY486_0038020 | Fam31 | 72.6 | MET | 18.4 | EPI | 3.91 | MET | |
| TvY486_0040710 | Fam31 | 94.1 | BSF | 27.6 | EPI | 3.13 | BSF | |
| TvY486_0042250 | Fam31 | 102.9 | BSF | 31.7 | EPI | 2.89 | BSF | |
| TvY486_0042880 | Fam31 | 118.3 | BSF | 9.6 | EPI | 13.10 | BSF | |
| TvY486_0044520 | Fam31 | 114.5 | MET | 31.7 | EPI | 3.44 | MET | |
| TvY486_0045910 | Fam31 | 120.3 | BSF | 10.3 | EPI | 12.56 | BSF | |
| TvY486_0002120 | Fam32 | 377.9 | BSF | 40.8 | EPI | 10.31 | BSF | |
| TvY486_0004160 | Fam32 | 503.7 | BSF | 53.8 | EPI | 10.41 | BSF | |
| TvY486_0010310 | Fam32 | 155.8 | BSF | 29.3 | EPI | 5.82 | BSF | |
| TvY486_0025570 | Fam32 | 193.1 | BSF | 5.3 | MET | 26.77 | BSF | |
| TvY486_0027130 | Fam32 | 285.5 | BSF | 32.7 | EPI | 9.78 | BSF | |
| TvY486_0042890 | Fam32 | 225.2 | BSF | 69.7 | EPI | 2.86 | BSF | |
| TvY486_0043780 | Fam32 | 421.4 | BSF | 43.9 | EPI | 10.64 | BSF | |
| TvY486_0001120 | Fam34 | 24.7 | BSF | 154.1 | MET | 6.96 | MET | |
| TvY486_0004310 | Fam34 | 177.4 | BSF | 61.9 | EPI | 2.31 | BSF | |
| TvY486_0004790 | Fam34 | 421.5 | BSF | 2256.7 | MET | 5.86 | MET/EPI | |
| TvY486_0007010 | Fam34 | 380.0 | BSF | 90.4 | EPI | 4.29 | BSF | |
| TvY486_0008750 | Fam34 | 84.4 | BSF | 25.6 | EPI | 2.95 | BSF | |
| TvY486_0009950 | Fam34 | 981.2 | BSF | 27.7 | EPI | 26.48 | BSF | |
| TvY486_0013520 | Fam34 | 46.6 | BSF | 233.9 | MET | 5.42 | MET | |
| TvY486_0017330 | Fam34 | 15.9 | BSF | 380.9 | MET | 20.97 | MET | |
| TvY486_0018370 | Fam34 | 317.9 | BSF | 81.8 | MET | 3.83 | BSF | |
| TvY486_0019090 | Fam34 | 9.1 | BSF | 261.1 | MET | 23.52 | MET | |
| TvY486_0020950 | Fam34 | 244.7 | BSF | 26.4 | MET | 10.34 | BSF | |
| TvY486_0021420 | Fam34 | 403.2 | BSF | 145.4 | EPI | 2.17 | BSF | |
| TvY486_0022140 | Fam34 | 23.7 | BSF | 185.0 | MET | 8.77 | MET | |
| TvY486_0024340 | Fam34 | 2178.6 | BSF | 96.6 | EPI | 20.21 | BSF | |
| TvY486_0029710 | Fam34 | 41.0 | BSF | 174.2 | MET | 4.35 | MET | |
| TvY486_0029720 | Fam34 | 50.6 | BSF | 667.5 | MET | 13.84 | MET | |
| TvY486_0030370 | Fam34 | 333.3 | BSF | 73.7 | EPI | 4.74 | BSF | |
| TvY486_0032770 | Fam34 | 52.9 | BSF | 617.6 | MET | 12.56 | MET | |
| TvY486_0034300 | Fam34 | 26.2 | BSF | 143.1 | MET | 6.00 | MET | |
| TvY486_0035000 | Fam34 | 131.5 | BSF | 41.0 | EPI | 2.82 | BSF | |
| TvY486_0035610 | Fam34 | 227.1 | BSF | 81.5 | MET | 2.18 | BSF | |
| TvY486_0035970 | Fam34 | 285.4 | BSF | 31.5 | EPI | 10.10 | BSF | |
| TvY486_0039910 | Fam34 | 465.4 | BSF | 2354.6 | MET | 5.47 | MET/EPI | |
| TvY486_0044510 | Fam34 | 21.1 | BSF | 392.5 | MET | 17.81 | MET | |
| TvY486_0900430 | Fam34 | 859.1 | BSF | 44.0 | EPI | 18.37 | BSF | |
| TvY486_0004880 | Fam35 | 0.3 | BSF | 31.0 | MET | 43.79 | MET | |
| TvY486_0006390 | Fam35 | 1.0 | BSF | 151.1 | MET | 51.87 | MET | |
| TvY486_0010560 | Fam35 | 3.9 | BSF | 539.8 | MET | 50.36 | MET | |
| TvY486_0011320 | Fam35 | 58.0 | MET | 8.0 | EPI | 8.12 | MET | |
| TvY486_0011480 | Fam35 | 1.5 | BSF | 451.0 | MET | 67.73 | MET | |
| TvY486_0014750 | Fam35 | 0.7 | BSF | 564.1 | MET | 92.02 | MET | |
| TvY486_0016780 | Fam35 | 4.1 | BSF | 50.9 | MET | 13.22 | MET | |
| TvY486_0030340 | Fam35 | 1.2 | BSF | 206.6 | MET | 55.65 | MET | |
| TvY486_0030570 | Fam35 | 1.6 | BSF | 394.5 | MET | 63.36 | MET | |
| TvY486_0039920 | Fam35 | 178.2 | MET | 25.3 | EPI | 7.93 | MET | |
| TvY486_0041300 | Fam35 | 2.1 | BSF | 769.3 | MET | 72.56 | MET | |
| TvY486_0044010 | Fam35 | 4.1 | BSF | 669.1 | MET | 53.79 | MET | |
| TvY486_0504020 | Fam35 | 0.8 | BSF | 319.7 | MET | 75.03 | MET | |
| TvY486_0004900 | Fam36 | 122.8 | BSF | 27.1 | EPI | 4.76 | BSF | |
| TvY486_0019680 | Fam36 | 245.7 | BSF | 26.9 | EPI | 10.19 | BSF | |
| TvY486_0034550 | Fam36 | 48.4 | BSF | 139.7 | MET | 2.34 | MET | |
| TvY486_0039550 | Fam36 | 136.2 | BSF | 21.4 | EPI | 7.11 | BSF | |
| TvY486_0008720 | Fam37 | 63.7 | BSF | 16.8 | EPI | 3.68 | BSF | |
| TvY486_0010370 | Fam37 | 112.0 | BSF | 31.3 | MET | 3.38 | BSF | |
| TvY486_0037640 | Fam37 | 107.2 | BSF | 37.4 | MET | 2.31 | BSF | |
| TvY486_0044820 | Fam37 | 86.5 | BSF | 15.0 | MET | 6.38 | BSF | |
| TvY486_0012580 | Fam38 | 40.9 | BSF | 13.8 | EPI | 2.47 | BSF | |
| TvY486_0018310 | Fam38 | 79.2 | BSF | 13.0 | MET | 6.79 | BSF | |
| TvY486_0019140 | Fam38 | 106.4 | BSF | 19.3 | MET | 6.05 | BSF | |
| TvY486_0038940 | Fam38 | 96.2 | BSF | 31.7 | MET | 2.56 | BSF | |
| TvY486_0045330 | Fam38 | 234.9 | BSF | 36.6 | MET | 7.18 | BSF | |
| TvY486_0014250 | Fam39 | 364.1 | BSF | 82.2 | EPI | 4.61 | BSF | |
| TvY486_0036690 | Fam39 | 136.8 | BSF | 35.2 | MET | 3.84 | BSF | |
| TvY486_0041320 | Fam39 | 22.0 | BSF | 6.7 | EPI | 2.95 | BSF | |
| TvY486_0044730 | Fam39 | 59.8 | BSF | 19.5 | EPI | 2.61 | BSF | |
| TvY486_1101450 | Fam39 | 53.8 | BSF | 183.7 | MET | 3.14 | MET | |
| TvY486_0012130 | Fam42 | 85.4 | BSF | 28.7 | EPI | 2.48 | BSF | |
| TvY486_0012790 | Fam42 | 90.8 | BSF | 10.1 | EPI | 10.03 | BSF | |
| TvY486_0017880 | Fam42 | 130.3 | BSF | 26.3 | EPI | 5.32 | BSF | |
| TvY486_0019770 | Fam42 | 17.8 | BSF | 1.8 | EPI | 10.99 | BSF | |
| TvY486_0027860 | Fam42 | 2.0 | BSF | 0.2 | EPI | 11.25 | BSF | |
| TvY486_0028850 | Fam42 | 146.1 | BSF | 16.4 | MET | 9.96 | BSF | |
| TvY486_0039500 | Fam42 | 194.4 | BSF | 37.7 | EPI | 5.60 | BSF | |
| TvY486_1109320 | Fam42 | 109.0 | BSF | 30.5 | EPI | 3.38 | BSF | |
| TvY486_0002350 | Fam43 | 0.2 | BSF | 5.1 | MET | 19.71 | MET | |
| TvY486_0017520 | Fam43 | 6.4 | MET | 0.8 | EPI | 8.51 | MET | |
| TvY486_0017890 | Fam43 | 2.7 | BSF | 1245.9 | MET | 78.16 | MET | |
| TvY486_0021360 | Fam43 | 0.5 | BSF | 129.6 | MET | 64.82 | MET | |
| TvY486_0027480 | Fam43 | 9.6 | BSF | 5992.2 | MET | 86.31 | MET | |
| TvY486_0031050 | Fam43 | 4.2 | MET | 0.5 | EPI | 9.41 | MET | |
| TvY486_0040930 | Fam43 | 57.1 | MET | 8.3 | EPI | 7.79 | MET | |
| TvY486_0044590 | Fam43 | 5.5 | BSF | 1708.8 | MET | 68.43 | MET | |
| TvY486_0005330 | Fam44 | 116.6 | BSF | 505.9 | MET | 4.48 | MET | |
| TvY486_0012520 | Fam44 | 62.3 | BSF | 266.6 | MET | 4.40 | MET | |
| TvY486_0027940 | Fam44 | 59.5 | BSF | 277.7 | MET | 4.94 | MET | |
| TvY486_0042450 | Fam44 | 40.3 | BSF | 162.7 | MET | 4.06 | MET | |
| (b) | Gene ID | CSP ID | Peptide | iBAQ: | FC | Max. stage | ||
| count | BSF | EPI | MET | |||||
| TvY486_0013520 | Fam34 | 5 | 3598.7 | 1887.8 | 20547.2 | 10.88 | MET | |
| TvY486_0032770 | Fam34 | 19 | 2030.8 | 817.7 | 7244.1 | 8.86 | MET | |
| TvY486_0029720 | Fam34 | 3 | 15149 | 10247.4 | 90551.7 | 8.84 | MET | |
| TvY486_0039910 | Fam34 | 5 | 76451 | 293863.7 | 95768.8 | 3.84 | MET | |
| TvY486_0036040 | Fam34 | 11 | 91.91 | 103.0 | 966.1 | 10.51 | MET | |
| TvY486_0034300 | Fam34 | 17 | 534.44 | 840.1 | 1175.6 | 2.2 | MET | |
| TvY486_0041300 | Fam35 | 4 | 5943 | 4140.9 | 27129.1 | 6.55 | MET | |
| TvY486_0039920 | Fam35 | 3 | 275.32 | 783.5 | 6109.4 | 22.19 | MET | |
| TvY486_0017890 | Fam43 | 4 | 1711.5 | 2652.5 | 52918.0 | 30.92 | MET | |
| TvY486_0027480 | Fam43 | 6 | 16573 | 11934.9 | 237265.1 | 19.88 | MET | |
| TvY486_0044590 | Fam43 | 5 | 41982 | 58347.1 | 1211175 | 28.85 | MET | |
| TvY486_0012520 | Fam44 | 8 | 2370.2 | 2182.6 | 165.9 | 14.28 | BSF |
NB: For transcripts, significant differential expression is estimated from pairwise comparisons of life cycle stages using Cufflinks. For peptides, iBAQ abundance is compared across all three stages simultaneously. q < 0.05 in all cases. Developmental stages are abbreviated with BSF (bloodstream form), EPI (epimastigote), MET (metacyclic).
Fig 4. Differential expression of Trypanosoma vivax-specific genes belonging to Cell Surface Phylome families 27–45.
The maximum likelihood phylogenies of 19 CSP families unique to T. vivax are shown at left. Node labels are omitted for clarity. Each terminal tip corresponds to a locus. Transcript abundance and peptide abundance are shown adjacent to each tip by horizontal bars color-coded by stage. Transcripts or peptides that also showed significant differential expression, (as defined in the text), are indicated by bold bars.
The best supported cases for developmental regulation concern the metacyclic-specific expression of Fam34, 35 and 43. We observed 34 distinct Fam34 transcripts and 24 of these were differentially expressed; 12 in the metacyclic and another 12 in the bloodstream form. However, the proteomic evidence indicates more selective developmental regulation; of 11 Fam34 proteins that were observed, five were preferentially expressed and all in the metacyclic stage (FC between 2.2–10.8).
Of 17 distinct Fam35 transcripts, 11 were differentially expressed; all were significantly more abundant in the metacyclic stage (FC between 13.2–92.0). Proteomic data support this view; of the six Fam35 peptides observed, all were most abundant in the metacyclic stage and two significantly so (TvY486_0041300 (FC = 6.55) and TvY486_0039920 (FC = 22.19)).
We observed seven distinct Fam43 transcripts and of these five were differentially expressed, all most abundant in the metacyclic (FC between 19.7–86.3). All three Fam43 peptides that were observed were preferentially expressed in the metacyclic stage (FC between 19.8–30.9). Hence, these results indicate that the putative T. vivax-specific gene families are (mostly) genuine protein-coding sequences, and are often developmentally regulated at the transcript and (where observed) protein level.
Expression of ‘Fam50’ genes
Fam50 is a CSP gene family that includes the BARP genes of T. brucei and the GARP and CESP genes of T. congolense, known to be preferentially expressed on their respective cell surfaces during the insect stages [25, 38–40], as well as various, currently uncharacterized, genes that may also be transcribed preferentially during insect stages [41]. The genomic complement of Fam50 genes in T. vivax is smaller and less diverse than those of the other species, which may reflect the simpler existence of T. vivax in the tsetse fly [26]. Our transcriptomic data include all 17 T. vivax Fam50 genes, 13 of which are transcribed most abundantly in the insect stages (see S1 Table). Six transcripts are significantly more abundant in the epimastigote or metacyclic stage relative to bloodstream forms (Table 3a) and one of these was confirmed by the proteomic data (i.e. TvY486_0012620). In total, five proteins were detected and all were differentially expressed (Table 3b); four were most abundant in the insect (FC between 2.2 and 4.2). A single protein (corresponding to TvY486_0001140) was significantly more abundant in the bloodstream form. Thus, while transcriptomic data seems to be a poor predictor of differential expression of Fam50 proteins, perhaps suggesting the highly dynamic promotion and repression of Fam50 variants, there is good evidence for developmental regulation of both Fam50 transcripts and peptides, largely in preference for the insect stages and so consistent with observations in other species.
Table 3. Preferential expression of (a) transcripts and (b) peptides belonging to Fam50 (BARP-like genes) in specific life cycle stages.
| a) | Gene ID | FPKM: | FC | Max. stage | ||
| BSF | EPI | MET | ||||
| TvY486_0000390 | 383.5 | 2159.8 | 1826.9 | 6.22 | EPI | |
| TvY486_0012620 | 481.9 | 2456.5 | 2497.4 | 5.52 | EPI/MET | |
| TvY486_0044170 | 381.3 | 2190.3 | 1869.5 | 6.36 | EPI | |
| TvY486_1000710 | 299.1 | 1777.0 | 1465.8 | 6.61 | EPI | |
| TvY486_1001160 | 439.7 | 1936.2 | 2077.4 | 4.57 | EPI/MET | |
| TvY486_1114940 | 347.6 | 2018.1 | 2138.0 | 6.44 | EPI/MET | |
| b) | Gene ID | iBAQ: | FC | Max. stage | ||
| BSF | EPI | MET | ||||
| TvY486_0001140 | 693301.4 | 163534.9 | 100466.4 | 6.90 | BSF | |
| TvY486_0012620 | 135508.0 | 265088.1 | 292408.3 | 2.16 | EPI/MET | |
| TvY486_0016400 | 71617.0 | 159017.0 | 98083.8 | 2.22 | EPI | |
| TvY486_0016530* | 179679.6 | 584272.8 | 139906.5 | 4.18 | EPI | |
| TvY486_1114940 | 11074.5 | 40556.9 | 17551.1 | 3.66 | EPI |
NB: Developmental stages are abbreviated with BSF (bloodstream form), EPI (epimastigote), MET (metacyclic). Differential expression is described by fold change (FC) in epimastigotes relative to bloodstream forms. q < 0.05 in all cases. * Note that these two coding sequences are near-identical and so cross-mapping of reads could affect their FPKM values. * Note that peptides mapping to this gene could equally map to two other gene models (TvY486_1001150/TvY486_1001160) that are identical at this position.
Expression of VSG-like genes
The bloodstream forms of African trypanosomes are defined partly by the expression of a VSG coat on the cell surface. In our analysis, we observed 89 distinct VSG transcripts with q < 0.05 (S1 Table), of which 61 were most abundant in bloodstream forms; however, most of these were observed at very low levels. There were 12 transcripts displaying significant preferential expression in the bloodstream stage (FC between 2.5–110.9; Table 4a). In our proteomic analysis we recorded nine distinct VSG sequences, of which three are represented by a single peptide and so unquantified (S2 Table). Of the remaining six (Table 4b), three were most abundant in bloodstream forms, including the two putative active VSG and a third sequence (TvY486_0000810, or identical paralog) that was expressed at a much lower level but still preferentially in bloodstream forms (FC = 5.7). A fourth VSG was expressed preferentially in the metacyclic stage (TvY486_0027560; FC = 2.8). Finally, two VSG were most abundant in the epimastigote; one of these (TvY486_0001860; FC = 4.3) was differentially expressed and the second nearly so (TvY486_0041140; FC = 2.1; q = 0.066). Notably, these low abundance VSG expressed in epimastigotes belong to a T. vivax-specific VSG-like family (Fam25), which is not seen in other African trypanosomes.
Table 4. Preferential expression of (a) transcripts and (b) peptides belonging to variant surface glycoprotein-like genes.
| a) | Gene ID | CSP ID | BSF | EPI | MET | FC | Max. stage | |
| TvY486_0009580 | Fam24 | 34180.1 | 245.0 | 301 | 109.2 | BSF | ||
| TvY486_0018880 | Fam24 | 26001.7 | 20.1 | 20.8 | 106.7 | BSF | ||
| TvY486_0002880 | Fam23 | 22.7 | 2.5 | 3.20 | 10.0 | BSF | ||
| TvY486_0027380 | Fam23 | 201.1 | 30.1 | 37.3 | 7.5 | BSF | ||
| TvY486_0028170 | Fam23 | 212.4 | 32.3 | 40.0 | 7.3 | BSF | ||
| TvY486_0025310 | Fam23 | 188.6 | 36.4 | - | 5.6 | BSF | ||
| TvY486_0019720 | Fam23 | 107.2 | 23.9 | 38.0 | 4.6 | BSF | ||
| TvY486_0006330 | Fam23 | 147.1 | 33.9 | 42.1 | 4.4 | BSF | ||
| TvY486_0039540 | Fam23 | 848.1 | 203.8 | - | 4.2 | BSF | ||
| TvY486_0010070 | Fam23 | 69.3 | 18.2 | 21.2 | 3.7 | BSF | ||
| TvY486_0011440 | Fam23 | 54.0 | 16.6 | 16.2 | 2.9 | BSF | ||
| TvY486_0043550 | Fam23 | 97.3 | 30.1 | - | 2.8 | BSF | ||
| TvY486_0040490 | Fam23 | 111.9 | 36.9 | 36.7 | 2.5 | BSF | ||
| Gene ID | CSP ID | Peptide | iBAQ: | FC | Max. stage | |||
| b) | count | BSF | EPI | MET | ||||
| TvY486_0018880 | Fam24 | 15 | 22283050 | 817644.1 | 454910.3 | 48.9 | BSF | |
| TvY486_0009580 | Fam24 | 22 | 217715149 | 2295147 | 1824989 | 119.3 | BSF | |
| TvY486_0000810* | Fam23 | 2 | 51810.2 | 21467.7 | 9041.9 | 5.7 | BSF | |
| TvY486_0001860 | Fam25 | 1 | 44940.7 | 113671.8 | 26300.0 | 4.3 | EPI | |
| TvY486_0027560# | Fam24 | 1 | 44757.3 | 54295.2 | 124747.7 | 2.7 | MET | |
| TvY486_0041140 | Fam25 | 1 | 49076.9 | 117452 | 61305.4 | 2.3 | EPI |
NB: For transcript abundance, details are provided for pairwise comparisons that resulted in significant differences (in some cases involving a single gene in two comparisons). Developmental stages are abbreviated with BSF (bloodstream form), EPI (epimastigote), MET (metacyclic). Differential expression is described by fold change (FC). q < 0.05 in all cases. Peptide count refers to the total number of unique peptides observed for a given protein. Note that peptides mapping to * could equally map to other, identical gene models: TvY486_0000330; TvY486_0028900; TvY486_0002800; TvY486_0003200; TvY486_0005340; TvY486_0005830; TvY486_0008280; TvY486_0031250; TvY486_0031440; TvY486_0039630; TvY486_0042400. Also peptides mapping to # could equally map to these other, identical gene models: TvY486_0009200; TvY486_0015630; TvY486_0016270; TvY486_0031530; TvY486_0031920; TvY486_0031980; TvY486_0042360
Discussion
We have produced transcriptomes and proteomes for three different developmental forms of T. vivax, and identified the transcripts and peptides that are significantly enriched in each. These data provide the first profile of global gene expression and developmental regulation throughout the complete T. vivax life cycle. The profile suggests a situation broadly similar to that already observed in T. brucei and T. congolense, though with significant distinctions, not least further evidence for developmental regulation of species-specific cell surface glycoproteins in both the vertebrate and insect stages of T. vivax.
Such are their sensitivities, transcriptomic methods typically provide much greater coverage of the genome than do proteomic methods. This might be more pronounced for trypanosomatids since they constitutively express all genes within polycistronic transcripts and regulate protein expression post-transcriptionally [27]. Thus, 85.1% of T. vivax genes are represented in our transcriptome, but few transcripts are unique to a particular life stage and the proportion of differentially expressed transcripts is only 11.2%. By contrast, the proteome represents only 16.3% of all genes, but in 798 cases where differential expression could be assessed, 74.6% of peptides show significant differential expression and these are unique to one life stage. It may be that developmentally regulated proteins are also particularly abundant, certainly this is true for the components of stage-specific cell surface coats, and this would cause differentially expressed peptides to be overrepresented within the proteome. Previous proteomic studies for T. brucei have reported more proteins than we have found here for T. vivax (i.e. 3553 [21] and 3458 [22]) but a smaller proportion with significant differential expression, i.e. 24.8%/39.2% respectively. A proteomic analysis of T. congolense identified 1291 proteins, of which 21.5% displayed significant differential expression [13]. Hence, it may be that greater sensitivity would reduce the proportion of cases showing differential expression if low abundance proteins are more likely to be constitutively expressed.
Previously, comparison of the T. vivax genome sequence with those of T. brucei and T. congolense demonstrated that there are more than 2000 genes that are only present in T. vivax [11]. Their specificity, and typically the absence of any recognizable protein domains, make these genes obscure. Nonetheless, they appear to be genuine, since we found a lack of protein-coding evidence in only a few cases. One defining feature is that they comprise multi-copy gene families (Fam27-45 of the CSP) that are thought to be expressed on the cell surface based on the presence of a putative signal peptide, transmembrane domain and/or glycerophosphosinositol anchor in their predicted protein sequence [26]. Comparative genomics also showed that T. vivax lacked procyclin, the canonical cell surface glycoprotein of T. brucei and T. congolense insect stages [42]. This is consistent with T. vivax lacking a procyclic stage in the insect mid-gut and raises the question of what coats the T. vivax surface if not procyclin. Clearly, the abundant T. vivax-specific gene families offer plausible candidates for the role, but it could also be filled by Fam50; which has been shown to include various surface glycoproteins expressed during the insect stages of T. brucei and T. congolense [26].
Certainly, our data indicate that multiple Fam50 proteins are expressed in T. vivax and preferentially expressed in the epimastigote, while several transcripts (belonging to different loci) were significantly enriched in both epimastigotes and metacyclics. The fact that the transcripts and peptides are not derived from the same loci may suggest that different Fam50 genes became activated in the period between our preparation of RNA and protein. This is not the situation we observe for VSG, for which the identity of enriched transcripts and peptides largely match, suggesting that regulation of Fam50 gene expression is highly dynamic with multiple isoforms being promoted and repressed over short intervals. The presence of transcripts in the metacyclic stage at levels comparable to the epimastigote levels may be an artefact (i.e. residual epimastigotes in the metacyclic preparation), since Fam50 peptides in metacyclics are sparse and comparable in abundance to bloodstream forms. In short, the expression of multiple Fam50 proteins in the T. vivax epimastigote supports the view derived from T. brucei and T. congolense that this is a conserved family of glycoproteins performing diverse roles in the insect stages of the life cycle. Given that BARP in T. brucei and CESP in T. congolense are cell-surface glycoproteins [25, 40], Fam50 homologs are therefore probably a prominent component of the epimastigote cell surface in T. vivax.
Although Fam50 is preferentially expressed in epimastigotes, other multi-copy families, including the various T. vivax-specific cases, seldom are. This study has confirmed that most T. vivax-specific gene families are expressed. In the cases of Fam33, 40, 41 and 45, which should now be discounted from the CSP, the apparent lack of transcription raises the question of what function these repetitive non-coding sequences might perform. Three T. vivax-specific gene families (Fam34, 35 and 43) are very strongly enriched in the metacyclic stage, which is intriguing because in T. brucei and T. congolense the metacyclic coat is characterized by VSG. While metacyclic VSG are replaced by other VSG upon differentiation into bloodstream forms, and so are temporally distinct, there is no metacyclic-specific cohort of VSG sequences [8, 11]. VSG may also be present on T. vivax metacyclics, since we observed a low abundance VSG protein preferentially expressed in metacyclics (TvY486_0027560). However, assuming that Fam34, 35 and 43 are expressed on the cell surface as predicted, it is clear that the infective form of T. vivax has a qualitatively different surface architecture to the other species, with a considerable non-VSG component.
The same could be claimed for bloodstream forms also. Three families show exclusive enrichment in bloodstream forms at the transcript level (Fam28-30), though without proteomic support. This could indicate that our analysis lacked the sensitivity to detect them, perhaps because in bloodstream forms the superabundant VSG dominates the sequencing effort, making the detection of lower abundance proteins less effective than for either metacyclic or epimastigote. The presence of numerous non-VSG surface proteins might account for the observation that the T. vivax VSG coat is less dense than that of T. brucei [43], and VSG comprise a smaller proportion of cell surface-expressed T. vivax transcripts [14]. Assuming that their surface role is correct and they are confirmed as having preferential expression in bloodstream forms, these families are particularly interesting they have properties as surface antigens that could be targeted for vaccine development. Immune responses to the VSG do not provide lasting and comprehensive protection because of antigenic variation and the considerable structural diversity of the VSG repertoire. By contrast, Fam28-30 number not more than 30 paralogs, are less structurally diverse, and multiple transcripts are expressed at relatively high abundance, indicating that these families are not subject to antigenic variation, if monoallelic expression is a diagnostic feature of that process.
The VSG genes themselves present an expression profile typical of other African trypanosomes. VSG expression is regulated to produce a succession of structural variants that can evade specific immune responses but also prevent exposure of the total VSG structural repertoire to the host immune system, which would lead to a comprehensive immune response. Thus, VSG genes are expressed in a monoallelic fashion from the highly regulated context of a dedicated VSG expression site [44]. In their analysis of expressed sequence tags (EST) from different T. congolense life stages, Helm et al. (2009) recorded 13 distinct VSG transcripts in metacyclic cells, with the most abundant comprising 24% of the total number, and 26 distinct VSG transcripts in bloodstream forms, with the most abundant contributing 62% of the total [12]. This supports the established experimental model in which most individuals of a T. congolense population express the same active VSG, while a few individuals express a range of low abundance alternatives. In fact, when combined, the 12 least abundant VSG EST were only 0.5% of all VSG transcripts in bloodstream forms [12]. Similarly in T. brucei, Jensen et al. (2009) identified in a microarray-based study cohorts of less abundant transcripts in addition to the known, active VSG, some of which were expressed most abundantly in the insect stages [16]. In contrast, a previous analysis of VSG transcripts in T. vivax using 454 sequencing technology concluded that only one VSG was expressed [14].
Proteomic analyses have presented a similar picture. In T. congolense, 11 different VSGs were identified across all life stages [13]. Four were confidently associated with the metacyclic stage while two others were significantly enriched in bloodstream forms (including the known, active VSG). In proteomic comparisons of procyclic and bloodstream forms of T. brucei, one proteome identified 10 canonical VSGs [22], while another only two [21], although different T. brucei strains were used. These did not include the active VSG because neither study used the reference strain (927), and so the active VSG did not map to a VSG gene in the reference genome. Consequently, the 10 VSGs identified by Butter et al. (2013) are all low abundance alternatives, represented by few peptides (< 7) and achieving poor coverage (< 9%) [22]. Three of these low abundance VSGs were preferentially expressed in procyclic forms [22]. Taking these previous data together, their obvious methodological variations notwithstanding, low abundance alternatives to the dominant VSG are observed at both the transcript and protein levels in both T. brucei and T. congolense. The role, if any, of these ‘accessory’ VSGs is unclear; some are very likely metacyclic VSGs and it is known that expression of these can continue several days after transmission [45] and so could be present in bloodstream forms. Alternatively, ‘accessory’ VSGs may simply be rare antigens expressed by low frequency subpopulations, or produced by inefficiency in the mechanism silencing inactive VSG expression sites.
In contrast to the previous result [14], we observed several low abundance VSGs in T. vivax consistent with the expression profiles of VSG observed in T. brucei and T. congolense. The two dominant VSG sequences were superabundant at both transcript and peptide levels. Therefore the identity of the active VSG remained constant in the period between RNA and protein preparation, meaning that this is unlikely to represent a transition between two VSGs and that T. vivax strain IL1392 probably a mixture of parasites expressing one of two different VSGs. Both of these active VSG belong to Fam24, the subtype homologous to canonical b-type VSG in T. brucei and T. congolense [11]. In a similar fashion to T. congolense, the less abundant VSG in T. vivax may represent metacyclic VSGs. One VSG, TvY486_0027560, may be a metacyclic VSG in this strain as it was preferentially expressed in the metacyclic form (its transcript was not recorded). Finally, two VSG-like sequences belonging to Fam25, a T. vivax-specific subtype [11], are preferentially expressed at low levels in epimastigotes. The roles of Fam25 and 26 genes remain mysterious, and there is no definitive evidence that they encode variant antigens.
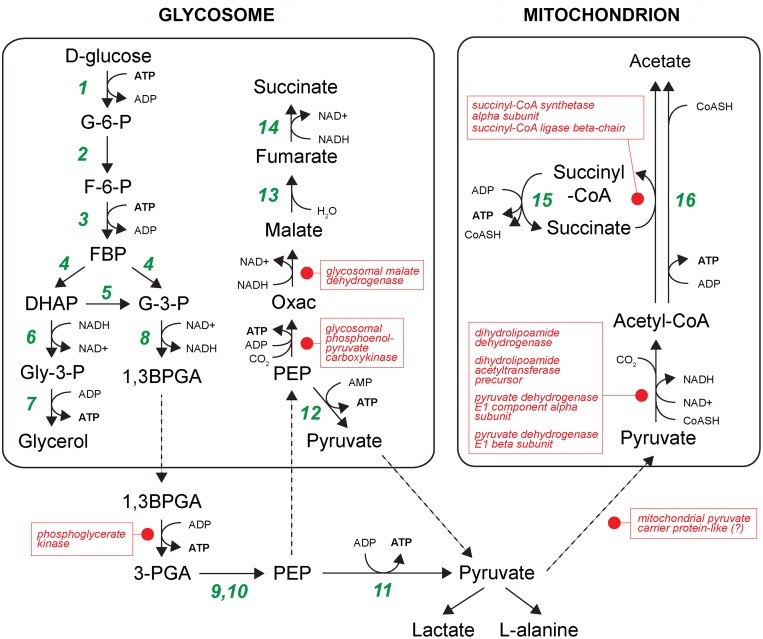
Beyond the differences in genetic repertoire that are evident from comparative genomics, it is presumed that differences in the regulation of conserved genes will contribute to phenotypic differences between African trypanosomes. The cohort of conserved genes identified in Figs 2 and 3 that are regulated conversely in T. vivax relative to T. brucei (and probably T. congolense) indicate that this is so. In the bloodstream stage, African trypanosomes exclusively employ glycolysis to exploit abundant glucose in host plasma to generate ATP via substrate-level phosphorylation in the glycosome [46]. In the tsetse fly, where glucose is limited but amino acids such as proline are present in the host hemolymph, the parasites generate ATP through gluconeogenesis and oxidative phosphorylation in the mitochondrion [46]. In this regard, T. vivax is consistent; all glycolytic enzymes are preferentially expressed in the bloodstream form, where they are among the most abundant transcripts and peptides, and all components of the electron transfer chain are preferentially expressed in the epimastigote (S3 and S4 Tables). The species differences highlighted in Fig 3 concern the metabolic steps linking glycolysis and events in the mitochondrion, i.e. pyruvate metabolism (Fig 5).
Fig 5. Energy metabolism in African trypanosomes, noting the position of enzymes with T. vivax-specific developmental regulation.
Glycolysis takes place within a specialized organelle, the glycosome, after which further substrate level phosphorylation takes place through the conversion of phosphoenolpyruvate ultimately to succinate in the glycosome, and through the conversion of pyruvate into acetate in the mitochondrion. Points marked with red dots and labels shaded red refer to proteins that are preferentially expressed during the vertebrate stage of T. vivax but in the insect stages of T. brucei (after Besteiro et al. 2005). Note that we could not differentiate between cytosolic and glycosomal phosophoglycerate kinase isoforms using our proteomic data. Abbreviations: 1,3BPGA, 1,3-bisphosphoglycerate; CoASH, coenzyme A; DHAP, dihydroxyacetone phosphate; F-6-P, fructose 6-phosphate; FBP, fructose 1,6-bisphosphate; G-3-P, glyceraldehyde 3-phosphate; G-6-P, glucose 6-phosphate; GLU, glutamate; Gly-3-P, glycerol 3-phosphate; Oxac, oxaloacetate; PEP,phosphoenolpyruvate; 3-PGA, 3-phosphoglycerate; SucCoA, succinyl-CoA. Enzymes are: 1) hexokinase: 2) glucose-6-phosphate isomerase; 3) phosphofructokinase; 4) aldolase; 5) triose-phosphate isomerase; 6) glycerol-3-phosphate dehydrogenase; 7) glycerol kinase; 8) glyceraldehyde-3-phosphate dehydrogenase; 9) phosphoglycerate mutase; 10) enolase; 11) pyruvate kinase; 12) pyruvate phosphate dikinase; 13) glycosomal fumarase; 14) NADH-dependent fumarate reductase; 15) acetate:succinate CoA-transferase; 16) possibly acetyl-CoA synthetase.
Experimental evidence indicates that T. brucei produces ATP during its insect stage by further substrate-level phosphorylation in the glycosome, by catabolizing phosphoenolpyruvate (PEP), and in the mitochondrion by catabolizing pyruvate. This results in insect forms excreting succinate and acetate, while bloodstream forms excrete pyruvate [47]. Accordingly, the enzymes for converting PEP into succinate and pyruvate into acetate are preferentially expressed in the procyclic form of T. brucei [21, 22]. Fig 5 describes the points in this pathway where differential expression is reversed in T. vivax. We see that enzymes for the catabolism of PEP, such as glycosomal malate dehydrogenase and glycosomal phosphoenolpyruvate carboxykinase, and for the conversion of pyruvate to acetate, i.e. multiple components of the pyruvate dehydrogenase complex and of the succinyl-CoA synthetase complex, are significantly more active in the bloodstream form than in the epimastigote. Additionally, the fumarase responsible for reaction 13 in Fig 5, while upregulated in procyclic form T. brucei, is constitutively expressed in T. vivax (TvY486_1105200; FC = 0.04). However, the final enzyme in the pathway (NADH-dependent fumarate reductase; reaction 14) is preferentially expressed in the insect stages in both species. We speculate that some other genes in Table 1 support this function; for example, TvY486_0901260, which possesses a mitochondrial pyruvate carrier protein domain homologous to mt1 in Humans, which is required to import pyruvate across the inner mitochondrial membrane [48], and TvY486_0702860, which encodes a bacterial-type nitro-FMN oxidoreductase that might serve to regenerate NAD+ [49].
Thus, we would predict that T. vivax excretes fumarate, acetate and perhaps succinate in its bloodstream stage rather than in the insect. It is not clear why T. vivax would benefit from pyruvate metabolism in the bloodstream when substrate-level phosphorylation using glucose should suffice. However, in the insect stage, when the parasite remains in the proboscis and without access to the hemolymph, it could be that such metabolism serves little purpose. Therefore, this may reflect a lack of upregulation in the epimastigote rather than adaptive upregulation in the bloodstream form, illustrating how life cycle variation has affected the regulation of energy metabolism in these organisms.
The first global perspective on gene expression in T. vivax has confirmed that a broadly similar process of developmental regulation occurs in all African trypanosome species. However, subtle differences, for instance in energy metabolism and putative cell surface molecules, offer new insights into the molecular basis for the life cycle differences that exist between species. Beyond the background of conservation, this study has confirmed the presence of numerous T. vivax-specific gene families and shown that these are developmentally regulated, indicating that the surface of T. vivax differs quite substantially from the model derived from other African trypanosomes.
Supporting Information
(DOCX)
Leading log-fold change in the first dimension is plotted on the x-axis and the second dimension is on the y-axis. Distances here correspond to leading log-fold-changes between replicates in each pairwise comparison of life stages: BSF vs EPI (left), BSF vs MET (centre) and EPI vs MET (right). These plots demonstrate that replicates cluster by life stage, reflecting the consistency in transcript abundance among replicates of the same stage.
(DOCX)
Normalized protein abundance levels across different samples were plotted to determine the principle axes of abundance variation. The first principle component is plotted on the x-axis and the second is plotted on the y-axis. The mass of grey numbers in the background refer to individual data points (proteins). Data points derived from individual replicates collected in each life stage are summed and represented by coloured dots: red (BSF), green (MET) and blue (EPI). The coloured dots cluster by life stage reflecting the consistency in expression profile provided by replicates of each condition.
(DOCX)
The number of comparisons possible for each life stage comparison is given in brackets.
(DOCX)
Genes without a non-zero value in all three conditions have been removed.
(XLSX)
(XLSX)
Significance is defined as p < 0.05 and FC ≥ 2.
(XLSX)
Significance is defined as q < 0.05 and FC ≥ 2.
(XLSX)
Data Availability
All cDNA sequence data are available from the European Nucleotide Archive (http://www.ebi.ac.uk/ena), accession number ERP001753. Details of the transcriptomic experiments are also available from the Array Express website (https://www.ebi.ac.uk/arrayexpress/), accession number E-ERAD-100. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD001617.
Funding Statement
This work was funded by Wellcome Trust (www.wellcometrust.org.uk) Grants WT 097826/Z/11/A and WT 098051. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Capewell P, Cooper A, Clucas C, Weir W, MacLeod A (2014) A co-evolutionary arms race: trypanosomes shaping the human genome, humans shaping the trypanosome genome. Parasitology, 12 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoare CA (1972) The Trypanosomes of Mammals, Blackwell, Oxford, 749 pp. [Google Scholar]
- 3. Roeder PL, Scott JM, Pegram RG (1984) Acute Trypanosoma vivax infection of Ethiopian cattle in the apparent absence of tsetse. Trop Anim Health Prod 16(3):141–7. [DOI] [PubMed] [Google Scholar]
- 4. Desquesnes M, Dia ML (2004) Mechanical transmission of Trypanosoma vivax in cattle by the African tabanid Atylotus fuscipes . Vet Parasitol 119(1):9–19. [DOI] [PubMed] [Google Scholar]
- 5. Jones TW, Dávila AM (2001) Trypanosoma vivax—out of Africa. Trends Parasitol 17(2):99–101. [DOI] [PubMed] [Google Scholar]
- 6. Osório AL, Madruga CR, Desquesnes M, Soares CO, Ribeiro LR, Costa SC. (2008) Trypanosoma (Duttonella) vivax: its biology, epidemiology, pathogenesis, and introduction in the New World—a review. Mem Inst Oswaldo Cruz 103(1):1–13. [DOI] [PubMed] [Google Scholar]
- 7. Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H et al. (2005) The genome of the African trypanosome Trypanosoma brucei . Science 309:416–422. [DOI] [PubMed] [Google Scholar]
- 8. Jackson AP, Sanders M, Berry A, McQuillan J, Aslett MA, et al. (2009) The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human african trypanosomiasis. PLoS Negl Trop Dis 4: e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goodhead I, Capewell P, Bailey JW, Beament T, Chance M, et al. (2013) Whole-genome sequencing of Trypanosoma brucei reveals introgression between subspecies that is associated with virulence. MBio 4(4). pii: e00197–13. 10.1128/mBio.00197-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sistrom M, Evans B, Bjornson R, Gibson W, Balmer O, et al. (2014) Comparative genomics reveals multiple genetic backgrounds of human pathogenicity in the Trypanosoma brucei complex. Genome Biol Evol 6(10):2811–2819. 10.1093/gbe/evu222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jackson AP, Berry A, Aslett M, Allison HC, Burton P, et al. (2012) Antigenic diversity is generated by distinct evolutionary mechanisms in African trypanosome species. Proc Nat Acad Sci USA, 109:3416–3421. 10.1073/pnas.1117313109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Helm JR, Hertz-Fowler C, Aslett M, Berriman M, Sanders M, et al. (2009) Analysis of expressed sequence tags from the four main developmental stages of Trypanosoma congolense . Mol Biochem Parasitol 168:34–42. 10.1016/j.molbiopara.2009.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eyford BA, Sakurai T, Smith D, Loveless B, Hertz-Fowler C, et al. (2011) Differential protein expression throughout the life cycle of Trypanosoma congolense, a major parasite of cattle in Africa. Mol Biochem Parasitol 177:116–125. 10.1016/j.molbiopara.2011.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greif G, Ponce de Leon M, Lamolle G, Rodriguez M, Piñeyro D, et al. (2013) Transcriptome analysis of the bloodstream stage from the parasite Trypanosoma vivax . BMC Genomics 14:149 10.1186/1471-2164-14-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koumandou VL, Natesan SK, Sergeenko T, Field MC. (2008) The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics 9:298 10.1186/1471-2164-9-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jensen BC, Sivam D, Kifer CT, Myler PJ, Parsons M (2009) Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei . BMC Genomics 10:482 10.1186/1471-2164-10-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Queiroz R, Benz C, Fellenberg K, Hoheisel JD, Clayton C (2009) Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics 10:495 10.1186/1471-2164-10-495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA (2010) Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res 38:4946–4957. 10.1093/nar/gkq237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Veitch NJ, Johnson PC, Trivedi U, Terry S, Wildridge D, et al. (2010) Digital gene expression analysis of two life cycle stages of the human-infective parasite, Trypanosoma brucei gambiense reveals differentially expressed clusters of co-regulated genes. BMC Genomics 11:124 10.1186/1471-2164-11-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Capewell P, Monk S, Ivens A, Macgregor P, Fenn K, et al. (2013) Regulation of Trypanosoma brucei total and polysomal mRNA during development within its mammalian host. PLoS One 8(6):e67069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Urbaniak MD, Guther ML, Ferguson MA (2012) Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS One 7: e36619 10.1371/journal.pone.0036619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Butter F, Bucerius F, Michel M, Cicova Z, Mann M, Janzen CJ (2013) Comparative proteomics of two life-cycle stages of stable isotope-labeled Trypanosoma brucei reveals novel components of the parasite's host adaptation machinery. Mol Cell Proteomics 12:172–179. 10.1074/mcp.M112.019224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gunasekera K, Wüthrich D, Braga-Lagache S, Heller M, Ochsenreiter T (2012) Proteome remodelling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics 13:556 10.1186/1471-2164-13-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Richardson JP, Beecroft RP, Tolson DL, Liu MK, Pearson TW (1988) Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol Biochem Parasitol 31(3):203–16. [DOI] [PubMed] [Google Scholar]
- 25. Urwyler S, Studer E, Renggli CK, Roditi I (2007) A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei . Mol Microbiol 63(1):218–228. [DOI] [PubMed] [Google Scholar]
- 26. Jackson AP, Allison HC, Barry JD, Field MC, Hertz-Fowler C, Berriman M (2013) A cell-surface phylome for African trypanosomes. PLoS Negl Trop Dis 7: e2121 10.1371/journal.pntd.0002121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kramer S (2012) Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol Biochem Parasitol 181(2):61–72. 10.1016/j.molbiopara.2011.10.002 [DOI] [PubMed] [Google Scholar]
- 28. D'Archivio S, Medina M, Cosson A, Chamond N, Rotureau B, et al. (2011) Genetic engineering of Trypanosoma (Duttonella) vivax and in vitro differentiation under axenic conditions. PLoS Negl Trop Dis 5(12):e1461 10.1371/journal.pntd.0001461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chamond N, Cosson A, Blom-Potar MC, Jouvion G, D'Archivio S, et al. (2010) Trypanosoma vivax infections: pushing ahead with mouse models for the study of Nagana. I. Parasitological, hematological and pathological parameters. PLoS Negl Trop Dis 4(8):e792 10.1371/journal.pntd.0000792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hirumi H, Nelson RT, Hirumi K (1983) Complete cyclic development of Trypanosoma vivax in vitro . J Protozool 30: 6A. [Google Scholar]
- 31. Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nature Methods 9 (4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goecks J., Nekrutenko A, Taylor J and The Galaxy Team (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11(8):R86 10.1186/gb-2010-11-8-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (2010) 26 (1): 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, et al. (2011) Global quantification of mammalian gene expression control. Nature 473(7347):337–242. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- 36. Conesa A, Götz S (2008) Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008: 619832 10.1155/2008/619832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pagano A, Letourneur F, Garcia-Estefania D, Carpentier JL, Orci L, Paccaud JP (1999) Sec24 proteins and sorting at the endoplasmic reticulum. J Biol Chem 274 (12):7833–7840. [DOI] [PubMed] [Google Scholar]
- 38. Ruepp S, Kurath U, Renggli CK, Brun R, Roditi I (1999) Glutamic acid/alanine-rich protein from Trypanosoma congolense is the functional equivalent of 'EP' procyclin from Trypanosoma brucei . Mol Biochem Parasitol 98(1):151–156. [DOI] [PubMed] [Google Scholar]
- 39. Bütikofer P, Vassella E, Boschung M, Renggli CK, Brun R, et al. (2002) Glycosylphosphatidylinositol-anchored surface molecules of Trypanosoma congolense insect forms are developmentally regulated in the tsetse fly. Mol Biochem Parasitol 119:7–16. [DOI] [PubMed] [Google Scholar]
- 40. Sakurai T, Sugimoto C, Inoue N (2008) Identification and molecular characterization of a novel stage-specific surface protein of Trypanosoma congolense epimastigotes. Mol Biochem Parasitol 161(1):1–11. 10.1016/j.molbiopara.2008.05.003 [DOI] [PubMed] [Google Scholar]
- 41. Savage AF, Cerqueira GC, Regmi S, Wu Y, El Sayed NM, Aksoy S (2012) Transcript expression analysis of putative Trypanosoma brucei GPI-anchored surface proteins during development in the tsetse and mammalian hosts. PLoS Negl Trop Dis 6(6):e1708 10.1371/journal.pntd.0001708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Acosta-Serrano A, Vassella E, Liniger M, Kunz Renggli C, et al. (2001) The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc Natl Acad Sci USA 98(4):1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vickerman K (1976) The Diversity of the kinetoplastid flagellates In Lumsden WHR, Evans DA, editors. Biology of the Kinetoplastida. Academic Press: London/New York/San Francisco: pp. 1–34. [Google Scholar]
- 44. Horn D, McCulloch R (2010) Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr Opin Microbiol 13:700–705. 10.1016/j.mib.2010.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Crowe JS, Barry JD, Luckins AG, Ross CA, Vickerman K (1983) All metacyclic variable antigen types of Trypanosoma congolense identified using monoclonal antibodies. Nature 306:389–391. [DOI] [PubMed] [Google Scholar]
- 46. Ginger ML, Fairlamb AH, Opperdoes FR (2007) Comparative genomics of trypanosome metabolism In: Barry JD, McCulloch R, Mottram JC, Acosta-Serrano A, editors. African Trypanosomes: After the Genome. Wymondham: Horizon Bioscience; pp. 373–416. [Google Scholar]
- 47. Besteiro S, Barrett MP, Rivière L, Bringaud F (2005) Energy generation in insect stages of Trypanosoma brucei: metabolism in flux. Trends Parasitol 21(4):185–191. [DOI] [PubMed] [Google Scholar]
- 48. Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, et al. (2012) A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337(6090):96–100. 10.1126/science.1218099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Oliveira IM, Henriques JA, Bonatto D (2007) In silico identification of a new group of specific bacterial and fungal nitroreductases-like proteins. Biochem Biophys Res Commun 355(4):919–925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Leading log-fold change in the first dimension is plotted on the x-axis and the second dimension is on the y-axis. Distances here correspond to leading log-fold-changes between replicates in each pairwise comparison of life stages: BSF vs EPI (left), BSF vs MET (centre) and EPI vs MET (right). These plots demonstrate that replicates cluster by life stage, reflecting the consistency in transcript abundance among replicates of the same stage.
(DOCX)
Normalized protein abundance levels across different samples were plotted to determine the principle axes of abundance variation. The first principle component is plotted on the x-axis and the second is plotted on the y-axis. The mass of grey numbers in the background refer to individual data points (proteins). Data points derived from individual replicates collected in each life stage are summed and represented by coloured dots: red (BSF), green (MET) and blue (EPI). The coloured dots cluster by life stage reflecting the consistency in expression profile provided by replicates of each condition.
(DOCX)
The number of comparisons possible for each life stage comparison is given in brackets.
(DOCX)
Genes without a non-zero value in all three conditions have been removed.
(XLSX)
(XLSX)
Significance is defined as p < 0.05 and FC ≥ 2.
(XLSX)
Significance is defined as q < 0.05 and FC ≥ 2.
(XLSX)
Data Availability Statement
All cDNA sequence data are available from the European Nucleotide Archive (http://www.ebi.ac.uk/ena), accession number ERP001753. Details of the transcriptomic experiments are also available from the Array Express website (https://www.ebi.ac.uk/arrayexpress/), accession number E-ERAD-100. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD001617.
All cDNA sequence data are available from the European Nucleotide Archive (http://www.ebi.ac.uk/ena), accession number ERP001753. Details of the transcriptomic experiments are also available from the Array Express website (https://www.ebi.ac.uk/arrayexpress/), accession number E-ERAD-100. The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD001617.