Abstract
Triple-negative breast cancer (TNBC) has the lowest 5-year survival rate of invasive breast carcinomas, and currently there are no approved targeted therapies for this aggressive form of the disease. The androgen receptor (AR) is expressed in up to one third of TNBC and we find that all AR+ TNBC primary tumors tested display nuclear localization of AR, indicative of transcriptionally active receptors. While AR is most abundant in the “luminal AR (LAR)” molecular subtype of TNBC, here, for the first time, we use both the new-generation anti-androgen enzalutamide and AR knockdown to demonstrate that the other non-LAR molecular subtypes of TNBC are critically dependent on AR protein. Indeed, AR inhibition significantly reduces baseline proliferation, anchorage-independent growth, migration, and invasion and increases apoptosis in four TNBC lines (SUM159PT, HCC1806, BT549, and MDA-MB-231), representing three non-LAR TNBC molecular subtypes (mesenchymal-like, mesenchymal stem–like, and basal-like 2). In vivo, enzalutamide significantly decreases viability of SUM159PT and HCC1806 xenografts. Furthermore, mechanistic analysis reveals that AR activation upregulates secretion of the EGFR ligand amphiregulin (AREG), an effect abrogated by enzalutamide in vitro and in vivo. Exogenous AREG partially rescues the effects of AR knockdown on proliferation, migration, and invasion, demonstrating that upregulation of AREG is one mechanism by which AR influences tumorigenicity. Together, our findings indicate that non-LAR subtypes of TNBC are AR dependent and, moreover, that enzalutamide is a promising targeted therapy for multiple molecular subtypes of AR+ TNBC.
Introduction
Triple-negative breast cancer (TNBC) constitutes 10% to 20% of invasive breast carcinomas and has the lowest 5-year survival rate compared with other breast cancer subtypes (1). 12% to 28% of patients with TNBC achieve a pathological complete response following neoadjuvant chemotherapy and have a good prognosis (2, 3). However, patients with TNBC and residual disease have a significantly worse overall survival than patients with non-TNBC subtypes and residual disease (2). The discrepancy in survival between patients with TNBC and non-TNBC with residual disease is exacerbated by the absence of effective targeted therapy for TNBC. TNBC lacks estrogen receptor (ER) and progesterone receptor (PR) expression as well as HER2 amplification and thus is unresponsive to traditional endocrine- or HER2-directed therapies that improve overall survival in other breast cancer subtypes. Although TNBC lacks the hormone receptors traditionally associated with breast cancer, many TNBCs express other hormone receptors, including the glucocorticoid receptor (4) and androgen receptor (AR). AR, a ligand-activated nuclear hormone transcription factor (5), is expressed in 12% to 36% of TNBC (6–9).
A defining role for AR and AR-regulated genes in the molecular biology and classification of breast cancer was established by microarray profiling studies of invasive breast carcinomas, including TNBC (10–14). Lehmann and colleagues characterized TNBC as a heterogeneous disease with seven molecular subtypes, including unstable, basal-like 1, basal-like 2, mesenchymal-like, mesenchymal stem–like (MSL), immunomodulatory, and luminal AR (LAR). The LAR subtype is similar to previously characterized molecular apocrine tumors (12, 13, 15) and its gene expression profile and chromatin-binding patterns mimic luminal, ER+ breast cancer, despite being ER-negative (11, 14). Within the TNBC molecular subtypes, LAR TNBC has the highest AR expression (16) and thus preclinical research has predominately focused on the efficacy of AR-targeted therapy using LAR cell lines as models of AR+ TNBC.
Our group and others have demonstrated that the LAR cell line MDA-MB-453 is sensitive to androgens in vitro (17, 18) and in vivo (17). Xenograft studies with AR antagonists have also demonstrated that LAR SUM185PE, CAL-148, and MDA-MB-453 cell lines are sensitive to bicalutamide (14) or enzalutamide (17). Although there are strong preclinical data to suggest that LAR TNBC subtypes may benefit from AR-targeted therapy, other TNBC molecular subtypes express AR and may also benefit from treatment with AR antagonists.
A phase II trial of bicalutamide in ER−/PR−/AR+ metastatic breast cancer demonstrated a 19% clinical benefit rate (19), indicating that AR antagonists may be an effective targeted therapy for some patients with AR+ TNBC. A phase II trial (NCT01889238) of the newer generation AR antagonist enzalutamide, which blocks AR nuclear localization and is thus less likely to act as a partial agonist, is underway in TNBC. While the inclusion criteria for the current phase II trial of enzalutamide is 1% AR+ staining, most in vitro studies have focused on AR in LAR TNBC cell line models with very high AR expression and little is known about the role of AR or efficacy of enzalutamide in TNBC with lower AR expression. We hypothesized that non-LAR, AR+ TNBC may also critically depend on AR and could benefit from treatment with enzalutamide. Our study indicates that multiple subtypes of AR+ TNBC depend on AR for proliferation, migration, and invasion, and tumor growth in vivo and provides promising preclinical data on the efficacy of enzalutamide in TNBC with low AR expression.
Materials and Methods
Cell culture
All cell lines were authenticated by short tandem repeat analysis and tested negative for Mycoplasma in July 2014. Molecular subtypes of TNBC cell lines used in the present study were previously categorized by Lehmann and colleagues (14). SUM159PT cells were purchased from the University of Colorado Cancer Center Tissue Culture Core (Aurora, CO) in August 2013 and were grown in Ham/F-12 with 5% FBS, penicillin/streptomycin, hydrocortisone, insulin, HEPES, and L-glutamine supplementation. MDA-MB-231 (MDA231) cells were purchased from the ATCC in August 2008 and were grown in minimum essential media with 5% FBS, penicillin/streptomycin, HEPES, L-glutamine, nonessential amino acids, and insulin supplementation. HCC1806 cells were obtained from the laboratory of Dr. Haihua Gu in 2011 and propagated in RPMI-1640 with 10% FBS and penicillin/streptomycin. BT549 cells, purchased from the ATCC in 2008, were grown in RPMI-1640 with 10% FBS, penicillin/streptomycin, and insulin. All crystal violet assays were conducted in 5% charcoal-stripped serum to directly study the effect of DHT on cellular proliferation or transcription respectively. All other experiments were performed in full serum, as described above, with the exception of migration assays that were performed in serum-free conditions to prevent cellular proliferation.
SUM159PT-TGL and HCC1806-TGL cells were generated by stable retroviral transduction with a SFG-NES-TGL vector, encoding a triple fusion of thymidine kinase, GFP, and luciferase and sorted for GFP. SUM159PT, HCC1806, BT549, and MDA231 AR knockdown cells were generated by lentiviral transduction of shRNAs targeting AR (pMISSION VSV-G, Sigma Aldrich), including AR shRNA 3715 (shAR15) and AR shRNA 3717 (shAR17). Lentiviral transduction of pMISSION shRNA NEG (shNEG) was used as a nontargeting control. Plasmids were purchased from the University of Colorado Functional Genomics Core Facility.
Cellular assays and reagents
Cells were treated with 10 μmol/L enzalutamide (Medivation), 10 nmol/L DHT (Sigma-Aldrich), and 1 μg/mL recombinant human amphiregulin (AREG; R&D Systems). A total of 10 μmol/L enzalutamide approximates the IC50 of the 4 cell lines studied (data not shown) and is a clinically achievable concentration. Circulating plasma Cmax values for enzalutamide and its active metabolite (N-desmethyl enzalutamide) are 16.6 μg/mL (23% CV) and 12.7 μg/mL (30% CV), respectively (enzalutamide package insert Exposure Rationale), which is equivalent to approximately 60 μmol/L total active drug in plasma at steady state. Androgen concentrations have been previously examined in breast cancer (20), and intratumoral DHT concentrations (249 pg/g) were significantly higher than in blood. The DHT concentration of the present study is consistent with other in vitro studies of DHT in breast cancer (18, 21) and approximates levels of circulating testosterone in obese, postmenopausal women (22), as well as DHT levels in FBS used during routine tissue culture propagation (23).
Migration and invasion scratch wound assays were performed with or without BD Matrigel (BD Biosciences), respectively, per the manufacturer’s instructions and scanned with the Incucyte ZOOM apparatus (Essen BioSciences). When an attractant was required for invasion, Transwell invasion assays were performed with BD BioCoat Matrigel Invasion Chambers (BD Biosciences) per the manufacturer’s protocol. Caspase-3/7 fluorescent reagent (Essen BioSciences) was used at a dilution of 1:1,000 and normalized to cell count (apoptotic index), following the manufacturer’s protocol, to assess apoptosis in vitro. The Amphiregulin Human ELISA Kit (Abcam) was used to measure extracellular AREG concentrations per the manufacturer’s protocol.
For crystal violet assays, cells were fixed in 10% formalin, rinsed in PBS, and stained with 5% crystal violet. Crystal violet was then dissolved in 10% acetic acid and measured at 540λ. MTS assays were performed with the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega) according to the manufacturer’s protocol. Proliferation assays were also performed using the Incucyte ZOOM imaging system (Essen BioSciences). Soft agar assays were performed in 0.5% bottom and 0.25% top layer agar (Difco Agar Noble, BD Biosciences).
Tumor studies
Xenograft experiments were approved by the University of Colorado Institutional Animal Care and Use Committee (IACUC protocol 83614(01)1E). All animal experiments were conducted in accordance with the NIH Guidelines of Care and Use of Laboratory Animals. A total of 106 SUM159PT-TGL or 500,000 HCC1806-TGL cells were mixed with Matrigel (BD Biosciences) and bilaterally injected into the mammary fat pads of female, athymic nu/nu mice (Taconic). Tumor burden was assessed by luciferase activity and caliper measurements [tumor volume was calculated as volume = (length × width2)/2]. Once tumors were established, mice were randomized into groups based on the total tumor burden as measured by in vivo imaging. Mice were administered enzalutamide in their chow (~a 50 mg/kg daily dose). Enzalutamide was mixed with ground mouse chow (Research Diets Inc.) at 0.43 mg/g chow. The feed was irradiated and stored at 4°C before use. Mice in the control group received the same ground mouse chow but without enzalutamide. All mice were given free access to enzalutamide-formulated chow or control chow during the study period. Mice were euthanized by carbon dioxide asphyxiation followed by cervical dislocation, and the tumors and mammary glands were harvested.
Histology
Tissues were fixed in 10% neutral-buffered formalin, and tissue processing and paraffin embedding were performed by either the UCDenver Tissue Biobanking and Processing Core or the UCH Anatomic Pathology Laboratory. Hematoxylin and eosin (H&E) stains were purchased from Anatech Ltd. and used per the manufacturer’s instructions.
Archival formalin-fixed, paraffin-embedded primary breast tumors designated as hormone receptor–negative and HER2 ≤ 10% were collected under the Institutional Review Board protocol Molecular and Cellular Predictors of Breast Cancer (#10-0755) from 130 women diagnosed at Massachusetts General Hospital (Partners) between 1977 and 1993. Although many samples were originally defined as hormone receptor–negative by radioimmunoassay, all samples were reevaluated by immunohistochemistry and hormone receptor–negative was defined as <1% positive staining for ER and PR. Slides were immunostained for AR as described below and evaluated for the percentage and intensity of AR expression.
Immunohistochemistry
Slides were deparaffinized in a series of xylenes and ethanols, and antigens were heat retrieved in 10 mmol/L citrate buffer, pH 6.0. Antibodies used were AR clone 441 (Dakocytomation) and AREG (HPA008720, Sigma Aldrich). Envision horseradish peroxidase (Dakocytomation) was used for detection.
In situ hybridization
Terminal deoxynucleotidyl transferase–mediated dUTP nick-end labeling (TUNEL) staining for apoptosis was performed using the ApopTag Plus Peroxidase In Situ Apoptosis Detection kit (Millipore), per the manufacturer’s instructions.
Immunoblotting
Whole-cell protein extracts (50 μg) were denatured, separated on SDS-PAGE gels, and transferred to polyvinylidene fluoride membranes. After blocking in 3% BSA in TBS–Tween, membranes were probed overnight at 4°C. Primary antibodies used include: AR (PG-21, 1:500 dilution; EMD Millipore), TOPO1 (C-21, 1:100 dilution; Santa Cruz Biotechnology, Inc.), p44/42 MAPK (4695, 1:1,000; Cell Signaling Technology), phospho p44/42 MAPK (9101S, 1:500; Cell Signaling Technology), and α-tubulin (clone B-5-1-2, 1:30,000 dilution; Sigma Aldrich). Following secondary antibody incubation, results were detected using Western Lighting Chemiluminescence Reagent Plus (Perkin Elmer).
Cellular fractionation
Cellular fractionation was performed using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce Biotechnology) as per the manufacturer’s instructions.
Real-time quantitative PCR
RNA was isolated by TRIzol (Invitrogen), and cDNA was synthesized from 1 μg total RNA, using M-Mulv reverse transcriptase enzyme (Promega). SYBR green quantitative gene expression analysis was performed using the following primers: AREG forward, 5′-CGAACCACAAATACCTGGCTA-3′ and AREG reverse, 5′-TCCATTTTTGCCTCCCTTTT-3′; βACTIN forward, 5′-CTGTCCA-CCTTCCAGCAGATG-3′ and βACTIN reverse, 5′-CGCAACTAAG-TCATAGTCCGC-3′; and RPL13A forward, 5′-CCTGGAGGAGAA-GAGGAAAGAGA-3′ and RPL13A reverse, 5′-TTGAGGACCTC-TGTGTATTTGTCAA-3′. Relative gene expression was calculated using the comparative cycle threshold method and values were normalized to β-ACTIN or RPL13A.
Statistical significance
Statistical significance was evaluated using a 2-tailed Student t test or ANOVA with GraphPad Prism software. P ≤ 0.05 was considered statistically significant.
Results
AR is expressed in 22% of TNBC patient tumors and in multiple molecular subtypes of TNBC
We examined 130 primary breast cancers designated as ER and PR negative, HER2 ≤ 10% positive [negative by College of American Pathology (CAP) and FDA criteria] for the presence of AR. In this group of tumors, 22% showed nuclear staining (range, 1%–100%). The presence of AR-positive tumor nuclei correlated with older patients using the Pearson Product Moment correlation (r = 0.383, P < 0.0001). There was a modest correlation between tumors from older patients with lower measures of proliferation (MIB-1: r = −0.230, P = 0.0121 and mitoses/10 hpf: r = −0.204, P = 0.0255). Representative images with a range of AR expression are displayed in Fig. 1A. Nuclear AR expression indicates that AR may be transcriptionally active in AR+ TNBC. Our findings are consistent with earlier studies which have reported AR protein expression in 12% to 36% of TNBC (6–9).
Figure 1.
AR expression and nuclear localization in TNBC patient samples and cell lines. A, representative immunohistochemistry (IHC) of AR protein expression (brown) in TNBC patient samples. Photomicrographs represent a 400× magnification. B, Western blotting for AR expression in a panel of TNBC cell lines representing LAR, basal-like 2 (BL2), MSL, and mesenchymal-like (ML; ref. 14) subtypes of TNBC. The prostate cancer cell line LNCaP is shown as a positive control for AR. C, nuclear–cytoplasmic fractionation of TNBC cell lines grown in 5% charcoal-stripped serum for 48 hours and following a 3-hour treatment with vehicle control (Veh), enzalutamide (ENZ), and/or DHT. Topoisomerase I (TOPO1) is a loading control for the nuclear fraction and α-TUBULIN is a loading control for the cytosolic fraction.
Previously, Lehmann and colleagues reported that TNBC is a heterogeneous disease with the highest AR mRNA and protein expression within the LAR molecular subtype of TNBC (14). However, AR is also expressed in cell lines representing the basal-like 1 and 2 (BL1, BL2), mesenchymal-like (ML), and MSL TNBC molecular subtypes (Fig. 1B) and may also present an opportunity for targeted therapy in these subtypes. In non-LAR TNBC cell lines, treatment with DHT increased nuclear localization of full-length AR, whereas enzalutamide, which blocks AR nuclear localization (17, 24, 25), inhibited this effect (Fig. 1C and Supplementary Fig. S1A). These findings demonstrate that AR nuclear localization is inhibited by enzalutamide in AR+ TNBC and that AR is expressed in cell lines representing multiple molecular subtypes of TNBC in addition to the LAR subtype.
AR inhibition decreases baseline proliferation and increases apoptosis in AR+ TNBC
AR inhibition was studied in 4 cell lines representing non-LAR TNBC subtypes including SUM159PT (MSL), HCC1806 (BL2), BT549 (ML), and MDA231 (MSL). By crystal violet staining, DHT increased baseline proliferation of the SUM159PT cell line and enzalutamide significantly decreased ligand-mediated and baseline proliferation in charcoal-stripped serum (Fig. 2A, P < 0.01). Interestingly, enzalutamide decreased baseline proliferation of HCC1806, BT549, and MDA231, but DHT did not increase proliferation in these cell lines when grown in charcoal-stripped serum. Enzalutamide also increased caspase-3/7 activity compared with vehicle control in SUM159PT, HCC1806, and BT549 (Fig. 2B, P < 0.001). Increased apoptosis was not observed in MDA231. In soft agar, enzalutamide significantly decreased colony formation compared with vehicle control in full serum conditions (Fig. 2C), suggesting that enzalutamide decreases anchorage-independent growth and may decrease tumorigenicity in vivo. Finally, SUM159PT cells were transduced with the ARF876L mutation that confers resistance to enzalutamide. Compared with the parental cells, expression of ARF876L prevented the growth-inhibitory effects of enzalutamide, indicating that the effects of enzalutamide are due to AR (Supplementary Fig. S1B).
Figure 2.
Enzalutamide decreases proliferation and anchorage-independent growth and increases apoptosis in multiple TNBC molecular subtypes. A, crystal violet assay of TNBC cell lines treated with vehicle control (Veh), enzalutamide (ENZ), and/or DHT in 5% charcoal-stripped serum for 5 to 10 days. B, apoptotic index of nuclear red SUM159PT, HCC1806 and BT549 cell lines treated with Veh (open circle) or enzalutamide (solid square) and green fluorescent caspase-3/7 reagent and imaged on the Incucyte ZOOM (Essen BioSciences). C, soft agar assays of TNBC cell lines treated with Veh or enzalutamide in full serum, stained with nitro blue tetrazolium, and quantified using pixel contrast analysis.*, P < 0.05; **, P < 0.01; ***, P < 0.001; error bars, SD.
To confirm that the effects of enzalutamide are specific to AR inhibition, we examined the effects of shRNAs specifically targeting AR (shAR15, shAR17) compared with a nontargeting control (shNEG). Transduction of shRNAs targeting AR decreased full-length AR protein expression and significantly reduced proliferation in an MTS assay in non-LAR TNBC cell lines (Fig. 3A and B and Supplementary Fig. S1C). By crystal violet assay, AR knockdown significantly inhibited baseline and ligand-mediated proliferation of SUM159PT cells indicating that the shRNAs are effectively targeting AR (Fig. 3C). The SUM159PT cell line was chosen for this assay because DHT increases its baseline proliferation in vitro. AR knockdown also increased apoptosis in all 4 cell lines as measured by cleaved caspase-3 activity (Fig. 3D).
Figure 3.
AR knockdown inhibits baseline and ligand-mediated proliferation and increases apoptosis in TNBC. A, Western blotting of TNBC cell lines infected with shRNAs targeting AR (shAR15, shAR17) compared with a nontargeting control (shNEG) on day 3. B, MTS assays of transduced TNBC cell lines. C, crystal violet assay of transduced SUM159PT shNEG or shAR15/shAR17 cells treated 1 week with vehicle control (Veh), enzalutamide (ENZ), and/or DHT. D, changes in apoptosis in AR knockdown cells measured with cleaved caspase reagent (Essen BioSciences) and normalized to cell count (apoptotic index) at 42 hours. Staurosporine (SSP) was used a positive control for apoptosis. *, P < 0.05; ***, P < 0.001 by ANOVA; error bars, SD.
Enzalutamide decreases tumor viability in vivo
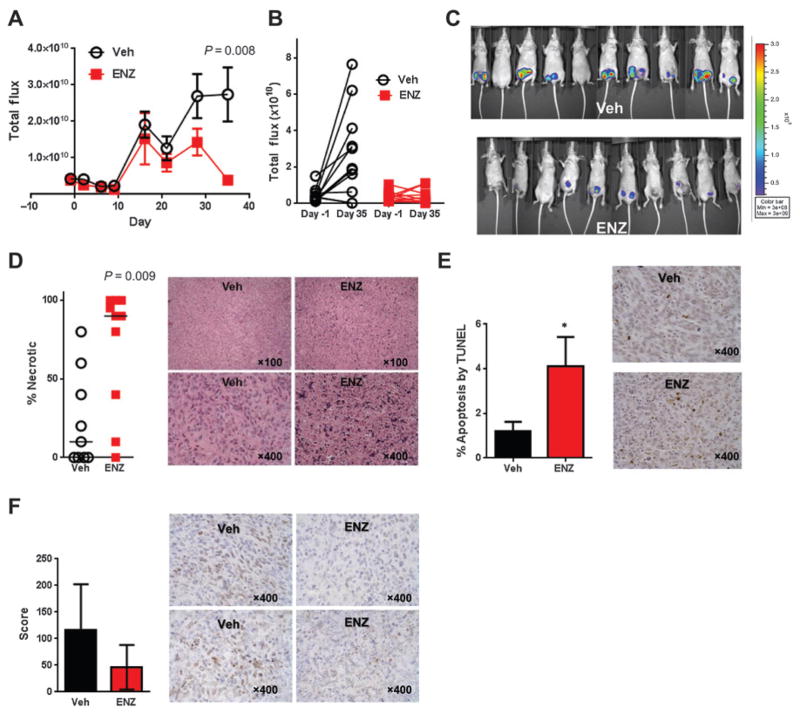
Luciferase-tagged SUM159PT-TGL cells, representing the MSL TNBC subtype, were bilaterally injected into the mammary fat pads of immunocompromised mice and treated with enzalutamide or vehicle control (Veh) following randomization when the tumors reached 50 mm3 (day -1, Supplementary Fig. S2A and S2B). Enzalutamide significantly decreased luciferase activity on day 35 (P = 0.008, Fig. 4A–C). While no significant differences in caliper measurements or tumor weights were found between treatment groups (Supplementary Fig. S3A–S3C), H&E staining demonstrated that the median percentage of necrotic tumor was 90% in the enzalutamide treatment group compared with 10% in Veh xenografts (P = 0.009, Fig. 4D). The percentage of necrotic tissue of H&E-stained sections was determined in a blinded fashion by a board-certified pathologist (P. Jedlicka). Enzalutamide-treated xenografts also exhibited a 4-fold increase in TUNEL staining (P = 0.04, Fig. 4E) and a 2-fold decrease in AR score (score = intensity range 0 to 3 × percent nuclear positivity, P = 0.07, Fig. 4F) compared with vehicle-treated controls.
Figure 4.
Enzalutamide (ENZ) decreases cellular viability and increases necrosis and apoptosis in SUM159PT xenografts. A, total flux growth curve of SUM159PT nude mice xenografts. Mice were randomized at day −1 and treatment was initiated on day 0. P value represents a 2-tailed t test comparing total flux between groups on day 35 and error bars represent SEM. B, change in total flux between randomization and day 35, by mouse. C, luminescent overlay of Veh and enzalutamide-treated mice. D, percent necrotic tissue by H&E staining. Horizontal bars represent median percentage necrotic tissue. P value represents a 2-tailed t test comparing percent necrosis between groups on day 35. Photomicrographs depict examples of tumor xenograft H&E staining showing viable tumor (Veh) and necrotic tumor (ENZ). E, TUNEL staining for apoptosis. Photomicrographs depict examples of TUNEL staining. F, AR nuclear score (score = intensity range 0 to 3× % positive) by IHC. Photomicrographs depict examples of AR staining in SUM159PT xenografts. *, P < 0.05; error bars, SEM.
As in the SUM159PT xenograft study, luciferase-tagged HCC1806-TGL cells, which represent the BL2 TNBC subtype, were bilaterally injected into the mammary fat pads of immunocompromised mice and treated with enzalutamide or vehicle following randomization (Supplementary Fig. S2C and S2D). Enzalutamide significantly decreased luciferase activity on days 10 and 14 (P < 0.001, Supplementary Fig. S4A–S4C). HCC1806 xenografts grew at a faster rate than SUM159PT xenografts, resulting in early termination of the study on day 14 and a high degree of necrosis in both treatment groups. However, by H&E staining, enzalutamide-treated xenografts displayed increased necrosis (Supplementary Fig. S4D). Enzalutamide-treated xenografts also exhibited increased TUNEL staining compared with vehicle-treated controls (P = 0.04, Supplementary Fig. S4E). No significant differences in caliper measurements or tumor weights were found between treatment groups (Supplementary Fig. S3D–S3F). In summary, our results show that enzalutamide decreases cellular viability while increasing necrosis and apoptosis in vivo in 2 non-LAR molecular subtypes of TNBC in addition to the LAR MDA-MB-453 cell line previously reported (17).
AR inhibition alters cellular morphology and decreases migration and invasion
AR knockdown altered cellular morphology of BT549 and MDA231 cells in 3D Matrigel (BD Biosciences) culture from stellate to round (Fig. 5A). In a scratch wound assay, AR knockdown significantly decreased migration compared with a nontargeting control in 4 AR+ non-LAR cell lines (Fig. 5B). Scratch wound assays were conducted in serum-starved, attractant-free conditions and over a short time course to minimize potential confounding effects of AR knockdown on cell proliferation. MDA231 and BT549 cell lines invade through Matrigel without an attractant and AR knockdown in these cell lines inhibited invasion (Fig. 5C). Changes in cellular morphology and decreased migration and invasion were next examined in BT549 cells treated with enzalutamide. In 3D Matrigel, cellular morphology was altered from predominately stellate to predominately round (Fig. 5D), and migration (Fig. 5E, left) and invasion (Fig. 5E, right) were significantly inhibited by enzalutamide. In identical serum-starved conditions, BT549 control wells treated with enzalutamide and cleaved caspase reporter (Essen BioSciences) exhibited no changes in proliferation or apoptosis (Supplementary Fig. S5), demonstrating that AR influences migration independently of proliferation or apoptosis. At this concentration of enzalutamide, no significant changes in migration were observed in other non-LAR cell lines tested.
Figure 5.
AR inhibition decreases migration and invasion of TNBC cells. A, cellular morphology (200×) of BT549 cells transduced with a nontargeting control (shNEG) compared with a shRNA targeting AR (shAR15) in 3D Matrigel culture. Arrow, stellate cellular morphology. B, migration scratch wound assay of TNBC cell lines with AR knockdown under serum-starved conditions. C, scratch wound assay of TNBC AR knockdown cell lines invading through Matrigel. D, changes in cellular morphology of BT549 cells treated with vehicle (Veh) or enzalutamide (ENZ) in 3D Matrigel culture (200×). E, migration (left) and invasion (right) assays of BT549 cells treated with or without enzalutamide. **, P < 0.01; ***, P < 0.001 by the t test at the final time point.
Amphiregulin is regulated by AR in TNBC and rescues decreased proliferation and migration associated with AR inhibition
By microarray and AR chromatin immunoprecipitation of an immortalized human prostate epithelial cell line, Bolton and colleagues identified amphiregulin (AREG) as an AR-regulated gene (26). AREG is required for mammary ductal morphogenesis and is the predominant EGF receptor (EGFR) ligand during mammary gland development (27). To date, AR is not known to regulate AREG in breast cancer or normal breast tissue. However, within TNBC, AR expression correlates with activated EGFR (28). We thus hypothesized that AR may regulate AREG in TNBC.
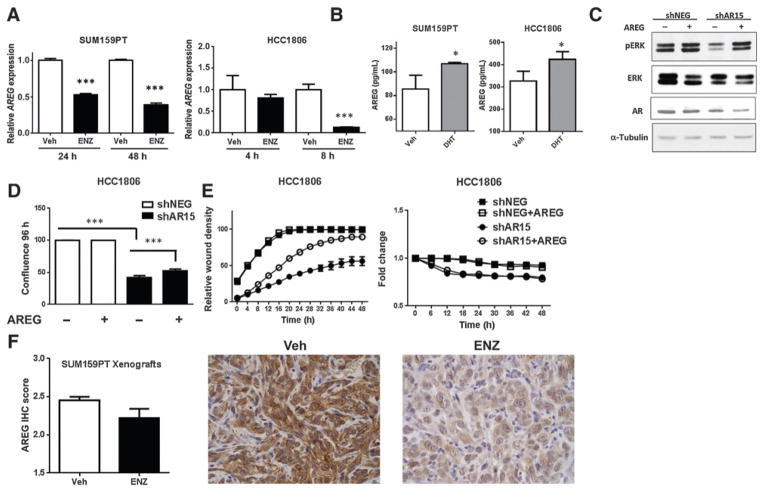
By quantitative real-time PCR (qRT-PCR), treatment with enzalutamide decreased AREG mRNA expression by 2-fold in SUM159PT and 4-fold in HCC1806 (P < 0.001, Fig. 6A). At the protein level, treatment with DHT significantly increased secreted AREG by ELISA in both SUM159PT and HCC1806 (P < 0.05, Fig. 6B). EGFR activation by AREG induces multiple downstream signaling pathways including MAPK (16). Compared with non-targeting controls, AR knockdown decreased endogenous phosphorylation of ERK, and exogenous AREG rescued this effect in HCC1806 (Fig. 6C). These results were recapitulated in the SUM159PT cell line (Supplementary Fig. S6A).
Figure 6.
AR regulation of amphiregulin mediates baseline proliferation and migration of TNBC. A, quantitative real-time PCR (qRT-PCR) for amphiregulin (AREG) in SUM159PT cells and HCC1806 cells treated with enzalutamide (ENZ) in full serum. B, ELISA for extracellular AREG in SUM159PT and HCC1806 cell lines treated with vehicle (Veh) or dihydrotestosterone (DHT) for 48 and 72 hours, respectively. C, Western blotting of HCC1806 shNEG and shAR15 cells treated for 30 minutes with exogenous human recombinant AREG. D, proliferation assay of HCC1806 shNEG and shAR15 cells in the absence or presence of exogenous AREG. E, migration (left) and proliferation (right) assays of HCC1806 cells treated with or without exogenous AREG in identical, serum-starved conditions. F, AREG protein expression by IHC in SUM159PT xenografts. P = 0.04 using a 1-tailed t test. Photomicrographs depict representative AREG staining (400×). *, P < 0.05; ***, P < 0.001 by the t test.
Given our data suggesting that AR regulates AREG which activates the MAPK signaling pathway with key roles in proliferation, migration, and invasion (29), we next tested whether exogenous AREG would rescue the phenotypes associated with AR inhibition. As in Fig. 3B, AR knockdown significantly inhibited proliferation of HCC1806 compared with nontargeting controls, and the addition of exogenous AREG partially rescued this effect (P < 0.0001, Fig. 6D). Similarly, AR knockdown decreased migration of HCC1806 cells (as in Fig. 5B) and exogenous AREG partially rescued this effect (Fig. 6E, left) without altering proliferation in these serum-free conditions (Fig. 6E, right). Exogenous AREG also partially rescued proliferation and invasion in SUM159PT (Supplementary Fig. S6B and S6C). Enzalutamide-treated SUM159PT xenografts displayed decreased AREG expression compared with vehicle controls (Fig. 6F). Together, these data indicate that AR regulation of AREG is one mechanism by which AR effects proliferation, migration, and invasion in AR+ TNBC.
Discussion
Compared to patients with non-TNBC, patients with TNBC with residual disease following chemotherapy have a significantly worse overall survival (2). The poor prognosis of patients with TNBC is due, in part, to a lack of effective targeted therapy. However, AR is expressed in up to a third of patients with TNBC (6–9) and represents an opportunity for targeted therapy. Indeed, if AR-targeted therapy is effective in AR+ TNBC, it would represent the first effective targeted therapy for this aggressive breast cancer subtype and would greatly benefit this population of women. Previous studies focused on the role of AR in the high AR-expressing, LAR molecular subtype of TNBC and found that this subtype was responsive to bicalutamide, whereas the non-LAR subtypes were less responsive or nonresponsive (14). In contrast, we find that multiple non-LAR subtypes (mesenchymal-like, MSL, and basal-like 2) with relatively low AR expression critically depend on AR for proliferation, migration, and invasion and that even those previously found to be resistant to bicalutamide are sensitive to the new-generation anti-androgen enzalutamide in vitro and in vivo.
In TNBC cell lines representing the “mesenchymal-like,” “mesenchymal stem–like,” and “basal-like” molecular subtypes (14), pharmacologic inhibition of AR with enzalutamide and AR knockdown decreased proliferation and anchorage-independent growth and increased apoptosis. Thus, AR may be required for optimal baseline proliferation even though DHT does not increase proliferation in all AR+ TNBC cell lines. The discordance between baseline inhibition and lack of ligand-mediated proliferation in some cell lines may indicate that the mechanism by which AR mediates proliferation is nontranscriptional or less ligand-dependent in the non-LAR cell lines. Interestingly, MDA231 cells were less sensitive to enzalutamide by soft agar and caspase-3/7 assays and a recent study suggests that this may be due to expression of AR variant 3, which lacks the ligand-binding domain, in this cell line (30).
Decreased viability and increased apoptosis by AR inhibition in vitro was recapitulated in SUM159PT and HCC1806 xenografts in nude mice. Although other groups have suggested that AR inhibition could promote survival through activation of PI3K signaling (31), enzalutamide significantly decreased survival in both wild-type (HCC1806) and PIK3CA-mutant (SUM159PT) cell lines. Sensitivity of SUM159PT xenografts to enzalutamide contrasts previous work demonstrating that bicalutamide did not inhibit tumor volume (14). The discrepancy in results may be due to differences in the mechanisms of action of the two AR antagonists. Bicalutamide permits AR nuclear localization and binding to chromatin, recruiting corepressors rather than coactivators, whereas enzalutamide inhibits nuclear localization and DNA binding (32). Bicalutamide has partial agonist effects in prostate cancer (33) and thus may also have partial agonist effects in TNBC. However, it should be noted that enzalutamide significantly increased tumor necrosis but did not decrease tumor volume according to caliper measurements. Thus, an increase in necrosis may not have been apparent in the bicalutamide study by measurement of tumor volume alone.
A phase II clinical trial of bicalutamide in AR+/ER−/PR− metastatic breast cancer reported a 19% clinical benefit rate and a 12-week longer median progression-free survival (19). Of note, the study included HER2-amplified patients and required 10% AR-positive staining for trial eligibility. However, bicalutamide has partial agonist effects (33) and patients with prostate cancer who acquire resistance to bicalutamide often respond to enzalutamide (34), suggesting that enzalutamide may be a more effective antagonist in TNBC. The results of the present study are promising and timely as a phase II clinical trial is currently testing the efficacy of enzalutamide in AR+ TNBC (NCT01889238). Our finding that non-LAR subtypes also critically depend on AR indicates that patients with relatively low AR expression may also benefit from AR-targeted therapy. Indeed, the trial has recently expanded patient eligibility to 1% AR+ staining, which may improve the number of patients eligible for treatment.
In vitro, AR inhibition altered cellular morphology and decreased migration and invasion suggesting that AR+ TNBC is also dependent on AR for these functions. Extensive evidence suggests that advanced, metastatic prostate cancer is causally related to continued AR activation (5), and recent prostate and bladder cancer studies demonstrate that AR regulates multiple metastasis-promoting genes (35–37). In breast cancer, initial surgically resected breast cancer metastases retain nuclear AR expression as in the primary tumor (38). Interestingly, breast cancer metastases, including those in patients with TNBC, also have significantly increased AR phosphorylation (39), indicative of active receptors.
Multiple studies have demonstrated that AR expression is associated with an overall favorable prognosis in breast cancer including the TNBC subtype (8, 40–42). However, this is not surprising because like ER, AR is indicative of a more well-differentiated form of the disease but may still drive tumor growth and therefore serve as a rational therapeutic target. High AR expression may be indicative of a more luminal, well-differentiated, less aggressive tumor, and this confers a good prognosis. Future studies are needed to further characterize the role of AR in breast cancer metastasis and determine if AR-targeted therapy will reduce metastatic burden in preclinical models of TNBC.
Treatment with AREG, an EGFR ligand with critical roles in normal mammary gland development, partially rescued decreased proliferation, migration, and invasion resulting from AR knockdown. Thus, AR regulation of AREG may be one mechanism by which AR mediates these critical functions and AR antagonists may also indirectly target EGFR signaling. Recent clinical trials demonstrate that treatment with the anti-EGFR antibody cetuximab in addition to chemotherapy may benefit patients with metastatic breast cancer (43, 44). In vitro data suggest that combined AR antagonist and EGFR or ERK1/2 inhibitors may be effective in TNBC (28). Exogenous AREG only partially rescued phenotypes associated with AR inhibition indicating that additional AR-regulated genes are likely involved in these phenotypes.
Although extensive genomic studies to identify novel therapeutic strategies have expanded our knowledge of the diverse molecular biology of TNBC, there are currently no effective targeted therapies for patients with TNBC. The present study demonstrates that multiple molecular subtypes of TNBC depend on AR for critical cancer phenotypes including viability, migration, and invasion. Despite heterogeneity among tumors, hormone receptor–targeted therapies have greatly improved the prognosis of multiple hormone-related malignancies and exploiting AR dependence with AR-targeted therapies may ultimately improve TNBC patient prognosis.
Supplementary Material
Acknowledgments
The authors acknowledge the Shared Resources at University of Colorado Denver Anschutz Medical Campus and the Cancer Center NCI Support Grant (P30CA046934).
Grant Support
The study was funded by the DOD BCRP Clinical Translational Award BC120183 (to J.K. Richer and A. Elias).
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
A. Elias has received other commercial research support from Medivation and Astellas. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: V.N. Barton, N.C. D’Amato, A. Elias, J.K. Richer
Development of methodology: V.N. Barton, N.C. D’Amato, N.S. Spoelstra, B.M. Jacobsen, J.K. Richer
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): V.N. Barton, M.A. Gordon, N.S. Spoelstra, R.E. Heinz, J.K. Richer
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): V.N. Barton, P. Jedlicka, B.M. Jacobsen, J.K. Richer
Writing, review, and/or revision of the manuscript: V.N. Barton, N.C. D’Amato, M.A. Gordon, N.S. Spoelstra, A. Elias, B.M. Jacobsen, J.K. Richer
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): V.N. Barton, H.T. Lind
Other (animal husbandry): B. L. Babbs
References
- 1.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 2.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–81. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 3.Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384:164–72. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 4.Skor MN, Wonder EL, Kocherginsky M, Goyal A, Hall BA, Cai Y, et al. Glucocorticoid receptor antagonism as a novel therapy for triple-negative breast cancer. Clin Cancer Res. 2013;19:6163–72. doi: 10.1158/1078-0432.CCR-12-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lonergan PE, Tindall DJ. Androgen receptor signaling in prostate cancer development and progression. J Carcinog. 2011;10:20. doi: 10.4103/1477-3163.83937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses’ Health Study. Mod Pathol. 2011;24:924–31. doi: 10.1038/modpathol.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mrklic I, Pogorelic Z, Capkun V, Tomic S. Expression of androgen receptors in triple negative breast carcinomas. Acta Histochem. 2013;115:344–8. doi: 10.1016/j.acthis.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Thike AA, Yong-Zheng Chong L, Cheok PY, Li HH, Wai-Cheong Yip G, Huat Bay B, et al. Loss of androgen receptor expression predicts early recurrence in triple-negative and basal-like breast cancer. Mod Pathol. 2014;27:352–60. doi: 10.1038/modpathol.2013.145. [DOI] [PubMed] [Google Scholar]
- 9.Safarpour D, Pakneshan S, Tavassoli FA. Androgen receptor (AR) expression in 400 breast carcinomas: is routine AR assessment justified? Am J Cancer Res. 2014;4:353–68. [PMC free article] [PubMed] [Google Scholar]
- 10.Guedj M, Marisa L, de Reynies A, Orsetti B, Schiappa R, Bibeau F, et al. A refined molecular taxonomy of breast cancer. Oncogene. 2012;31:1196–206. doi: 10.1038/onc.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson JL, Macarthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG, et al. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011;30:3019–27. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C, et al. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene. 2006;25:3994–4008. doi: 10.1038/sj.onc.1209415. [DOI] [PubMed] [Google Scholar]
- 13.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–71. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stern KA, Place TL, Lill NL. EGF and amphiregulin differentially regulate Cbl recruitment to endosomes and EGF receptor fate. Biochem J. 2008;410:585–94. doi: 10.1042/BJ20071505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cochrane DR, Bernales S, Jacobsen BM, Cittelly DM, Howe EN, D’Amato NC, et al. Role of the androgen receptor in breast cancer and preclinical analysis of enzalutamide. Breast Cancer Res. 2014;16:R7. doi: 10.1186/bcr3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, Imai Y, et al. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 2011;20:119–31. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gucalp A, Tolaney S, Isakoff SJ, Ingle JN, Liu MC, Carey LA, et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic breast cancer. Clin Cancer Res. 2013;19:5505–12. doi: 10.1158/1078-0432.CCR-12-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Recchione C, Venturelli E, Manzari A, Cavalleri A, Martinetti A, Secreto G. Testosterone, dihydrotestosterone and oestradiol levels in postmenopausal breast cancer tissues. J Steroid Biochem Mol Biol. 1995;52:541–6. doi: 10.1016/0960-0760(95)00017-t. [DOI] [PubMed] [Google Scholar]
- 21.Peters AA, Ingman WV, Tilley WD, Butler LM. Differential effects of exogenous androgen and an androgen receptor antagonist in the peri-and postpubertal murine mammary gland. Endocrinology. 2011;152:3728–37. doi: 10.1210/en.2011-1133. [DOI] [PubMed] [Google Scholar]
- 22.Gallicchio L, Macdonald R, Wood B, Rushovich E, Helzlsouer KJ. Androgens and musculoskeletal symptoms among breast cancer patients on aromatase inhibitor therapy. Breast Cancer Res Treat. 2011;130:569–77. doi: 10.1007/s10549-011-1611-2. [DOI] [PubMed] [Google Scholar]
- 23.Sedelaar JP, Isaacs JT. Tissue culture media supplemented with 10% fetal calf serum contains a castrate level of testosterone. Prostate. 2009;69:1724–9. doi: 10.1002/pros.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–90. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belikov S, Oberg C, Jaaskelainen T, Rahkama V, Palvimo JJ, Wrange O. FoxA1 corrupts the antiandrogenic effect of bicalutamide but only weakly attenuates the effect of MDV3100 (Enzalutamide) Mol Cell Endocrinol. 2013;365:95–107. doi: 10.1016/j.mce.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–17. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aupperlee MD, Leipprandt JR, Bennett JM, Schwartz RC, Haslam SZ. Amphiregulin mediates progesterone-induced mammary ductal development during puberty. Breast Cancer Res. 2013;15:R44. doi: 10.1186/bcr3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuenca-Lopez MD, Montero JC, Morales JC, Prat A, Pandiella A, Ocana A. Phosphokinase profile of triple negative breast cancer and androgen receptor signaling. BMC Cancer. 2014;14:302. doi: 10.1186/1471-2407-14-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–90. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 30.Hu DG, Hickey TE, Irvine C, Wijayakumara DD, Lu L, Tilley WD, et al. Identification of androgen receptor splice variant transcripts in breast cancer cell lines and human tissues. Hormones Cancer. 2014;5:61–71. doi: 10.1007/s12672-014-0171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehmann BD, Bauer JA, Schafer JM, Pendleton CS, Tang L, Johnson KC, et al. PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors. Breast Cancer Res. 2014;16:406. doi: 10.1186/s13058-014-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osguthorpe DJ, Hagler AT. Mechanism of androgen receptor antagonism by bicalutamide in the treatment of prostate cancer. Biochemistry. 2011;50:4105–13. doi: 10.1021/bi102059z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kemppainen JA, Wilson EM. Agonist and antagonist activities of hydro-xyflutamide and Casodex relate to androgen receptor stabilization. Urology. 1996;48:157–63. doi: 10.1016/s0090-4295(96)00117-3. [DOI] [PubMed] [Google Scholar]
- 34.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 35.Yin JJ, Liu YN, Tillman H, Barrett B, Hewitt S, Ylaya K, et al. AR-regulated TWEAK-FN14 pathway promotes prostate cancer bone metastasis. Cancer Res. 2014;74:4306–17. doi: 10.1158/0008-5472.CAN-13-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacob S, Nayak S, Fernandes G, Barai RS, Menon S, Chaudhari UK, et al. Androgen receptor as a regulator of ZEB2 expression and its implications in epithelial-to-mesenchymal transition in prostate cancer. Endocr Relat Cancer. 2014;21:473–86. doi: 10.1530/ERC-13-0514. [DOI] [PubMed] [Google Scholar]
- 37.Jing Y, Cui D, Guo W, Jiang J, Jiang B, Lu Y, et al. Activated androgen receptor promotes bladder cancer metastasis via Slug mediated epithelial-mesenchymal transition. Cancer Lett. 2014;348:135–45. doi: 10.1016/j.canlet.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 38.Cimino-Mathews A, Hicks JL, Illei PB, Halushka MK, Fetting JH, De Marzo AM, et al. Androgen receptor expression is usually maintained in initial surgically resected breast cancer metastases but is often lost in end-stage metastases found at autopsy. Hum Pathol. 2012;43:1003–11. doi: 10.1016/j.humpath.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren Q, Zhang L, Ruoff R, Ha S, Wang J, Jain S, et al. Expression of androgen receptor and its phosphorylated forms in breast cancer progression. Cancer. 2013;119:2532–40. doi: 10.1002/cncr.28092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters KM, Edwards SL, Nair SS, French JD, Bailey PJ, Salkield K, et al. Androgen receptor expression predicts breast cancer survival: the role of genetic and epigenetic events. BMC Cancer. 2012;12:132. doi: 10.1186/1471-2407-12-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agoff SN, Swanson PE, Linden H, Hawes SE, Lawton TJ. Androgen receptor expression in estrogen receptor-negative breast cancer. Immunohistochemical, clinical, and prognostic associations. Am J Clin Pathol. 2003;120:725–31. doi: 10.1309/42F0-0D0D-JD0J-5EDT. [DOI] [PubMed] [Google Scholar]
- 42.Luo X, Shi YX, Li ZM, Jiang WQ. Expression and clinical significance of androgen receptor in triple negative breast cancer. Chin J Cancer. 2010;29:585–90. doi: 10.5732/cjc.009.10673. [DOI] [PubMed] [Google Scholar]
- 43.Baselga J, Gomez P, Greil R, Braga S, Climent MA, Wardley AM, et al. Randomized phase II study of the anti-epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple-negative breast cancer. J Clin Oncol. 2013;31:2586–92. doi: 10.1200/JCO.2012.46.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nechushtan H, Vainer G, Stainberg H, Salmon AY, Hamburger T, Peretz T. A phase 1/2 of a combination of Cetuximab and Taxane for “triple negative” breast cancer patients. Breast. 2014;23:435–8. doi: 10.1016/j.breast.2014.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.