Abstract
Background
Protein tyrosine phosphatase receptor type D (PTPRD) is a putative tumor suppressor in several cancers including head and neck squamous cell carcinoma (HNSCC). STAT3 is a frequently hyperactivated oncogene in HNSCC. As STAT3 is a direct substrate of PTPRD, we sought to determine the genetic or epigenetic alterations of PTPRD that contribute to overactive STAT3 in HNSCC.
Methods
We analyzed data from The Cancer Genome Atlas (TCGA) and our previous whole-exome sequencing study and summarized the mutation, methylation, and copy number status of PTPRD in HNSCC and other cancers. In vitro studies involved standard transfection and MTT protocols, as well as methylation-specific PCR.
Results
Our findings indicate that PTPRD mutation, rather than methylation or copy number alteration, is the primary mechanism by which PTPRD function is lost in HNSCC. We demonstrate that overexpression of wild-type PTPRD in HNSCC cells significantly inhibits growth and STAT3 activation while PTPRD mutants do not, suggesting that mutation may lead to loss of function and subsequent hyper-phosphorylation of PTPRD substrates, especially STAT3. Importantly, we determined that HNSCC cells harboring an endogenous PTPRD mutation are more sensitive to STAT3 blockade than PTPRD wild-type cells. We additionally found that PTPRD mRNA expression does not correlate with pSTAT3 expression, suggesting that alterations that manifest through altered mRNA expression, including hypermethylation and gene copy number alterations, do not significantly contribute to STAT3 overactivation in HNSCC.
Conclusion
PTPRD mutation, but not methylation or copy number loss, may serve as a predictive biomarker of sensitivity to STAT3 inhibitors in HNSCC.
Introduction
Protein tyrosine phosphatase receptor type D (PTPRD) is a member of the receptor-type protein tyrosine phosphatase (PTPR) family. PTPRs are membrane-integral enzymes that catalyze the removal of phosphate groups from specific proteins, thereby impacting cell signaling. Dysregulation of PTPR signaling by genetic and/or epigenetic mechanisms may therefore ultimately lead to cancerous phenotypes. [1] Several PTPR family members, including PTPRD, have been reported to function as tumor suppressors where loss of function alterations may drive tumor growth. [2,3] Genetic events including mutation, gene deletion, or epigenetic silencing may lead to decreased phosphatase activity of PTPRs and enhanced oncogenic signaling. [1]
We recently reported the cumulative mutation profile of the PTPR gene family in cancer with a focus on PTPRT mutation leading to STAT3 activation in head and neck squamous cell carcinoma (HNSCC). [4] Our analysis revealed that 15 solid tumor types harbored mutations of at least one PTPR gene. PTPRD is one of the most commonly mutated PTPR family members in HNSCC, and PTPRD has been reported to function as a tumor suppressor in this malignancy. [5] PTPRD is also mutated, deleted, or hyper-methylated in glioblastoma (GBM), while the gene is unmethylated and expressed in normal brain tissue. [6] Furthermore, PTPRD mutations were found to be associated with increased expression of phosphorylated STAT3, a direct PTPRD substrate, in GBM. In addition to GBM, 13% and 25% of HNSCC tumors analyzed in the above study harbored PTPRD mutation or promoter methylation, respectively. Homozygous deletion of PTPRD has additionally been reported in laryngeal cancer, suggesting that genetic aberrations affecting PTPRD function may be a common event across many cancers. [5]
These cumulative findings led us to hypothesize that genetic and/or epigenetic alteration of PTPRD may contribute to enhanced signaling and growth in HNSCC where key components of the pathway may serve as plausible therapeutic targets. Here, we summarize the genetic and epigenetic profile of PTPRD in HNSCC from The Cancer Genome Atlas (TCGA) and our prior HNSCC mutational landscape study. [7] We then tested the consequences of PTPRD alterations found in human HNSCC tumors in relevant preclinical models to assess STAT3 activity and sensitivity to STAT3 inhibition.
Materials and Methods
Cell culture, drug treatment, and transfection
All HNSCC cell lines were genotypically verified as previously described. [4] Cal27 cells were obtained from ATCC (Manassas, VA). PE/CA-PJ34clone12 and PE/CA-PJ49 cells were obtained from Sigma-Aldrich (St. Louis, MO). 686LN cells were obtained from Georgia Chen at MD Anderson Cancer Center (Houston, TX). Cal27 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Mediatech, Inc., Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Gemini Bio-Products, West Sacramento, CA). 686LN cells were cultured in DMEM/F12 (Life Technologies, Grand Island, NY) supplemented with 10% FBS. PE/CA-PJ34clone12 and PE/CA-PJ49 were cultured in Iscove's Modification of DMEM (Mediatech Inc., Manassas, VA) supplemented with 10% FBS and 2 mM L-glutamine (Life Technologies, Grand Island, NY). All cells were maintained in an incubator at 37°C and 5% CO2. JSI-124 (Calbiochem, Billerica, MA) was dissolved in DMSO. Transfection was performed with Lipofectamine 2000 (Life Technologies, Grand Island, NY) or FuGENE HD (Promega Corporation, Madison, WI) according to the manufacturer’s instructions with 4 μg DNA diluted in Opti-MEM (Life Technologies, Grand Island, NY).
Site-directed mutagenesis
PTPRD mutations (K1502M, S384R, T1100M and L1147F) were generated from the wild-type plasmid using the Phusion site-directed mutagenesis kit (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s protocol and confirmed by Sanger sequencing. Plasmid amplifications were performed using the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany) or the Hurricane Maxi Prep Kit (GerardBIOTECH, Oxford, OH) according to the manufacturer’s instructions.
Immunoblotting
Primary antibodies for pSTAT3 (Y705) and STAT3 were obtained from Cell Signaling Technology (Danvers, MA). β-tubulin primary antibody was purchased from Abcam (Cambridge, MA). Secondary antibodies were purchased from BioRad (Hercules, CA). Blots were quantitated by densitometry using ImageJ software.
MTT assay
MTT assays were performed by incubating cells in 5 mg/ml 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide in PBS at for 30 min at 37°C and 5% CO2. After incubation, the solution was aspirated and replaced with DMSO. Absorbance of this solution was measured at 570 nm on a spectrophotometer.
Methylation-specific PCR
Tumor and normal tissue were obtained under the auspices of an Institutional Review Board-approved protocol at the University of Pittsburgh. DNA was isolated from formalin-fixed paraffin embedded tissue using the QIAamp DNA FFPE Tissue Kit, and cell line DNA was isolating using the QIAamp DNA Mini Kit. Bisulfite conversion was performed using the EpiTect Bisulfite Kit and methylation-specific polymerase chain reaction (MSP) was conducted using EpiTect MSP Kit. All kits were used according to the manufacturer’s instructions (Qiagen, Hilden, Germany). MSP primers were designed using MethPrimer software. [8] For methylation, the forward and reverse primer sequences were GTTTTATTTTGGATTTTTGGAATC and TACCTAACGCGACCTAAACG, respectively. For unmethylation, the forward and reverse primer sequences were TATTTTGGATTTTTGGAATTGT and AACTACCTAACACAACCTAAACAAA, respectively. Primers were specific for the intended methylation status as determined using the EpiTect Control DNA Set (Qiagen, Hilden, Germany).
Data download and analysis
Mutation, copy number alteration, and RNA-Seq data were aggregated from the cBio Portal [9,10] and published reports. [6,7,9] DNA methylation data were obtained through the TCGA data matrix (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm). DNA and protein sequences and domain annotations were obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Reverse phase protein array data were obtained from The Cancer Proteome Atlas (http://app1.bioinformatics.mdanderson.org/tcpa/_design/basic/index.html). Statistical tests were performed using GraphPad Prism 5 software (GraphPad, La Jolla, CA).
Trypan Blue Exclusion
PE/CA-PJ34clone12 cells were plated on 6-well plates at 100,000 cells per well. After 24 hours, cells were transfected with vector control, wild-type PTPRD, or PTPRD mutants in triplicate using 4 μg of plasmid DNA, 12 μL FuGENE HD (Promega Corporation, Madison, WI), and 200 μL Opti-MEM (Life Technologies, Grand Island, NY) per well added directly to wells containing 3 mL complete medium. After 72 hours, cells were trypsinized and resuspended in PBS containing 0.2% trypan blue (MP Biomedicals, LLC, Santa Ana, CA). Each cell suspension was counted in four fields on a hemacytometer and the average was used to calculate the total number of cells present.
Results
PTPRD mutations occur frequently in HNSCC and other cancers
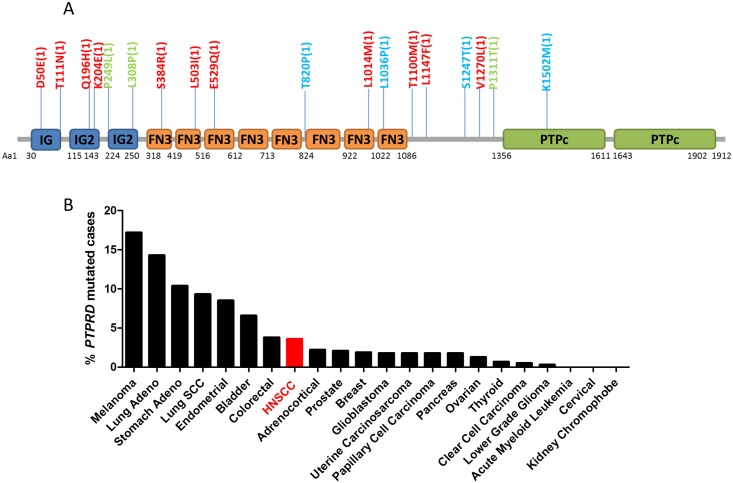
Somatic mutation is a common event leading to altered gene and protein function in cancer. Eighteen non-synonymous PTPRD mutations have been identified to date in HNSCC tumors. [6,7,9,10] These mutations appear scattered throughout the gene and protein sequence. (Fig 1A) They are all non-recurrent, with each occurring in a single HNSCC tumor, suggesting that these are likely be loss-of-function mutations. Of these 18 mutations, 12 occur in the extracellular domain, 1 is localized to the catalytic domain, and 5 are in the transmembrane region where no conserved domain has been identified. In addition to HNSCC, PTPRD mutations are also widely observed in other cancer types. (Fig 1B) In The Cancer Genome Atlas (TCGA) collection, PTPRD is most frequently mutated in cutaneous melanoma (59/344 cases, 17.2%), followed by lung adenocarcinoma (33/230 cases, 14.3%), and stomach adenocarcinoma (30/289 cases, 10.4%). [9,10] As in HNSCC, PTPRD mutations in these cancers do not appear to cluster into hotspot regions or residues, suggesting that loss of PTPRD function by somatic mutation is a common event across multiple cancer sites.
Fig 1. PTPRD mutations are common in HNSCC and other cancers.
(A) Localization of PTPRD protein mutations reported in HNSCC tumors. Red: TCGA tumors [9,10]; Green: from [6]; Blue: from [7]. IG, Immunoglobulin; IG2, Second immunoglobulin (Ig)-like domain of the receptor protein tyrosine phosphatase (RPTP)-F; FN3, Fibronectin type 3 domain; PTPc, Protein tyrosine phosphatase, catalytic domain. (B) Frequency of PTPRD mutations in human cancers as determined by TCGA, with HNSCC shown in red.
The only mutation detected in HNSCC that has been previously identified in another cancer is T1100M, which was reported in chronic lymphocytic leukemia. [11] Interestingly, several other mutation sites found in HNSCC have a different amino acid substitution reported in other cancers. For example, while K204E is observed in HNSCC, K204Q has been reported in esophageal cancer. [12] In addition, a review of the COSMIC database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) demonstrates detection of L503V in liver cancer (compared to L503I in HNSCC) and L1036M in colon adenocarcinoma (compared to L1036P in HNSCC). Ding et al additionally reported a mutation at this site (L1036Q) in lung adenocarcinoma. [13] Interestingly, while K1502M is the only catalytic domain mutation identified to date in HNSCC, TCGA has identified a K1502* nonsense mutation in lung adenocarcinoma. Together, these findings suggest that while the specific PTPRD mutations found to date in HNSCC are unique, the amino acid sites at which they occur may represent important residues that are susceptible to genetic alterations. Indeed, multiple sequence alignment analysis indicates that these residues are highly conserved across species, suggesting they may have a critical role in proper PTPRD function (analysis not shown).
HNSCC-derived PTPRD mutants lead to increased cell growth
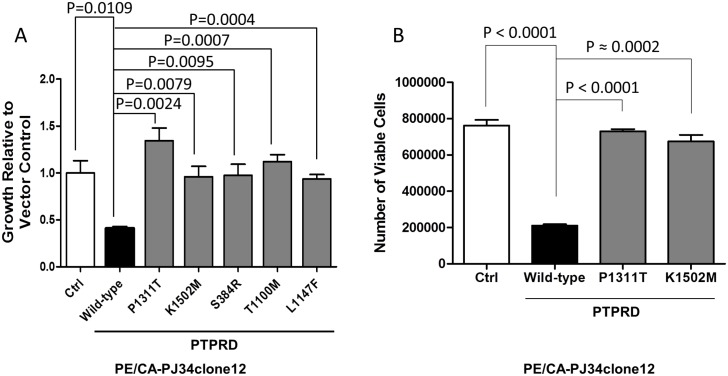
PTPRD has been reported to serve as a tumor suppressor in human cancer. [3,5,6] To determine the functional consequences of PTPRD mutation in HNSCC, we generated several representative HNSCC-derived PTPRD mutants by site-directed mutagenesis, including one mutation in the extracellular domain (S384R), one in the catalytic domain (K1502M), and two in the transmembrane region (T1100M and L1147F). Transient overexpression of these constructs in a HNSCC cell line with no endogenous PTPR family mutations (PE/CA-PJ34clone12) revealed that all of the PTPRD mutants tested led to increased growth as determined by MTT assay relative to PTPRD wild-type-transfected cells. (Fig 2A) MTT assay results were further confirmed with representative mutants by trypan blue exclusion assay in PE/CA-PJ34clone12 cells (Fig 2B). Collectively, these results suggest that PTPRD mutations inactivate the tumor suppressive function of PTPRD irrespective of their localization throughout the gene, leading to increased growth/proliferation in HNSCC cells.
Fig 2. HNSCC-derived PTPRD mutations lead to increased cell growth/proliferation in HNSCC cells.
(A) Cell growth was assessed by MTT assay 48 hrs after transfection and normalized to vector-transfected controls. One-way ANOVA P = 0.0007. Depicted P values represent the results of pairwise two-tailed unpaired t tests. The experiment was performed three times with similar results. (B) Cell proliferation was assessed by trypan blue exclusion assay 72 hrs after transfection. One-way ANOVA P < 0.0001. Depicted P values represent the results of pairwise two-tailed unpaired t tests.
PTPRD mutation is associated with increased pSTAT3 expression
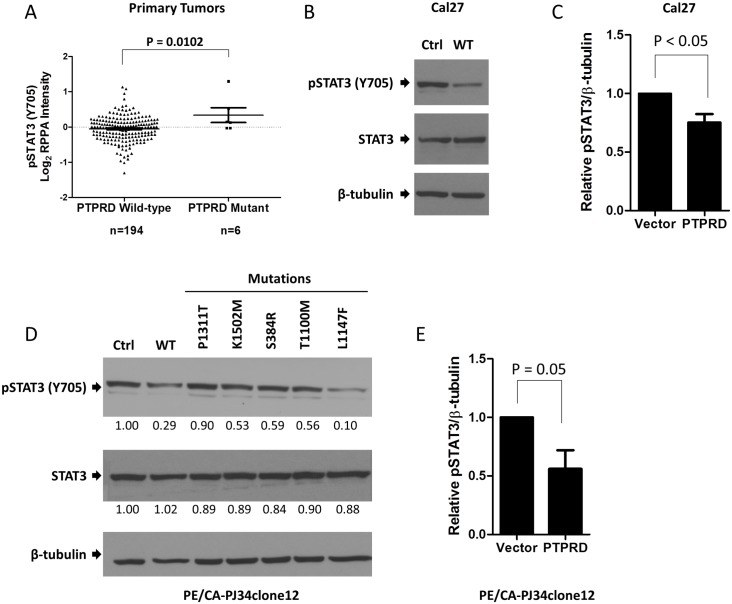
STAT3 is hyper-activated by constitutive phosphorylation of tyrosine 705 (Y705) in many cancer types, including HNSCC. [14,15] Importantly, pSTAT3 (Y705) is a known direct substrate of PTPRD. [6] To determine the effect of PTPRD mutation on STAT3 phosphorylation in HNSCC, we first examined 200 HNSCC tumors with both whole exome sequencing and RPPA analyses performed by TCGA and The Cancer Proteome Atlas (TCPA). HNSCC tumors with non-synonymous PTPRD mutations express significantly higher levels of pSTAT3 (Y705) relative to tumors with wild-type PTPRD (Fig 3A). Notably, PTPRD is the only PTPR family member for which this correlation is observed. To test the association between PTPRD mutation and pSTAT3 expression directly, we transfected an HNSCC cell line with known endogenous PTPRD mutation (Cal27 harboring mutation S387L) with wild-type PTPRD or vector control and found that overexpression of wild-type PTPRD leads to significantly decreased pSTAT3 expression (P ≤ 0.05). (Fig 3B and 3C) Furthermore, overexpression of mutant PTPRD in HNSCC cells without endogenous PTPR family mutations (PE/CA-PJ34clone12) leads to increased pSTAT3 (Y705) expression relative to PTPRD wild-type-overexpressing cells for nearly all mutations tested. (Fig 3D and 3E) Together, these results demonstrate that PTPRD mutation leads to increased STAT3 signaling in HNSCC cells and tumors.
Fig 3. PTPRD mutation is associated with increased pSTAT3 (Y705) expression in HNSCC tumors and cell lines.
(A) HNSCC tumors harboring PTPRD mutations express increased pSTAT3 (Y705) relative to PTPRD wild-type tumors as determined by whole exome sequencing and reverse-phase protein array (RPPA). Two-tailed unpaired t test. (B) Overexpression of wild-type PTPRD in a PTPRD-mutant cell line (Cal27) leads to decreased pSTAT3 (Y705) expression (C) Quantitation of three replicate experiments as shown in B. Two-tailed unpaired t test. (D) PTPRD-wild-type HNSCC cells (PE/CA-PJ34clone12) transiently overexpressing mutant PTPRD exhibit increased pSTAT3 (Y705) expression relative to wild-type-expressing cells. (E) Quantitation of three replicate experiments as shown in D. Two-tailed unpaired t test.
HNSCC cells with endogenous PTPRD mutation are more sensitive to STAT3 pathway inhibition
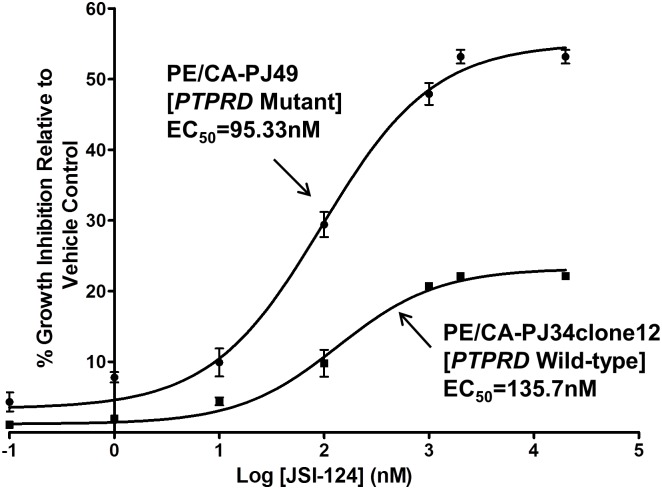
Since PTPRD mutations increase STAT3 activation in HNSCC, we next sought to determine if PTPRD mutation leads to increased sensitivity to STAT3 pathway inhibition. To test our hypothesis, we employed HNSCC cells that harbor an endogenous PTPRD mutation (PE/CA-PJ49). Treatment with the JAK/STAT inhibitor JSI-124 revels that these PTPRD mutant cells exhibit enhanced sensitivity relative to PTPR family wild-type cells (PE/CA-PJ34clone12), suggesting that PTPRD mutation may serve as a predictive biomarker for response to STAT3 pathway inhibitors (Fig 4).
Fig 4. HNSCC cells harboring an endogenous PTPRD mutation (PE/CA-PJ49) are more sensitive to the STAT3 pathway inhibitor JSI-124 relative to representative PTPRD WT HNSCC cells (PE/CA-PJ34clone12).
Cells were treated with increasing concentrations of JSI-124 for 24 hours followed by MTT assay. The experiment was performed three times with similar results.
PTPRD mRNA expression does not correlate with pSTAT3 expression
Promoter hypermethylation and gene copy number loss represent two additional mechanisms that may lead to loss of function of PTPRD via downregulation of mRNA expression. Importantly, PTPRD mRNA expression does not correlate with pSTAT3 expression in TCGA HNSCC samples, suggesting that methylation and copy number loss do not significantly contribute to STAT3 overactivation in HNSCC (S1A Fig). Indeed, a more detailed analysis reveals that while PTPRD methylation does correlate with mRNA expression as would be expected for any gene (S1B Fig), we observe no correlation between methylation and pSTAT3 expression (S1C Fig), nor do we observe any instances of aberrant promoter hypermethylation (defined here as a methylation level greater than three standard deviations above the mean methylation of the same genetic locus in organ-matched normal samples). In order to validate these findings, we performed methylation-specific PCR on an independent cohort of HNSCC tumors and found evidence of PTPRD promoter methylation in 75% of tumors analyzed (30/40) (representative analysis in S2 Fig). Importantly, we also observed PTPRD promoter methylation in five paired normal oral mucosa samples from these HNSCC patients (S3 Fig), further suggesting that the PTPRD methylation observed in HNSCC is not tumor-specific.
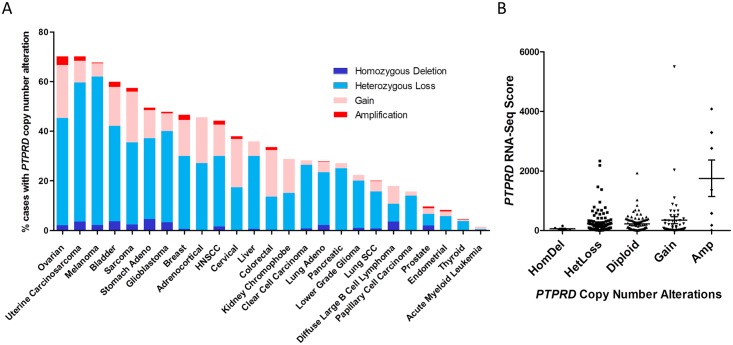
PTPRD copy number alterations occur to a substantial degree in HNSCC and other cancers (Fig 5A), where gene copy loss, particularly heterozygous loss, occurs more frequently than gain in HNSCC and all cancers analyzed with the exception of colorectal and cervical cancers. Importantly, we do not observe a significant correlation between copy number alterations and mRNA expression in HNSCC (Fig 5B). Together, these findings suggest that promoter hypermethylation and gene copy number loss do not significantly contribute to STAT3 overactivation in HNSCC.
Fig 5. PTPRD copy number alterations are frequent across cancers but are not associated with PTPRD mRNA expression in HNSCC.
(A) Copy number alteration of PTPRD in human cancers as determined by TCGA. (B) PTPRD copy number alterations do not correlate with altered PTPRD mRNA expression in HNSCC. A Jonckheere-Terpstra test was performed using StatXact software (Cytel, Cambridge, MA).
Discussion
STAT3 is an oncogene that is hyper-activated in many cancers, including HNSCC, where STAT3 hyper-activation is associated with decreased survival and emergent resistance to EGFR-targeted therapy. [9,16,17] Potential mechanisms that may result in increased STAT3 activation in cancer include increased signaling through upstream receptor or non-receptor tyrosine kinases such as EGFR and JAK, and/or inactivation of negative regulators of STAT3, including protein tyrosine phosphatases. [18,19] We recently reported that the receptor-type protein tyrosine phosphatase (PTPR) family is frequently mutated in HNSCC and other cancers and demonstrated that HNSCC-derived mutations of PTPRT induce STAT3 phosphorylation and drive HNSCC cell survival. [4] As PTPRD represents an additional receptor-like phosphatase that directly targets STAT3, we sought to determine if genetic or epigenetic loss of PTPRD function may contribute to STAT3 overactivation in HNSCC.
In the present study, we first identified 18 previously uncharacterized PTPRD mutations in HNSCC tumors. These mutations are all non-recurrent and are located throughout gene, a pattern that is consistent with that generally observed for tumor suppressor genes. We have shown that overexpression of wild-type PTPRD leads to growth suppression in HNSCC cells, while overexpression of representative mutants does not, thus establishing a functional consequence of PTPRD mutation in HNSCC models. These findings are consistent with those of previous in vitro studies across several cancer types, including glioblastoma, melanoma, colorectal cancer, and neuroblastoma, where wild-type PTPRD has been observed to suppress colony formation and cell growth as well as enhance apoptosis, while cancer-derived PTPRD mutants do not. [3,6,20] These cumulative findings suggest that PTPRD mutations in cancer lead to loss of its tumor suppressive function.
In order to determine if PTPRD mutation affects the activity of STAT3, a PTPRD substrate, we analyzed TCGA and TCPA data and found that HNSCC tumors with PTPRD mutations express significantly elevated pSTAT3 (Y705) relative to PTPRD-wild-type tumors. We previously reported a similar association between pSTAT3 (Y705) expression and mutation of a group of putative PTPR tumor suppressor genes, including PTPRD. [4] When each gene is considered individually rather than as a group, PTPRD is the only PTPR family member for which mutation is significantly associated with pSTAT3 (Y705) expression (S4 Fig). This result suggests that while the number of tumors harboring a mutation in any single PTPR family member may be insufficient to detect a significant difference in pSTAT3 (Y705) expression, PTPRD mutation in particular is uniquely associated with an increase in pSTAT3 (Y705) expression that is sufficiently large to detect statistical significance. Futhermore, overexpression of wild-type PTPRD in HNSCC cell lines harboring endogenous PTPRD mutations leads to downregulation of pSTAT3 (Y705), thus confirming that PTPRD regulates STAT3 activation in HNSCC models. While wild-type PTPRD leads to downregulation of pSTAT3 (Y705) in HNSCC cells, overexpression of most HNSCC-derived mutants does not alter pSTAT3 (Y705) expression relative to vector control, suggesting that these mutations lead to loss of function, but not in a dominant negative manner. In the case of the L1147F mutation tested herein, overexpression in HNSCC cells leads to increased growth, but not increased pSTAT3 (Y705) expression (see Figs 2A and 3D), indicating that certain mutations may lead to cancerous phenotypes in a STAT3-independent manner. These mutations may manifest through alternate mechanisms which may include extracellular interactions or alteration of relative affinities for alternate enzymatic substrates.
As PTPRD mutation leads to increased STAT3 activation in HNSCC, we next tested whether cells harboring a PTPRD mutation may be more sensitive to STAT3 pathway inhibition. Here we demonstrate that HNSCC cells with an endogenous PTPRD mutation are more sensitive to the JAK/STAT inhibitor JSI-124 than HNSCC cells harboring no PTPR family mutations, suggesting that HNSCC tumors with PTPRD mutations may be exquisitely sensitive to STAT3 inhibitors that are currently in preclinical and clinical development. In order to devise an optimal treatment strategy for HNSCC patients with PTPRD mutations, further study of additional PTPRD-interacting proteins that may serve as therapeutic targets may be warranted. In particular, PTPRD mutation may additionally confer sensitivity to aurora kinase A inhibitors currently in clinical development, where aurora kinase A phosphorylation and stability are normally regulated by PTPRD. [3]
An additional mechanism by which PTPRD function may be lost is through mRNA downregulation, including by promoter hypermethylation or gene copy number loss. First, we determined that PTPRD mRNA expression does not correlate with pSTAT3 expression in HNSCC, suggesting that these mechanisms are not likely to significantly contribute to STAT3 overactivation in HNSCC. It should be noted that this analysis may be complicated by the several cases in which little or no PTPRD mRNA was detected. Indeed, our overexpression studies in HNSCC cells suggest that expression of PTPRD would be expected to impact pSTAT3 (Y705) expression. Nonetheless, this correlation has not emerged in the HNSCC tumors analyzed to date.
A more detailed analysis of these mechanisms next revealed that the PTPRD promoter is not hypermethylated in HNSCC relative to organ-matched normal tissue, a finding we validated in an independent cohort of HNSCC tumor and matched normal pairs by methylation-specific PCR. The disparity between our present findings and previous reports of PTPRD promoter methylation in HNSCC is likely due to the prior lack of comparison between tumor and normal tissue. [6] The low level of PTPRD promoter methylation observed in HNSCC is additionally not associated with altered pSTAT3 (Y705) expression, further suggesting that this event does not significantly contribute to the cancer phenotype in HNSCC. A similar lack of aberrant PTPRD promoter hypermethylation has also been reported in cutaneous squamous cell carcinoma, suggesting this may be an uncommon event in multiple epithelial malignancies. [21]
Our analysis of TCGA data indicates that nearly half of HNSCC tumors harbor a copy number alteration of PTPRD and that copy number loss is more frequent than copy number gain in HNSCC and across nearly all cancers analyzed. PTPRD homozygous or heterozygous deletion has also been reported in cutaneous squamous cell carcinoma [21,22], GBM [6,20,23,24], lung cancer [25,26], neuroblastoma [27], metastatic melanoma [28,29], squamous cell carcinoma of the vulva [30], hepatocellular carcinoma [31], and laryngeal squamous cell carcinoma [5]. Importantly, no significant association was detected between PTPRD copy number alteration and PTPRD mRNA expression in HNSCC, suggesting that copy number loss is not the primary mechanism of loss of PTPRD function in HNSCC.
In conclusion, we have demonstrated herein that somatic mutation of PTPRD is the primary mechanism by which PTPRD loss of function occurs in HNSCC. We propose that nearly all of these mutations lead to loss of catalytic activity and hence reduced dephosphorylation of the substrate pSTAT3 (Y705). While promoter methylation and copy number loss are detectable at the PTPRD locus, these events are not associated with upregulation of pSTAT3 (Y705) expression. In contrast, somatic mutation of PTPRD leads to increased STAT3 activation in HNSCC tumors and cell lines, concomitant with increased cell growth and sensitivity to STAT3 pathway inhibition. These findings suggest that PTPRD mutation may represent a predictive biomarker for exquisite response to STAT3 targeted therapy in HNSCC.
Supporting Information
(A) PTPRD mRNA expression is not significantly associated with pSTAT3 (Y705) expression. n = 172, Pearson r = 0.05157, P = 0.5017, R2 = 0.002659. (B) PTPRD promoter methylation correlates with PTPRD mRNA expression. n = 172, Pearson r = -0.5242, P < 0.0001, R2 = 0.2747. (C) PTPRD promoter methylation is not significantly associated with pSTAT3 (Y705) expression. n = 211, Pearson r = 0.0008795, P = 0.9899, R2 = 7.735e-007.
(TIF)
Methylation was observed in 30/40 (75%) HNSCC tumors analyzed. M denotes primers amplifying methylated sequences, while U denotes primers amplifying unmethylated sequences.
(TIF)
Five HNSCC tumors and matched normal mucosa from the same patients were collected and analyzed by MSP. M denotes primers amplifying methylated sequences, while U denotes primers amplifying unmethylated sequences.
(TIF)
Whole exome sequencing and reverse-phase protein array data reveal that no other PTPR family mutations are significantly associated with pSTAT3 (Y705). P values represent the results of two-tailed unpaired t tests. N/A indicates insufficient sample size for calculation.
(TIF)
Data Availability
Genomic and proteomic data are available from The Cancer Genome Atlas (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm) and The Cancer Proteome Atlas (http://app1.bioinformatics.mdanderson.org/tcpa/_design/basic/index.html).
Funding Statement
This work was supported by National Institutes of Health (www.nih.gov) funding P50CA097190 and R01CA77308 to JRG, and F31DE024007 to NDP, the American Cancer Society (www.cancer.org) to JRG, the China Scholarship Council (http://en.csc.edu.cn) to YD, and the General Research Fund from the Research Grant Council of Hong Kong (http://www.ugc.edu.hk/) 17114814 and Seed Funding for Basic Research to VL.
References
- 1. Östman A, Hellberg C, Böhmer FD (2006) Protein-tyrosine phosphatases and cancer. Nature Reviews Cancer 6: 307–320. [DOI] [PubMed] [Google Scholar]
- 2. Nakamura M, Kishi M, Sakaki T, Hashimoto H, Nakase H, et al. (2003) Novel tumor suppressor loci on 6q22-23 in primary central nervous system lymphomas. Cancer research 63: 737–741. [PubMed] [Google Scholar]
- 3. Meehan M, Parthasarathi L, Moran N, Jefferies CA, Foley N, et al. (2012) Protein tyrosine phosphatase receptor delta acts as a neuroblastoma tumor suppressor by destabilizing the aurora kinase a oncogene. Mol Cancer 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lui VWY, Peyser ND, Ng PK-S, Hritz J, Zeng Y, et al. (2014) Frequent mutation of receptor protein tyrosine phosphatases provides a mechanism for STAT3 hyperactivation in head and neck cancer. Proceedings of the National Academy of Sciences 111: 1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Giefing M, Zemke N, Brauze D, Kostrzewska‐Poczekaj M, Luczak M, et al. (2011) High resolution ArrayCGH and expression profiling identifies PTPRD and PCDH17/PCH68 as tumor suppressor gene candidates in laryngeal squamous cell carcinoma. Genes, Chromosomes and Cancer 50: 154–166. 10.1002/gcc.20840 [DOI] [PubMed] [Google Scholar]
- 6. Veeriah S, Brennan C, Meng S, Singh B, Fagin JA, et al. (2009) The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proceedings of the National Academy of Sciences 106: 9435–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, et al. (2011) The mutational landscape of head and neck squamous cell carcinoma. Science 333: 1157–1160. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li L-C, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18: 1427–1431. [DOI] [PubMed] [Google Scholar]
- 9. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2: 401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling 6: pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, et al. (2013) Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152: 714–726. 10.1016/j.cell.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, et al. (2013) Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nature genetics 45: 478–486. 10.1038/ng.2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, et al. (2008) Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455: 1069–1075. 10.1038/nature07423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sen M, Thomas SM, Kim S, Yeh JI, Ferris RL, et al. (2012) First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer discovery 2: 694–705. 10.1158/2159-8290.CD-12-0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yin W, Cheepala S, Roberts JN, Syson-Chan K, DiGiovanni J, et al. (2006) Active Stat3 is required for survival of human squamous cell carcinoma cells in serum-free conditions. Molecular cancer 5: 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grandis JR, Drenning SD, Zeng Q, Watkins SC, Melhem MF, et al. (2000) Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proceedings of the National Academy of Sciences 97: 4227–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bowman T, Garcia R, Turkson J, Jove R (2000) STATs in oncogenesis. Oncogene 19: 2474–2488. [DOI] [PubMed] [Google Scholar]
- 18. Thomas SM, Grandis JR (2004) Pharmacokinetic and pharmacodynamic properties of EGFR inhibitors under clinical investigation. Cancer Treat Rev 30: 255–268. [DOI] [PubMed] [Google Scholar]
- 19. Suzuki A, Hanada T, Mitsuyama K, Yoshida T, Kamizono S, et al. (2001) CIS3/SOCS3/SSI3 plays a negative regulatory role in STAT3 activation and intestinal inflammation. J Exp Med 193: 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Solomon DA, Kim J-S, Cronin JC, Sibenaller Z, Ryken T, et al. (2008) Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer research 68: 10300–10306. 10.1158/0008-5472.CAN-08-3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lambert SR, Harwood CA, Purdie KJ, Gulati A, Matin RN, et al. (2012) Metastatic cutaneous squamous cell carcinoma shows frequent deletion in the protein tyrosine phosphatase receptor Type D gene. International Journal of Cancer 131: E216–E226. [DOI] [PubMed] [Google Scholar]
- 22. Purdie KJ, Lambert SR, Teh MT, Chaplin T, Molloy G, et al. (2007) Allelic imbalances and microdeletions affecting the PTPRD gene in cutaneous squamous cell carcinomas detected using single nucleotide polymorphism microarray analysis. Genes, Chromosomes and Cancer 46: 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ortiz B, Fabius AW, Wu WH, Pedraza A, Brennan CW, et al. (2014) Loss of the tyrosine phosphatase PTPRD leads to aberrant STAT3 activation and promotes gliomagenesis. Proceedings of the National Academy of Sciences 111: 8149–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ortiz B, White JR, Wu WH, Chan TA (2014) Deletion of Ptprd and Cdkn2a cooperate to accelerate tumorigenesis. Oncotarget 5: 6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kohno T, Otsuka A, Girard L, Sato M, Iwakawa R, et al. (2010) A catalog of genes homozygously deleted in human lung cancer and the candidacy of PTPRD as a tumor suppressor gene. Genes, Chromosomes and Cancer 49: 342–352. 10.1002/gcc.20746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, et al. (2005) Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer research 65: 5561–5570. [DOI] [PubMed] [Google Scholar]
- 27. Stallings RL, Nair P, Maris JM, Catchpoole D, McDermott M, et al. (2006) High-resolution analysis of chromosomal breakpoints and genomic instability identifies PTPRD as a candidate tumor suppressor gene in neuroblastoma. Cancer research 66: 3673–3680. [DOI] [PubMed] [Google Scholar]
- 28. Stark M, Hayward N (2007) Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Research 67: 2632–2642. [DOI] [PubMed] [Google Scholar]
- 29. Dutton‐Regester K, Aoude LG, Nancarrow DJ, Stark MS, O'Connor L, et al. (2012) Identification of TFG (TRK‐fused gene) as a putative metastatic melanoma tumor suppressor gene. Genes, Chromosomes and Cancer 51: 452–461. 10.1002/gcc.21932 [DOI] [PubMed] [Google Scholar]
- 30. Micci F, Panagopoulos I, Haugom L, Dahlback HSS, Pretorius ME, et al. (2013) Genomic aberration patterns and expression profiles of squamous cell carcinomas of the vulva. Genes, Chromosomes and Cancer 52: 551–563. 10.1002/gcc.22053 [DOI] [PubMed] [Google Scholar]
- 31. Nalesnik MA, Tseng G, Ding Y, Xiang G-S, Zheng Z-l, et al. (2012) Gene deletions and amplifications in human hepatocellular carcinomas: correlation with hepatocyte growth regulation. The American journal of pathology 180: 1495–1508. 10.1016/j.ajpath.2011.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) PTPRD mRNA expression is not significantly associated with pSTAT3 (Y705) expression. n = 172, Pearson r = 0.05157, P = 0.5017, R2 = 0.002659. (B) PTPRD promoter methylation correlates with PTPRD mRNA expression. n = 172, Pearson r = -0.5242, P < 0.0001, R2 = 0.2747. (C) PTPRD promoter methylation is not significantly associated with pSTAT3 (Y705) expression. n = 211, Pearson r = 0.0008795, P = 0.9899, R2 = 7.735e-007.
(TIF)
Methylation was observed in 30/40 (75%) HNSCC tumors analyzed. M denotes primers amplifying methylated sequences, while U denotes primers amplifying unmethylated sequences.
(TIF)
Five HNSCC tumors and matched normal mucosa from the same patients were collected and analyzed by MSP. M denotes primers amplifying methylated sequences, while U denotes primers amplifying unmethylated sequences.
(TIF)
Whole exome sequencing and reverse-phase protein array data reveal that no other PTPR family mutations are significantly associated with pSTAT3 (Y705). P values represent the results of two-tailed unpaired t tests. N/A indicates insufficient sample size for calculation.
(TIF)
Data Availability Statement
Genomic and proteomic data are available from The Cancer Genome Atlas (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm) and The Cancer Proteome Atlas (http://app1.bioinformatics.mdanderson.org/tcpa/_design/basic/index.html).