Abstract
Many cell types require the biophysical and biochemical cues within the 3D extracellular matrix (ECM) to exhibit their true physiologically relevant behavior. As a result, cell culture platforms have been evolving from traditional 2D petri-dish plates into 3D biomatrices, and there is a need for developing analytic tools to characterize 3D cell culture. The existing cell counting method, using a hemocytometer or coulter counter, requires that cells are suspended in fluids prior to counting. This poses a challenge for 3D cell culture as cells are embedded in a 3D biomatrix. We use a facile 3D cell counting method that overcomes this limitation and allows for in situ cell counting in a 3D cell culture using equipment that is commonly available in a biology lab. Using a breast tumor cell line, MDA-MB-231, as a model system, we demonstrated that MDA-MB-231 cells (1) grow slower within a 3D collagen matrix than on a 2D substrate for an extended growth time (a week) with a comparable, initial cell-to-cell distance, (2) their cell growth rate decreases with the increase of collagen concentration, showing a linear growth rate rather than an exponential growth rate. Further work using flow cytometry showed that the observed growth rate reduction was consistent with the retardation of the transition to S (synthesis) phase in the cell cycle. This work demonstrates the validity of the 3D cell counting method and the importance of cell-ECM interactions in cell proliferation.
Keywords: hemocytometer, 3D ECM, cell growth, cell cycle, collagen
Introduction
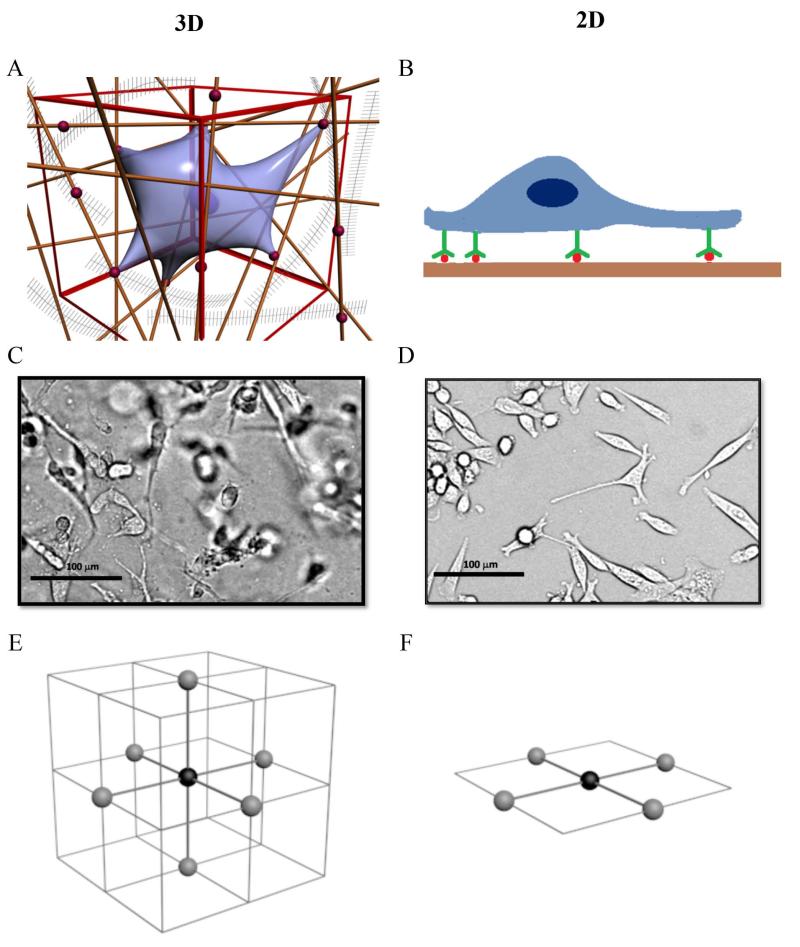
Biophysical cues (e.g. mechanical support from the 3D biomatrix architecture, adhesions to the ECM) and biochemical cues (e.g. matrix bound chemokines and growth factors) play instrumental roles in cellular behavior.1-4 It is now accepted that cells behave very differently when they are plated on a 2D substrate, in comparison to when they are embedded in a 3D extracellular matrix (ECM).5-7 When a cell is cultured within a 3D ECM (See Figures 1AC), the cell binds to the adhesive molecules such as fibronectin or laminin within the ECM, and is mechanically supported by the fiber network all around its surface. When a cell is plated on a 2D substrate (See Figures 1BD), the cell binds to the adhesive molecules on its basal side, and receive mechanical support from one side. It has been shown that cells display smaller focal adhesion complexes, and down-regulate their integrin binding in 3D cell culture than in 2D culture.5 It is no surprise that cells adhere,5 migrate,4, 6 and form micro-scale tissues2, 7 very differently in 3D than in 2D cell culture.
Figure 1. 3D versus 2D cell culture.
A: An illustration of a cell embedded in a 3D collagen matrix (reproduced from Wu and Swartz25 with the permission of ASME, originally adapted from Griffith and Swartz in Ref [1]). The straight lines represent collagen fibers, and red dots are adhesion molecules such as fibronectin. The cell adheres to the collagen matrix via integrin matrix binding and is supported by the collagen fiber architecture around its entire surface. B: An illustration of a cell cultured on a 2D substrate. The cell adheres to the substrate via integrin binding, and it is supported by the substrate only on its basal side. C: A micrograph of cells (MDA-MB-231) embedded in a 3D collagen gel. This image was taken using a bright field microscope, and note the dark spots are cells out of focus. D: A micrograph of cells (MDA-MB-231) plated on a well plate surface. The dimension of the images is 300 pixel × 250 pixel, which corresponds to 389 μm × 323 μm. Scale bar is 100 μm. E: An illustration showing 6 nearest cells (gray spheres) surrounding a cell (black sphere) of interest in a 3D cubic lattice. F: An illustration showing 4 nearest cells (gray spheres) surrounding a cell (black sphere) of interest in a 2D square lattice.
Matrix stiffness and cellular binding sites are critical role players in cell proliferation, because each cell needs to anchor itself within the ECM and generates sufficient traction to divide.8, 9 Although extensive studies have been carried out in investigating the roles of matrix stiffness and the adhesion in cell migration,6, 10-14 less is known about their roles in cell proliferation.8, 9, 12 Current understanding of the roles of biophysical cues in cell growth is largely derived from studies in 2D cell culture.12, 15 Growth dependence studies on substrate stiffness in a 2D cell culture revealed that glioma cells grew faster on stiffer substrates,12 while NIH/3T3 fibroblasts preferred to grow on soft substrates.15 Recent cell growth studies in 3D cell culture using synthetic biomaterials9, 16 revealed that integrin binding was necessary for fibroblast proliferation, and its growth rate was observed to decrease with the increase of the matrix stiffness of the PEG based synthetic gel.9 It should be noted that results of growth studies of fibroblasts in 3D are opposite to those from 2D cell culture.12
Moving 3D cell culture into the mainstream requires the development of advanced analytic tools for quantifying cellular behavior in 3D.1, 3, 13, 16-21 For cell growth curve measurements, conventional cell counting methods, such as hemocytometer or coulter counter, are not suitable for 3D cell culture. These methods require that the ECM component of the 3D cell culture be digested before cell counting, as a result, it is not possible to monitor the growth of the same population of cells in situ. The cost of the biomatrix, and the time consuming cell preparation procedure before cell counting limits the use of conventional cell counting methods in 3D cell culture.
To overcome limitations of the conventional cell counting method, we have developed a 3D in situ cell counting method for counting cells within a biomatrix using a bright field microscope, an x-y automated microscope stage and a commercial imaging software. This method allows us to count cells of the same population in situ, while it does not require the re-suspension of the cells before counting. Using this method, we measured the breast cancer cell line MDA-MB-231 growth curve in a 3D cell cultures with various collagen concentrations. This method is straightforward to implement and will find wide applications in the field of tissue engineering as the 3D cell culture becomes mainstream.
Materials and methods
2D and 3D cell culture
A malignant breast cancer cell line, MDA-MB-231, was obtained as a gift from National Institute of Health through the Nanobiotechnology Center (NBTC) at Cornell University. Cells were maintained in a growth medium composed of DMEM (Invitrogen, Carlsbad, CA) with 10% FBS (Atlanta Biologicals, Lawrenceville, GA) and Pen/Strep (100 units and 100 μg/ml respectively, Invitrogen) in a T75 flask (Corning, Lowell, MA) at a 37 °C, 5% CO2-controlled humidified incubator (Forma, Thermo Scientific, Asheville, NC). Collagen type I was extracted from rat tails (Pel-Freez, Rogers, AR) and stored at 5 mg/ml in 0.1% acetic acid at 4 °C. The properties of this in-house collagen were carefully characterized in reference by Cross et al.21 All the experiments were set up using cell cultures with 50-75% confluence in T75 flasks. For 3D cultures, cells were re-suspended in DMEM with 10% FBS and then mixed with 10× M199, 1N NaOH, and 0.5% collagen on ice. The final cell density was prepared after counting cells using a hemocytometer and the pH was adjusted with 1N NaOH at 7.2. Typically, 0.022 mL of 1N NaOH is needed for neutralizing 1 mL of the collagen stock. After the mixture was ready at 4 °C, pre-chilled multiwell plates (6-well plate, Corning) were placed on top of an ice block and then a droplet of 15 μl (or diameter ~3 mm) of the mixture was placed in a well. The multi-well plates were placed in a 37 °C, 5% CO2-controlled humidified incubator in an inverted position for 7 minutes. Then the collagen mixture was further incubated for additional 13 minutes in the upright position. In this way, we obtained homogenously distributed cells in the z-direction. After the collagen was gelled, 1.92 mL growth medium was added in each well. To avoid rapid evaporation, PBS was added in any empty space in the multi-well plates.
For 2D cell culture counting using a hemocytometer (Bright-Line Hemocytometer, Hausser Sci., Horsham, PA), we used duplicates of 6-well plates. At each time point, cells from one well were extracted using trypsin (Trypsin EDTA, Mediatech, Inc, Manassas, VA).
Image acquisition, processing and cell tracking
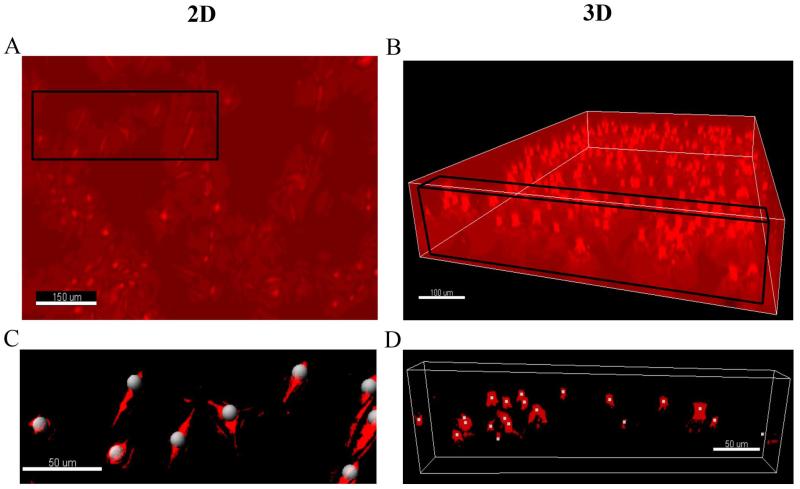
Images were taken at different time points using MetaMorph software (Molecular Devices, Inc., Sunnyvale, CA) with 10× objective lens (Olympus), a microscope (Olympus IX81, Center Valley, PA), and a CCD camera (ORCA-R2, Hamamatsu Photonics, Bridgewater, NJ). We used bright field microscopy for all the images taken. For 2D cell culture, an image of 1344 pixel × 1024 pixel (or 864 μm × 660 μm) was taken at every 12 hours for up to 7 days (See Figure 2A). For 3D cell culture, a Z-stack of 100 images at a 5-μm interval was acquired using an x-y automated microscope stage (MS-2000, Applied Scientific Instrumentation, Eugene, OR) and the same microscope setting described above. The images were reconstructed into a 3D image as shown in Figure 2B. Both 2D and 3D images were processed off-line using background subtraction and Gaussian filter. We used the Spot function in Imaris software (Bitplane, Zurich) to track cell positions. The tracking results were typically validated by visually inspecting the tracked spots as shown in Figure 2C-D. The total spot/cell number detected in each image was recorded for later analysis.
Figure 2. Illustration of the 3D in situ cell counting method.
A: An image of cells plated on the surface of a 6-well plate. Size of the image is 433 μm × 330 μm. B: An image of cell embeded within a collagen matrix. Size of the image is 864 μm × 660 μm × 400 μm; C: A close view of the 2D cell culture (see the black rectangular in A). The white dots are the tracked cells using a commercial software Imaris. Size of image is 175 μm × 80 μm; D: A close-up image of the 3D cell culture (See the black rectangular box in B). The white dots are the tracked cells. Size of the image is 389 μm × 20 μm × 500 μm. Note that the size of provided images for this illustration in A is different for the better visualization purpose from the size of the actual analyzed images described in Materials and Methods.
Data analysis
For live cell counting method, we took three images (size of 864.3 μm × 660.5 μm for 2D and 864.3 μm × 660.5 μm × 400 μm for 3D) at each time point from three different positions in one or two wells of the 6-well plate, and then digitally tracked the number of the cells in these three images. For 2D images, each image had cell numbers ranging from 73 to 605 cells; and for 3D images, 128 to 1108 cells during the cell culture period. The normalized cell number is the average cell numbers from the 3 images divided by the average cell numbers at t = 0. Error bars are standard deviation of the 3 data points. The entire experimental procedure was the same for 2D and 3D cell culture counting.
Cell cycle quantification
The collagen gel was digested with a 1 mg/mL collagenase (Sigma, St Louis, MO) solution. A million cells were collected, centrifuged, and re-suspended in 200 μL of cold propidium iodide hypotonic staining solution containing 50 μg/mL propidium iodine (Sigma), 1 μL/mL Triton X-100 (Sigma), and 1 mg/mL sodium citrate (Sigma). Cells were incubated at room temperature for 1 h and analyzed by flow cytometry (BD LSRII) using 488-nm excitation and collected through 550 long-pass dichroic and a 576/26 band-pass filters. Doublets were identified by a propidium iodide voltage pulse photomultiplier tube signal width versus area plot and excluded from the analysis.22
Results
The in situ cell counting method is validated against a conventional off-line hemocytometer
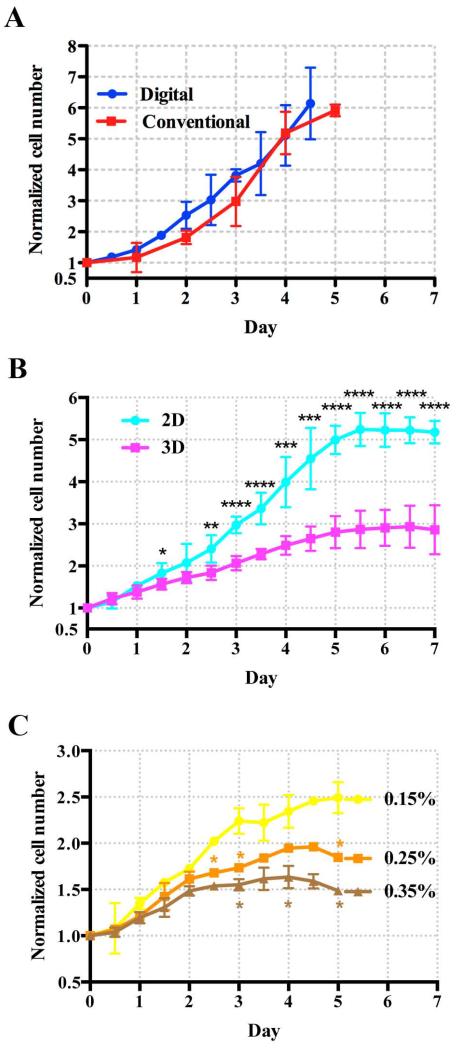
We first validated the in situ cell counting method against the conventional hemocytometer cell counting method. Figure 3A shows that cell population growth curves obtained from these two cell counting methods agree with each other within the experimental errors. In both cases, MDA-MB-231 breast cancer cells were cultured on the 2D substrates of the 6-well plates at an initial cell density of 6500 cells/cm2. Using the in situ cell counting method, we imaged cells of the same population through the entire experiment. For hemocytometer, a different sub-group of cells were extracted from the well plate for cell counting. The growth curves, represented by the normalized cell number (cell density divided by the initial cell density) versus time, are shown in Figure 3A. Both curves follow exponential growth pattern, with specific growth rate, μ = 0.468 (R2 = 0.999) for hemocytometer and 0.490 (R2 = 0.992) day−1 for 3D cell counting. Although the two growth curves agree with each other within experimental errors, there is a slight trend that the cell counts from the in situ counting method is higher than those from hemocytometer. This slight difference might be due to the cell loss during the cell extraction process that is necessary for the hemocytometer method.
Figure 3. Growth Curves of MDA-MB-231.
Normalized cell number is defined as the cell density divided by the initial cell density. A: Validation of the 3D in situ cell counting method against the conventional hemocytometer. The dots are data points from 3D cell counting method, and the squares are from the hemocytometer. The solid lines are for visualization purpose. The cells were seeded on the 2D substrate of a 6-well plate, and the initial cell seeding density was 6,500 cells/cm2 in both cases. B: The growth rate of MDA-MB-231 cells in 3D cell culture with 0.15% collagen (squares) is reduced when compared with that in 2D planar culture (dots). Initial cell seeding density for 2D cell culture was 23,000 cells/cm2 and that for 3D cell culture was 1 million cells/mL; The inter-cell spacing was about 67 μm for 2D and 100 μm for 3D cell culture. C: The growth of MDA-MB-231 cells in 3D cell culture is reduced as the concentration of collagen increases. The collagen concentrations are 0.15% (dots), 0.25% (squares) and 0.35% (triangles) respectively. Student t-test was applied at each time point (*: 0.01<p<0.05, **: 0.001<p<0.01, ***: p<0.001, ****: p<0.0001).
MDA-MB-231 cells grow faster on a 2D substrate than within a 3D collagen matrix
We then investigated the cell population growth on a 2D substrate versus within a 3D collagen matrix. We hypothesized that cells’ growth rate is modulated by the different mechanical environment provided by the 2D substrate and the 3D collagen matrix, in particular, the architectural support and adhesion molecule presentation.
Figure 3B shows that cells cultured on a 2D tissue culture plate grew faster than cells plated in a 0.15% collagen matrix. At the stationary phase, MDA-MB-231 cells in 0.15% collagen matrix (3D) reached 2.9±0.6-fold proliferation. However, for the conventional 2D cell culture, the final fold-change was 5.2±0.3, which is approximately 1.5 times higher than that in collagen matrix. This result indicates that 3D environment such as collagen matrix reduces proliferation rate of tumor cells, which is consistent with previous report using fluorescent cytometry.23 Interestingly, the growth kinetics in 3D followed a linear profile , rather than an exponential one. We note that there was no spheroid formed under the given conditions.
Different initial cell densities for 2D and 3D cell culture were used for the data presented in Figure 3A to minimize the difference of biochemical conditions due to the two cell culture conditions. It is known that cell growth rate depended on the initial cell seeding density, likely due to the cytokines and growth factors secreted by the cells. Although there have been comparison studies between 2D and 3D growth conditions, a careful consideration of a fair comparison is still lacking. To ensure that the chemical environment of the two cell cultures were identical at initial time point, we adjusted the initial average cell-cell distance (100 μm for 3D and 67 μm for 2D cell culture) such that the mitogenic factors each cell received from its nearest neighboring cells were similar for 2D and 3D cell culture. The rationale for this choice of differential initial cell – cell spacing in 2D and 3D is the following. We made four assumptions: (i) All the cells were located at the vertices of an equal distance grid either in 2D or 3D space (See Figure 1EF). (ii) Each cell received the same amount of mitogenic factors secreted by its nearest neighboring cells; (iii) The concentration of the mitogenic factors secreted by each cell peaked at the center of the cell and fell as 1/r, where r is the distance from the center of the cell. (iv) Only the secretions of the nearest neighbors were taken into consideration. We know that each cell in a 3D cell culture has 6 nearest neighboring cells (top, bottom, front, back, right and left, or see Figure 1E) while 4 for 2D cell culture (front, back, right and left, or see Figure 1F). Thus, to ensure that the concentration of the cytokines secreted by all the nearest neighbors in 2D and 3D at the location of the cell of interest to be the same, the ratio of the cell – cell distance in 2D and 3D needs to be 4:6. To choose the initial average cell-cell spacing in 3D, we first cultured cells under initial cell density of 25×103, 1×106, and 2×106 cells/mL within 0.15% collagen. We found that the initial density of 1×106 cells/mL required about a week for the cell culture to reach the saturation phase, which was a comparable number to the 2D cell culture growth curve. Therefore, the average cell-cell distance of 100 μm or a cell density of 106 cells/mL was chosen as the initial cell seeding density for 3D cell culture (1 cell/(100 μm)3). Subsequently, the average cell-cell distance in a 2D cell culture was determined to be 100 × (4/6) μm = 67 μm in order to ensure that each cell in 2D and in 3D receive the equal amount of mitogenic factors secreted by its nearest neighbors. This corresponds to the initial cell seeding density of 2.3×104 cells/cm2 (1 cell/(67 μm)2). We emphasize here that this is a first order of approximation to take into account differential transport properties of cytokines in 2D and 3D. A rigorous modeling using Fick’s law of diffusion is necessary to obtain the precise ratio. To ensure that the total number of cells within each well is the same for 2D and 3D cell culture, we placed 15 drops of 3D cell culture (each drop was 15 μL of cell embedded collagen), or a total of ~2.25×105 cells (15 × 15 μL × 106 cells/mL) in one well for the 3D cell culture. Note that the surface area of one well is 9.6 cm2, and the total number of cells in a well for 2D culture is 2.2×105 cells (9.6 cm2 × 2.3×104 cells/cm2).
Cell population growth rate decreases with the increase of collagen matrix concentration
To investigate how the collagen concentration affects cell proliferation, we grew cells in collagen biomatrix with various concentrations (0.15%, 0.25% and 0.35%). We note that 0.15% is the lowest concentration at which the collagen can be polymerized in a robust and reproducible way. We used initial cell seeding density of 106 cells/mL, 8 drops (each drop contained 15 μL cell culture) of cell culture in each well, and the 3D in situ cell counting methods in all three situations. Interestingly, the cell number fold changes at the saturation phase were 2.49±0.17, 1.96±0.04 and 1.64±0.12 for collagen concentration of 0.15%, 0.25% and 0.35% (See Figure 3C) respectively. This indicates that dense collagen matrix reduces cell population growth rate. When the proliferation in 0.15% collagen reached the saturation phase at day 3.5, the cell numbers were 1.4 times higher than those in 0.35% collagen were. Here, we monitor the cell proliferation with real time video imaging, and most cells were confirmed to be alive through active proliferation. The growth curves were fitted to a linear line, and the slopes of the lines were found to be 0.323 (R2 = 0.959), 0.278, (R2 = 0.998) and 0.232 (R2 = 0.992) day−1 for collagen concentrations of 0.15, 0.25, and 0.35% respectively.
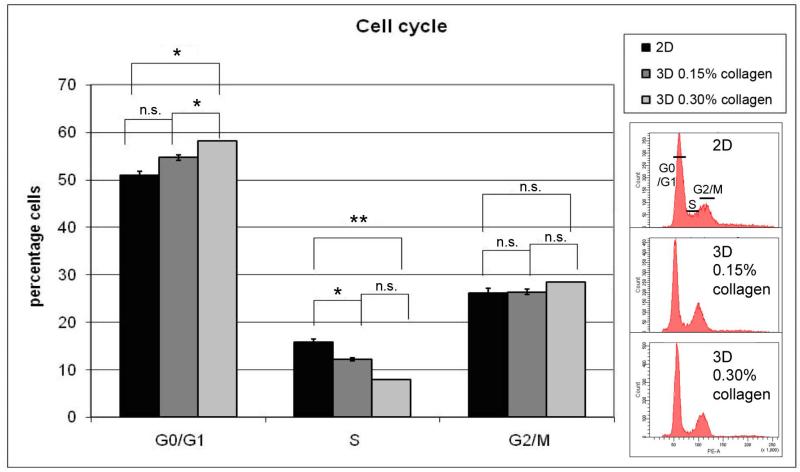
Collagen matrix enhances the number of cells in G0/1 phase
To understand the mechanism behind the reduced cell population growth rate, cell cycle phase distribution was analyzed. The MDA-MB-231 cell populations cultured in collagen matrix presented an enrichment in G0/G1 correlating with the percentage of collagen, as determined by DNA histograms generated by flow cytometry. As the growth was reduced, the percentage of G0/G1 cells increased and that of S decreased as shown in Figure 4. The percentage of G2/M phase did not change significantly. The graph represents the results of three repeats (Mean±SEM) with the paired 2-tailed t test. The data are consistent with a progressive retardation of G1 transit associated with increasing collagen in the matrix. We also note that another cell type, HL-60, showed the same general trend, a decrease of the proportion of the cells in S in 3D compared to 2D, either untreated or treated with an inducer of differentiation. This indicates that the finding is not specific just to a cell line.
Figure 4. Collagen matrix enhances the percentage of cells in G0/G1.
Cell cycle analysis was performed by flow cytometry using the hypotonic propidium iodide (PI) solution method. The cells were grown in three different conditions: 2D, 3D 0.15% collagen (low collagen), and 3D 0.30% collagen (high collagen). The results of 3 repeats (mean±SEM) are presented using a 2-tailed paired t test (*: 0.01<p<0.05, **: 0.001<p<0.01, n.s.: non-significant). Representative flow cytometry data showing distribution of relative DNA content of the cells are on the right.
Conclusion and discussions
We have developed a 3D in situ cell counting method for measuring cell population growth curves of 3D cell cultures using a bright field microscope and a commercially available software. This method overcomes the limitations of the conventional cell counting methods for the counting of cells in 3D ECM in that it allows for in situ live cell counting of the same cell population for an extended time period (days). It should be noted that the innovation here is the cell counting in situ, eliminating the step where cells need to be suspended in fluids prior to cell counting. This method can be adapted easily for cell counting of a 2D cell culture. We used a commercially available software for locating the cells. The development of a computer code to locate the cells is straightforward, which will cut down the cost of this method significantly.
Using the 3D cell counter, we found that MDA-MB-231 breast cancer cells grew faster when plated on a 2D substrate than within a 3D collagen biomatrix. Further more, cell growth rate decreased with the increase of the collagen concentration. Cell cycle analysis using flow cytometry revealed that the percentage of cell population in G0/G1 phase increased with the increase of collagen matrix. These results indicate that matrix density plays important roles in MDA-MB-231 cell proliferation.
It is interesting to note that the cell growth curve in 3D cell culture does not follow the typical exponential growth curve as in the case of 2D. This is consistent with previous report that growth curve was linear in 3D in comparison to 2D using human mesenchymal stem cells.24 In 2D, cell growth rate depends sensitively on the initial seeding density, and it reaches saturation phase once the cells reach confluence (also known as contact inhibition). In 3D, cells do not appear to be in contact with each other when reaching saturated phase. The average cell-cell distance in saturated phase from our experiments (See Figure 3B) was 30 μm for 2D cell culture, and 69 μm for 3D cell culture. This result is important because it reveals that contact inhibition may not be the reason for cell culture to reach saturation phase in 3D cell culture as it is for 2D.
It remains to be explored the exact mechanisms behind the reduced cell growth rate in high collagen concentrations. Possible causes can be stiff mechanical environment, the smaller pore size of the collagen network, and the more abundant binding sites presented to the cells within the dense collagen. It has been reported that inhibiting integrin β1 association with collagen enhanced MDA-MB-231 cell growth.23 In synthetic gel, integrin binding sites were required for sustaining fibroblast cell growth.9 It is likely that the relation of cell growth and integrin binding sites is biphasic, in a similar way to cell migration.10 For gel stiffness, it has been reported that cell growth was suppressed by the synthetic gel with high Young’s modulus. However, this may not be able to translate directly to natively derived gel, since the architecture of the cross-linked synthetic gel is distinctly different from the natively derived gels. Carefully designed experiments are needed to examine the individual mechanical cues, namely matrix stiffness, microstructure and matrix binding sites in cell proliferation. Furthermore, the next level of inquiry will be to link the mechano-sensing molecules directly with the biomechanical cues within the cellular environment.
Acknowledgement
BK, STZ and Wu thank support from the National Institute of Health (R21CA138366, R21RR025801). RPB and AY thank support from the National Cancer Institute (R01CA033505, R01CA152870). All authors thank support from the Cornell Center on the Microenvironment & Metastasis, a Physical Science Oncology Center (award number U54CA143876).
Footnotes
We do not have any conflict of interest.
Contributor Information
Beum Jun Kim, Dept. of Biological and Environmental Engineering, Cornell University, Ithaca, NY 14853.
Shuting Zhao, Dept. of Biological and Environmental Engineering, Cornell University, Ithaca, NY 14853.
Rodica P. Bunaciu, Dept. of Biomedical Sciences, Cornell University, Ithaca, NY 14853
Andrew Yen, Dept. of Biomedical Sciences, Cornell University, Ithaca, NY 14853.
Mingming Wu, Dept. of Biological and Environmental Engineering, Cornell University, Ithaca, NY 14853.
Literature Cited
- 1.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nature Reviews Molecular Cell Biology. 2006;7(3):211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 2.Hebner C, Weaver VM, Debnath J. Modeling morphogenesis and oncogenesis in three-dimensional breast epithelial cultures. Annual Review of Pathology-Mechanisms of Disease. 2008;3:313–339. doi: 10.1146/annurev.pathmechdis.3.121806.151526. [DOI] [PubMed] [Google Scholar]
- 3.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nature Reviews Molecular Cell Biology. 2007;8(10):839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 4.Cukierman E, Pankov R, Yamada KM. Cell interactions with three-dimensional matrices. Curr. Opin. Cell Biol. 2002;14(5):633–639. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 5.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294(5547):1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 6.Friedl P, Brocker EB. The biology of cell locomotion within three-dimensional extracellular matrix. Cellular and Molecular Life Sciences. 2000;57(1):41–64. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischbach C, Kong HJ, Hsiong SX, Evangelista MB, Yuen W, Mooney DJ. Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement. Proc Natl Acad Sci U S A. 2009;106(2):399–404. doi: 10.1073/pnas.0808932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park CC, Zhang H, Paravicini M, Gray JW, Baehner F, Park CJ, Bissell MJ. beta(1) integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer Research. 2006;66(3):1526–1535. doi: 10.1158/0008-5472.CAN-05-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bott K, Upton Z, Schrobback K, Ehrbar M, Hubbell JA, Lutolf MP, Rizzi SC. The effect of matrix characteristics on fibroblast proliferation in 3D gels. Biomaterials. 2010;31(32):8454–8464. doi: 10.1016/j.biomaterials.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 10.Zaman MH, Trapani LM, Siemeski A, MacKellar D, Gong HY, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. U. S. A. 2006;103(29):10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Collagen-based cell migration models in vitro and in vivo. Seminars in Cell & Developmental Biology. 2009;20(8):931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulrich TA, Pardo EMD, Kumar S. The Mechanical Rigidity of the Extracellular Matrix Regulates the Structure, Motility, and Proliferation of Glioma Cells. Cancer Research. 2009;69(10):4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gobin AS, West JL. Cell migration through defined, synthetic extracellular matrix analogues. Faseb Journal. 2002;16(3):751–753. doi: 10.1096/fj.01-0759fje. [DOI] [PubMed] [Google Scholar]
- 14.Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, Hirsch K, Keller M, Forster R, Critchley DR, Fassler R, Sixt M. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453(7191):51–5. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 15.Tan PS, Teoh SH. Effect of stiffness of polycaprolactone (PCL) membrane on cell proliferation. Materials Science & Engineering C-Biomimetic and Supramolecular Systems. 2007;27(2):304–308. [Google Scholar]
- 16.Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA, Rizzi SC. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials. 2010;31(32):8494–8506. doi: 10.1016/j.biomaterials.2010.07.064. [DOI] [PubMed] [Google Scholar]
- 17.Haessler U, Kalinin Y, Swartz MA, Wu M. An agarose-based microfluidic platform with a gradient buffer for 3D chemotaxis studies. Biomed. Microdev. 2009;11(4):827–35. doi: 10.1007/s10544-009-9299-3. [DOI] [PubMed] [Google Scholar]
- 18.Haessler U, Pisano M, Wu M, Swartz MA. Dendritic cell chemotaxis in 3D under defined chemokine gradients reveals differential response to ligands CCL21 and CCL19. Proc Natl Acad Sci U S A. 2011;108(14):5614–9. doi: 10.1073/pnas.1014920108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat. Methods. 2010;7(12):969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legant WR, Pathak A, Yang MT, Deshpande VS, McMeeking RM, Chen CS. Microfabricated tissue gauges to measure and manipulate forces from 3D microtissues. Proc. Natl. Acad. Sci. U. S. A. 2009;106(25):10097–10102. doi: 10.1073/pnas.0900174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cross VL, Zheng Y, Choi NW, Verbridge SS, Sutermaster BA, Bonassar LJ, Fischbach C, Stroock AD. Dense type I collagen matrices that support cellular remodeling and microfabrication for studies of tumor angiogenesis and vasculogenesis in vitro. Biomaterials. 2010;31(33):8596–8607. doi: 10.1016/j.biomaterials.2010.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunaciu RP, Yen A. Activation of the Aryl Hydrocarbon Receptor AhR Promotes Retinoic Acid-Induced Differentiation of Myeloblastic Leukemia Cells by Restricting Expression of the Stem Cell Transcription Factor Oct4. Cancer research. 2011;71(6):2371–80. doi: 10.1158/0008-5472.CAN-10-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu YH, Guo X, Brandt Y, Hathaway HJ, Hartley RS. Three-dimensional collagen represses cyclin E1 via beta 1 integrin in invasive breast cancer cells. Breast Cancer Res. Treat. 2011;127(2):397–406. doi: 10.1007/s10549-010-1013-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grayson WL, Ma T, Bunnell B. Human mesenchymal stem cells tissue development in 3D PET matrices. Biotechnol. Prog. 2004;20(3):905–912. doi: 10.1021/bp034296z. [DOI] [PubMed] [Google Scholar]
- 25.Wu MM, Swartz MA. Modeling Tumor Microenvironments In Vitro. J Biomech Eng-T Asme. 2014;136(2) doi: 10.1115/1.4026447. [DOI] [PMC free article] [PubMed] [Google Scholar]