SUMMARY
Human mitochondrial ribosomes are specialized in the synthesis of 13 proteins, which are fundamental components of the oxidative phosphorylation system. The pathway of mitoribosome biogenesis, the compartmentalization of the process and factors involved remain largely unknown. Here, we have identified the DEAD box protein DDX28 as an RNA granule component essential for the biogenesis of the mitoribosome large subunit (mt-LSU). DDX28 interacts with the 16S rRNA and the mt-LSU. RNAi-mediated DDX28 silencing in HEK293T cells does not affect mitochondrial mRNA stability, 16S rRNA processing or modification. However, it leads to reduced levels of 16S rRNA and mt-LSU proteins, impaired mt-LSU assembly, deeply attenuated mitochondrial protein synthesis and consequent failure to assemble oxidative phosphorylation complexes. Our findings identify DDX28 as essential during the early stages of mitoribosome mt-LSU biogenesis, a process that mainly takes place near the mitochondrial nucleoids, in the compartment defined by the RNA granules.
Keywords: DDX28, Mitochondrial translation, Mitochondrial ribosome assembly, RNA helicase, RNA granules
INTRODUCTION
Protein synthesis is universally catalyzed by the ribosome. Within mitochondria, ribosomes (mitoribosomes) are specialized in the synthesis of a small set of proteins encoded in the mitochondrial DNA (mtDNA). In humans, the mtDNA encodes 13 proteins, all subunits of the oxidative phosphorylation (OXPHOS) system enzymes, two ribosomal RNAs (12S and 16S rRNAs) and 22 tRNAs. Mitoribosomes sediment as 55S particles and consist of a 28S small subunit (mt-SSU), formed by a 12S rRNA and ~29 ribosomal proteins (MRPs), and a 39S large subunit (mt-LSU), formed by a 16S rRNA and ~50 MRPs (Christian and Spremulli, 2009; O’Brien, 1971). The three-dimensional structures of the human 39S large subunit at 3.4 Å resolution (Brown et al. 2014), of porcine mt-LSU at 4.9 Å and 3.4 Å resolution (Greber et al., 2014) and the 7-Å structure of the 28S small subunit (Kaushal et al., 2014) were recently determined by cryo-electron (cryo-EM) microscopy. They confirmed that the 55S ribosomes differ from bacterial (70S) and cytoplasmic ribosomes (80S) in their lower RNA:protein ratio, where significant amounts of RNA have been replaced by mitochondrion-specific proteins, and have provided a wealth of information (Brown et al., 2014; Greber et al., 2014).
The processes of mtDNA replication and expression occur in submitochondrial matrix compartments. The mtDNA and proteins involved in mtDNA metabolism form a nucleoprotein complex called the mitochondrial nucleoid (Bogenhagen et al., 2008; Holt et al., 2007). Analysis of bromouridine (BrU) labeling in mitochondria has identified BrU-positive foci in close proximity to the nucleoids (Iborra et al., 2004). These foci, termed RNA granules, contain ribosomal proteins (Jourdain et al., 2013) and ribosomal RNA-modifying enzymes (Lee et al., 2013). They also contain the protein GRSF1 (G-rich sequence binding factor 1) and ribonuclease P (RNase P, (Holzmann et al., 2008) and accumulate newly synthesized mRNAs to regulate their processing, storage, sorting or translation (Antonicka et al., 2013; Jourdain et al., 2013). Furthermore, GRSF1 depletion induces a defect in the assembly of the mitoribosomal SSU and marked attenutation of mitochondrial protein synthesis (Antonicka et al., 2013; Jourdain et al., 2013). More recent data has suggested that early steps of ribosome assembly could occur at or near the nucleoids (Bogenhagen et al., 2014). However, the possibility that the bulk of ribosome biogenesis could occur in the RNA granules remains to be explored.
Currently, little is known about how the mitoribosome is assembled and the factors involved. Biogenesis of cytoplasmic ribosomes requires more than 180 transacting proteins and bacterial ribosome assembly involves ~20 proteins, in addition to rRNA modifying enzymes (Shajani et al., 2011). To date, the proteins known to facilitate mitoribosome biogenesis, excluding rRNA modification enzymes and their co-factors, are only a few. They include several poorly-characterized conserved GTPases such as the mt-LSU assembly factors MTG1 and MTG2 as well as the mt-SSU assembly factor C4orf14 (Barrientos et al., 2003; He et al., 2012a; Kotani et al., 2013). In mammalian cells, two mitochondrial transcription factor family proteins play additional roles in mt-LSU assembly, perhaps coordinating transcription and ribosome biogenesis. The precise role of mTERFD1 (or mTERF3) in promoting mt-LSU biogenesis is not understood (Wredenberg et al., 2013), but mTERF4 forms a complex with the rRNA methyltransferase NSUN4 to promote its recruitment to the mt-LSU (Camara et al., 2011) in order to facilitate monosome assembly (Metodiev et al., 2014). The matrix AAA-protease system is involved in the processing of the mt-LSU protein MRPL32, allowing for its association with pre-ribosomal particles in the late stages of mt-LSU assembly (Nolden et al., 2005). Moreover, C7orf30, a member of the DUF143 family of ribosomal silencing factors, participates in mt-LSU assembly (Fung et al., 2013; Rorbach et al., 2012) by interacting with MRPL14 and promoting its incorporation into the mt-LSU (Fung et al., 2013).
Recently, we identified in Saccharomyces cerevisiae the DEAD-box protein Mrh4 as an RNA helicase that promotes late stages of mt-LSU assembly by facilitating remodeling of rRNA-protein interactions (De Silva et al., 2013). The best BLAST match to Mrh4 in the human proteome is DDX28, which displays RNase-sensitive ATPase activity in vitro (Valgardsdottir et al., 2001).
DDX28 was reported to have dual subcellular localization in monkey COS1 cells; whereas most DDX28 resides in mitochondria, a small portion was found in the nucleolus (Valgardsdottir et al., 2001). It was suggested that mitochondrial DDX28 is part of an RNA-protein complex interacting peripherally with the mitochondrial inner membrane (Valgardsdottir et al., 2004). GFP-fused DDX28 was detected in punctate structures, adjacent to mitochondrial nucleoids (Valgardsdottir et al., 2004). The function of DDX28 remains unknown.
In the present study, we report that DDX28 is a mitochondrial RNA granule component that interacts with the 16S rRNA and plays a role in mt-LSU biogenesis, acting at the early stages of assembly required for 16S rRNA stability. We propose that the RNA granules are factories where mitoribosome production occurs.
RESULTS AND DISCUSSION
DDX28 is a Conserved Mitochondrial Matrix Protein that Localizes to RNA Granules
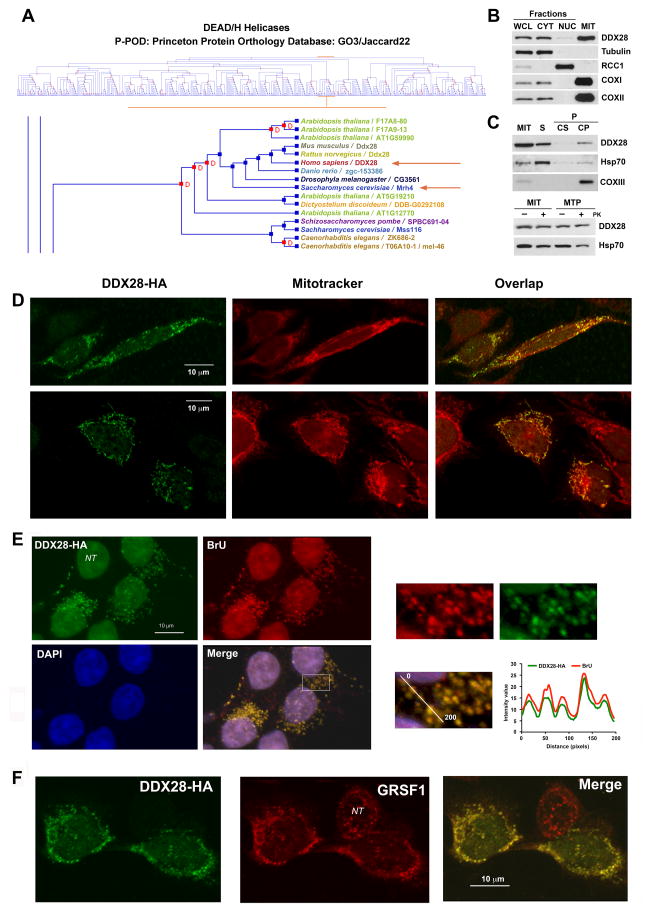
A cluster analysis of all identified DEAD/H helicases across species grouped yeast Mrh4 with human DDX28 (Fig 1A). We generated an antibody against a DDX28 peptide, which allowed detecting the protein in whole cell extracts by immunoblotting (Fig 1B). Analyses of nuclear and cytoplasmic fractions revealed that DDX28 is predominantly a mitochondrial protein, although traces were also detected in the nuclear fraction (Fig 1B). Using brief sonication, alkaline carbonate extraction and proteinase protection assays in mitochondria and mitoplasts, the ~60 kDa DDX28 protein was sub-localized as a soluble mitochondrial matrix protein (Fig 1C). Immunohistochemical studies showed that hemagglutinin (HA)-tagged DDX28 forms punctate structures in mitochondria but not in nuclei from HeLa (Fig 1D) and HEK293T cells (Fig 1E–F), similar to those previously shown in monkey COS1 cells (Valgardsdottir et al., 2004).
Figure 1. The Conserved Dead Box Protein DDX28 is Predominantly a Mitochondrial Matrix-Soluble Protein that accumulates in RNA granules.
(A) Cluster analysis of DEAD/H helicases highlighting the clustering of yeast Mrh4 and mammalian DDX28, obtained using the Princeton Protein Orthology Database (P-POD). A GO3/Jaccard22 graph is presented (http://ppod.princeton.edu) (Heinicke et al., 2007).
(B) Immunoblot analyses of DDX28 levels in HEK293T whole cell lysate (WCL), cytoplasmic (CYT) and nuclear (NUC) fractions as well as in isolated mitochondria (MIT). Antibodies against organelle-specific proteins were used as controls.
(C) Mitochondria isolated from HEK293T cells were fractionated into soluble (S) and membrane-bound (P) mitochondrial proteins by brief sonication and centrifugation. The pellet was submitted to alkaline extraction to allow the separation of the extrinsic proteins present in the supernatant (Cs) from the intrinsic proteins in the pellet (Cp). Equivalent volumes of each fraction were analyzed by immunoblotting using antibodies against DDX28, the matrix-soluble protein Hsp70 and the inner membrane intrinsic protein COXII. The lower panel represents a proteinase K protection assay in mitochondria (MIT) and mitoplasts (MTP) prepared by hypotonic swelling of mitochondria. The samples were analyzed by immunoblotting using antibodies against DDX28 and Hsp70.
(D–F) Immunofluorescence analysis of (D) HeLa cells with anti-DDX28 antibody and mito-tracker red to visualize the mitochondrial network. (E) Bromouridine-treated HEK293T cells with anti-HA and anti-bromouridine antibodies and DAPI to stain the nuclei. (F) HEK293T cells with anti-HA and anti-GRSF antibodies.
NT, non-transformed cell. In (A) and (C), the square marked in the “Merge” image was magnified and the graphs represent relative intensity of the fluorescence signal along the line in the magnified panel.
See also Figure S1.
Immunofluorescence studies on HEK293T cells overexpressing DDX28-HA showed that most DDX28-positive foci colocalize with bromouridine (BrU)-labeled RNA granules (Fig 1E) and GRSF1 (Fig 1F). On the contrary, most of DDX28-containing foci did not colocalize with mitochondrial nucleoids (Fig S1A). The small proportion of BrU-labeled RNA that colocalized with mtDNA (<5%) could indicate actively transcribing nucleoids, as proposed (Iborra et al., 2004).
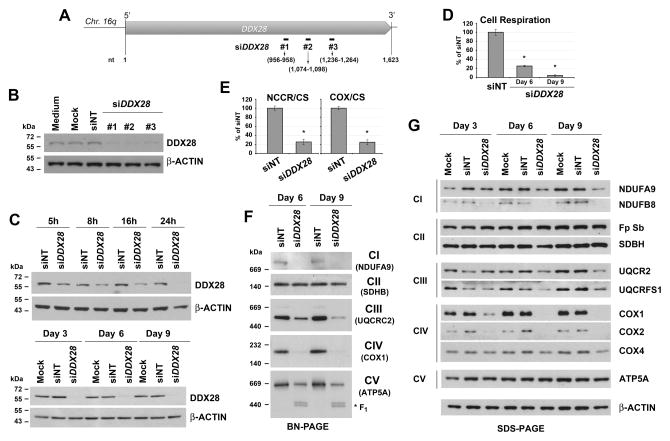
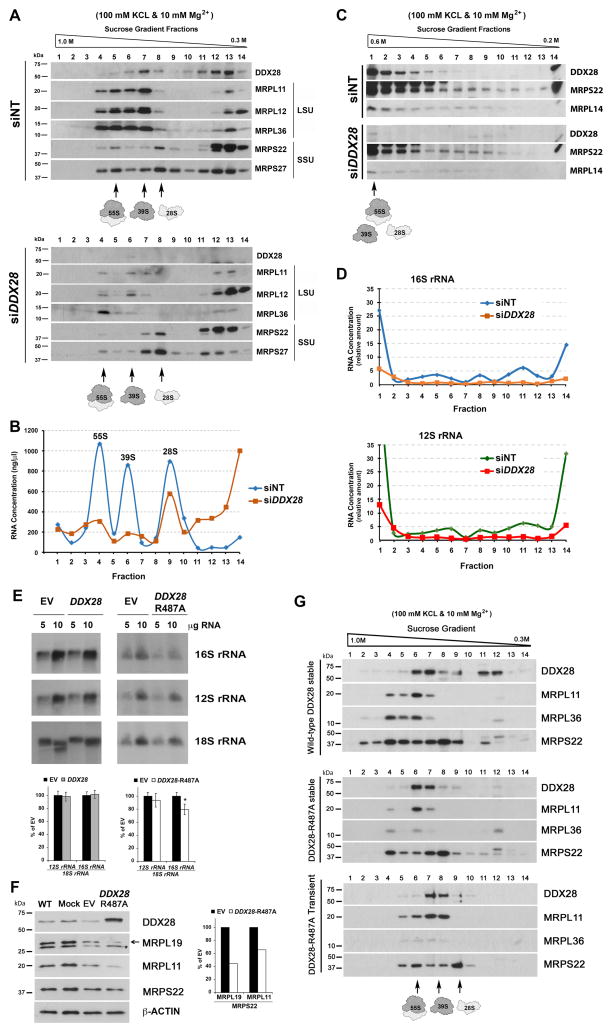
To analyze the role of DDX28 in RNA granule stability and in mitochondrial metabolism we conducted siRNA-specific (Invitrogen Stealth duplex siRNA) knockdown of endogenous DDX28 expression in cultured HEK293T cells. Compared with the non-targeting control oligonucleotides (siNT), transient transfection of three different DDX28-specific siRNA duplexes (siDDX28, Fig 2A) led to a marked reduction of DDX28 at day 3 (Fig 2B). Because the three siRNA sets had similar DDX28 silencing efficiency (Fig 2B), in all subsequent experiments we used set #1. A time-course analysis indicated that DDX28 protein levels were below 5% following 1, 3, 6 and 9 days of silencing (Fig 2C). As reported for GRSF1, mtDNA nucleoids and BrU-positive foci were still visible in siDDX28 cells (Fig S1B), indicating that DDX28 is not essential for formation of the granules.
Figure 2. DDX28 is Required for OXPHOS Assembly and Function.
(A) Scheme depicting the human DDX28 gene and the position of the oligonucleotides used for silencing experiments.
(B) Knock-down of DDX28 protein in HEK293T cells using three different siRNA sets (#1 to #3), verified by immunoblotting of whole cell extracts. An antibody against β-ACTIN was used as loading control. siNT is the non-targeting silencing control.
(C) Time course of DDX28 using the siRNA set # 1 analyzed as in panel (B).
(D) Cell respiration measured polarographically.
(E) Complex I+III (NADH cytochrome c reductase or NCCR) and Complex IV (cytochrome c oxidase or COX) activities measured spectrophotometrically in frozen-thawed cells and normalized by citrate synthase activity. In (D) and (E), error bars represent the mean ± SD of three independent repetitions. * denotes p < 0.05.
(F) Immunoblot analyses of mitochondrial OXPHOS complexes in whole control and siDDX28 cell extracts separated by Blue Native PAGE.
(G) Immunoblot analyses of the steady-state levels of OXPHOS enzyme subunits separated by SDS PAGE. An antibody against β-ACTIN was used as a loading control.
DDX28 is Essential for Mitochondrial OXPHOS Competence
To gain insight into the specific role of DDX28, we characterized mitochondrial functions in siDDX28 cells. The overall effect of DDX28 silencing was a decrease in endogenous cell respiration, which was approximately 25% and 5% of control cells following 6 and 9 days of silencing, respectively (Fig 2D). At day 9, CI+CIII and CIV activities were ~25% of control (Fig 2E). Blue Native-PAGE analyses showed that the steady-state levels of mitochondrial OXPHOS complexes I, III, IV and V, which contain mtDNA-encoded subunits, were markedly decreased whereas the level of complex II, formed exclusively by nucleus-encoded subunits, was not affected (Fig 2F), suggesting a primary defect in mtDNA content or expression. Consistently, examination of OXPHOS enzyme subunits by denaturing SDS-PAGE showed a decreased accumulation of complex I, III and IV components (Fig 2G). The mtDNA-encoded subunits (such as COX1 and COX2) were particularly affected whereas the nucleus-encoded subunits were more stable. This is particularly true for ATP5A, a subunit of the F1 portion of the ATPase, formed by only nucleus-encoded subunits, which assembles even in the absence of the FO segment (Fig 2F–G).
DDX28 is Required for Efficient Biogenesis and Function of the Mitochondrial Translation Machinery
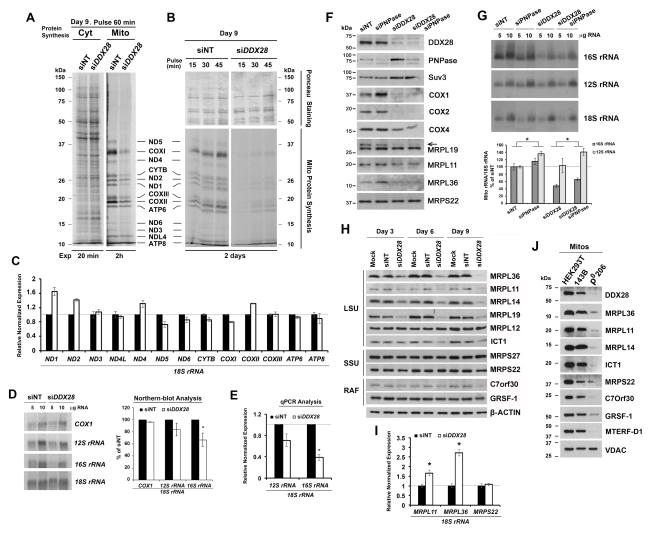
We next investigated whether the OXPHOS defect in siDDX28 cells was due to a primary lesion in mtDNA expression. In vivo protein synthesis analysis in the presence or absence of emetine, a cytoplasmic protein synthesis inhibitor, revealed that the respiratory defect was due to a marked decrease in mitochondrial protein synthesis, while cytoplasmic protein synthesis was not affected (Fig 3A–B).
Figure 3. DDX28 is Required for mtDNA Gene Expression.
(A) Metabolic labeling of cytoplasmic or mitochondrial translation products with [35S]-methionine (in the absence or presence of emetine, a cytoplasmic protein synthesis inhibitor). Mitochondrial translation products are indicated on the right.
(B) Kinetics of in vivo mitochondrial translation performed as in panel A. The upper portion of the membrane was Ponceau-stained for loading control.
(C–E) Steady-state levels of mitochondrial rRNA and mRNA levels in control and siDDX28 cells estimated by (D) Northern blotting or (C, E) qPCR. Error bars represent the mean ± SD of three independent repetitions. * denotes p < 0.05.
(F–G) Steady-state levels of the indicated proteins by immunoblotting (F) and of mitochondrial rRNAs by northern blotting (G) in siNT and siDDX28 cells, further silenced or not for PNPase.
(H) Immunoblot analyses of the steady-state levels of mitoribosome LSU and proteins and ribosomal assembly factors (RAF) in whole cell lysates from control HEK293T and siDDX28 cells.
(I) Steady-state levels of mRNAs for mitochondrial ribosomal proteins in siNT and siDDX28 cells estimated by qPCR. Error bars represent the mean ± SD of three independent repetitions. * denotes p < 0.05.
(J) Immunoblot analyses of the steady-state levels of mitoribosome LSU and SSU proteins and ribosomal assembly factors (RAF) in mitochondria (mitos) from HEK293T, 143B and 143B-206 rho0 (ρ0) cells. See also supplementary Fig S2.
The mitochondrial translation defect was not accounted for by a general loss of mtDNA, as assumed from the visualization of mitochondrial nucleoids by immunofluorescence and estimated by Southern-blot analyses (Fig S1B–C). We next used northern blot and qRT-PCR analyses to investigate the steady-state levels of mitochondrial transcripts. The mRNA levels in siDDX28 cells were in general the same as in non-silenced cells, although somehow elevated for ND2 and ND1 (Fig 3C). The 12S rRNA was slightly but not significantly lowered and the 16S rRNA was deeply and significantly decreased (Fig 3D–E), discarding a general defect in the synthesis of polycistronic transcripts and suggesting a ribosomal defect. Human mitochondrial RNA decay is mediated, at least in part, by the mitochondrial degradosome, consisting of a complex formed by the ribonuclease polynucleotide phosphorylase (PNPase) and the helicase hSuv3. Because silencing of PNPase increases the half-life of some mitochondrial transcripts (Borowski et al., 2013), we tested whether PNPase depletion in siDDX28 cells would stabilize the rRNAs. We first observed that DDX28 depletion enhances degradosome levels (Fig 3F) and as a consequence, PNPase depletion was only mild when compared with wild-type levels (Fig 3F). However, levels of 12S and 16S rRNA were modestly but significantly stabilized by PNPase silencing in both siNT and siDDX28 cells (Fig 3G). These results further support the notion that the absence of DDX28 renders the 16S rRNA unstable, probably because it fails to assemble with mt-LSU proteins.
In all systems, rRNAs are synthesized as precursors that undergo processing and modifications prior to becoming mature transcripts. These processes involve conformational changes, which in both Escherichia coli and S. cerevisiae are assisted by RNA helicases (Bohnsack et al., 2009; Srivastava and Schlessinger, 1988). Therefore, we tested a possible role of DDX28 in assisting the processing or modification of the 16S rRNA. Northern blot analysis (Fig 3D) showed a complete processing of the 16S rRNA in siDDX28 cells. Human 16S rRNA contains three sites for methylation of 2′-O-ribose residues, a Gm residue at G1145 (Lee and Bogenhagen, 2014) and a UmGm at U1369G1370 of the human 16S rRNA A-loop, which modifications are catalyzed by the 2′-O-ribose methyltransferases MRM1, MRM2 and MRM3, respectively (Lee and Bogenhagen, 2014; Rorbach et al., 2014). Decreased levels of these enzymes do not lead to 16S rRNA depletion but to aberrant ribosome assembly (Rorbach et al., 2014). Here, primer extension analyses revealed that the residual 16S rRNA detected in siDDX28 cells contains the three methylated residues (Fig S2A), discarding a role for DDX28 in assisting 16S rRNA modification.
Immunoblot analyses of siDDX28 cells showed marked decreases in the steady-state levels of several mt-LSU proteins (MRPL36, MRPL11, MRPL14, MRPL19 and ICT1 but not MRPL12), suggesting an mt-LSU assembly defect, whereas levels of the mt-SSU proteins (MRPS22 and MRP27) remained unchanged (Fig 3H). Noticeably, the steady-state level of C7orf30, the MRPL14 assembly chaperone (Fung et al., 2013), was also decreased in siDDX28 cells (Fig 3H). However, the stability of DDX28 was not affected by depletion of MRPL14, which attenuates C7orf30 levels, indicating that the stabilities of these three proteins are not interdependent (Fig S2B). Because a small portion of DDX28 was detected in the nucleus, the possibility existed that DDX28 could regulate either transcription of genes encoding for ribosomal proteins or the stability of these transcripts. However, while levels of the mt-SSU gene MRPS22 mRNA were not affected, as expected, those of the mt-LSU genes MRPL11 and MRPL36 were significantly increased (Fig 3I), suggesting the existence of a compensatory mechanism when the mt-LSU fails to assemble.
The accumulation of mt-LSU MRPs in siDDX28 mitochondria resembles the pattern observed in rho0 mitochondria (Fig 3J), devoid of mtDNA, in which the turnover of some of MRPs is enhanced in the absence of rRNAs, as reported in yeast (De Silva et al., 2013; Kaur and Stuart, 2011). Together, these results suggest a role for DDX28 in mt-LSU assembly or stability.
DDX28 Interacts with the mt-LSU
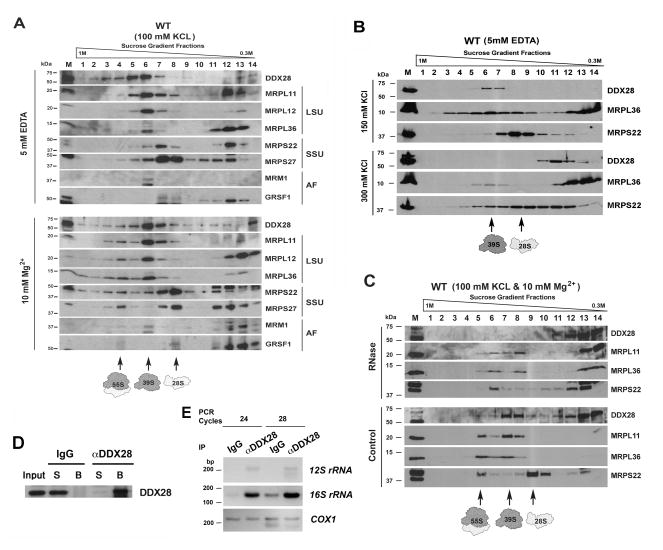
The hypothesis that DDX28 plays a role in mitoribosome assembly was further tested by examination of the native size of DDX28 by sucrose gradient sedimentation. The experiment was performed by using HEK293T mitochondrial protein extracts, prepared in the presence of 1% digitonin and increasing KCl concentrations. When the extraction buffer contained 100 mM KCl and 10 mM MgCl2 to preserve the integrity of the monosomes, DDX28 was found to co-sediment with dissociated mt-LSU but not with the mt-SSU or with the monosome (Fig 4A). When the extraction was performed in the presence of 5 mM EDTA to promote dissociation of the large and small ribosomal subunits, DDX28 was still found to co-sediment with mt-LSU proteins (Fig 4A and S3A). The co-sedimentation of DDX28 with the mt-LSU was salt-sensitive, disrupted by concentrations ≥300 mM KCl (Fig 4B). Incubation of the mitochondrial extracts with 600U RNase A to partially disrupt mitoribosomal integrity prior to loading the sucrose gradient brought DDX28 to sediment more slowly in the lighter fractions (Fig 4C). Furthermore, the co-sedimentation was also disrupted in cellular extracts prepared in the presence of 1 mM EDTA and 1% NP40. However, when the cells were incubated in the presence of the reversible crosslinker DSP (dithiobis[succinimidylpropionate]) prior to extraction with 1% NP40, a portion of DDX28 co-sedimented with mt-LSU proteins (Fig S3B). These results suggested that DDX28 interacts with the 39S mt-LSU and therefore it could play a role in its assembly and/or stability.
Figure 4. DDX28 Associates with the Large Mitoribosome Subunit.
(A–C) Sucrose gradient sedimentation analyses of DDX28 and mitoribosomal proteins from wild-type (WT) HEK293T mitochondrial extracts prepared in the presence of the indicated concentrations of (A) EDTA or MgCl2, (B) KCl, and (C) RNase A. The fractions were analyzed by immunoblotting using Abs against the indicated proteins.
(D) Immunoprecipitation of DDX28 from HEK293T mitochondrial extracts using an anti-DDX28 Image Ab (Sigma) as explained in the experimental procedures section.
(E) PCR analyses of reverse transcribed DDX28-co-immunoprecipitated RNA from UV-induced cross-linked mitochondria.
See also Fig S3.
We noticed that a minute fraction of the GRSF1 co-sediments with mt-SSU but not mt-LSU markers, consistent with a proposed role in mt-SSU biogenesis (Jourdain et al., 2013), although most of the protein accumulated in the top fractions of the gradients where a portion of DDX28 also accumulated (Fig 4A). The stability of GRSF1 is independent of DDX28 since either increased (Fig S4) or knockdown levels of DDX28 (Fig 3H) did not affect the levels of GRSF1. However, in rho0 cells, the levels of DDX28, several MRPs and C7orf30 and GRSF1 were markedly decreased (Fig 3J), indicating that accumulation of these proteins depends on the presence of mitochondrial transcripts.
DDX28 interacts with the 16S rRNA
Because DDX28 interacts with the mt-LSU, it would be expected to bind the 16S rRNA. To test this, highly purified WT mitochondria were subjected to UV-mediated protein-RNA crosslinking before disrupting them with 1% SDS, diluting the extract to final 0.05% SDS, proceeding to DDX28 immunoprecipitation using an efficient anti-DDX28 antibody (Fig 4G) with IgG as a negative control and isolation of the co-immunoprecipitated RNA. Following reverse transcription, PCR analysis showed that in both treated and control mitochondria, the 12S rRNA was poorly detected and the COX1 mRNA was not enriched in any sample. In contrast, 16S rRNA was significantly enriched in cross-linked samples immunoprecipitated with an anti-DDX28 Ab (Fig 4H), thus demonstrating an interaction of DDX28 with the 16S rRNA in vivo.
Depletion of DDX28 does not Induce Accumulation of mt-LSU Assembly Intermediates
An essential step in ribosome biogenesis is formation of the ribonucleoprotein particle, by means of assembling the MRPs with the rRNAs (Shajani et al., 2011). Therefore, we investigated whether any ribosome assembly intermediate, the potential DDX28 substrate, accumulates in siDDX28 mitochondria. Analyses in 0.3M – 1M sucrose gradients showed that the ~5% of residual DDX28 expressed in siDDX28 mitochondria co-sediments with the mt-LSU, and is enough to contribute to the formation of a functional subunit able to form a monosome with the mt-SSU (Fig 5A). These results are consistent with the residual protein synthesis detected in siDDX28 cells (Fig 3A–B). Importantly, the sucrose gradients failed to show the apparent accumulation of any large mt-LSU precursor. Among the proteins tested, MRPL11, MRPL36, MRPL14, MRPL19 and ICT1 are extensively degraded in siDDX28 cells (Fig 3H and 5), probably as a consequence of their inability to assemble. The non-assembled portion of the more stable MRPL12 sedimented in the top fractions of the gradient (Fig 5A), further indicating the absence of large assembly intermediates, at least containing the subunits tested. The RNA profile in the gradients using siDDX28 mitochondrial extracts show some residual rRNA on the 39S particle (Fig 5B). Expansion of the slower sedimenting portion of the gradient in a 0.2M – 0.6M gradient (Fig 5C) and analysis by qPCR of RNA purified from each fraction, failed to detect substantial amounts of 16S rRNA in any subassembly (Fig 5D). These data suggest that the substrate of DDX28 is either naked 16S rRNA or an early mt-LSU assembly intermediate.
Figure 5. Depletion of DDX28 does not Induce Accumulation of Large Mitoribosomal Subunit Assembly Intermediates and DDX28 helicase Activity is Required for its Function in vivo.
(A–B) Sedimentation analyses in 0.3M – 1M sucrose gradients of mitochondrial extracts prepared from HEK293T siNT and siDDX28 mitochondria in the presence of 1% digitonin, 100 mM KCl and 10 mM Mg2+. The fractions were used to analyze the distribution of DDX28 and ribosomal proteins by immunoblotting.
(B) The fractions in (A) were used to extract and measure total RNA concentration.
(C) Similar to (A) but using 0.2 M–0.6 M sucrose gradients.
(D) For each fraction in (C), total RNA was isolated, reverse transcribed and 16S and 12S levels quantified by qPCR. Values were normalized to the lowest for each cell line and plotted.
(E) Steady-state levels of mitochondrial rRNAs in HEK293T cells transfected with an empty vector (EV) or overexpressing either DDX28 or DDX28-R487A, estimated by northern blotting and quantified in the bottom panel. Error bars represent the mean ± SD of three independent repetitions. * denotes p < 0.05.
(F) Immunoblot analyses of the steady-state levels of DDX28, DDX28-R487A and ribosomal proteins and assembly factors in the indicated cell lines.
(G) Sucrose gradient sedimentation analyses of DDX28 and mitoribosomal proteins from mitochondrial extracts prepared from HEK293T cells stably or transiently expressing DDX28-R487A.
See also Fig S4.
The helicase activity of DDX28 is essential for its function
To investigate whether the helicase activity of DDX28 is necessary for its role in mitoribosome assembly, we created constructs to overexpress either wild-type DDX28 or a variant mutated in a conserved arginine in the DDX28 helicase domain to alanine (R487A). An equivalent mutation in the Spb4 helicase involved in yeast cytoplasmic ribosome assembly (Garcia-Gomez et al., 2011) or in the DbpA helicase required for bacterial ribosome assembly (Sharpe Elles et al., 2009) causes a dominant-negative phenotype when overexpressed in the corresponding wild-type strains. Overexpression of the DDX28-R487A allele, but not of the wild-type allele, induced a mild but clear mt-LSU assembly defect, as seen by lowered steady-state levels of 16S rRNA and some mt-LSU mitoribosomal proteins (Fig 5E and F). The residual mt-LSU proteins sediment in sucrose gradients with a profile similar to that in wild-type cells (Fig 5G), as seen for siDDX28 cells. We conclude that the helicase activity of DDX28 is necessary for its function in vivo.
DDX28 Interacts with mt-LSU Proteins and other RNA Granule Components
To determine the DDX28 interactome, we performed immunoprecipitation (IP) followed by mass spectrometry analyses or immunoblotting analyses using either wild-type mitochondria or mitochondria isolated from cells stably expressing HA-6xHis-tagged DDX28 (Fig S4A). These cells accumulate levels of DDX28-HA slightly higher than those of endogenous DDX28 (Fig S4B). In sucrose gradients, both proteins co-sedimented with the mt-LSU (Fig S4C), suggesting that the tagged protein is functional. Notably, the higher amount of total DDX28 did not affect the levels of mitoribosomal proteins (Fig S4B). Using a polyclonal anti-HA Ab for immunoprecipitation, the efficiency was ~50% (Fig S4D), while the anti-DDX28 Ab was more efficient (~95%) (Fig 4G). Our mass spectrometry analyses identified the target DDX28 as expected, plus four major groups of relevant proteins (Table S1): First, most mitoribosomal proteins (45 mt-LSU and 29 mt-SSU MRPs). Second, several previously identified factors involved in the biogenesis of mt-LSU (MTG1, C7orf30 and MTERF-D1) or mt-SSU (ERAL1), together with a set of potential mitoribosomal assembly/maintenance factors including proteases, GTPases and chaperones. Third, mitochondrial RNA metabolism proteins, including: GRSF1; LRPPRC, and SLIRP involved in the stability, polyadenylation and coordination of translation of mitochondrial mRNAs (Chujo et al., 2012; Ruzzenente et al., 2012); RNase P; the 16S methyltransferase RMTL1/MRM3; the pentatricopeptide repeat domain protein 1 (PTCD1) involved in the processing of mitochondrial polycistronic transcripts that contain leucine tRNA (Rackham et al., 2009); and the translational regulator PTCD3 (Davies et al., 2009). Fourth, a set of translation factors and most aminoacyl tRNA synthetases.
Notably, our analyses identified only two proteins previously associated with mitochondrial nucleoids: the helicase DHX30 that acts as a transcriptional regulator (Minczuk et al., 2011), and the single-stranded DNA-binding protein SSBP1, but not TFAM or any other major nucleoid component (Table S1). By means of immunodetection, several DDX28-coimmunoprecipitated relevant proteins were detected by western blot (Fig S4D). Relatively small amounts of MRPL36, MRPL14, C7orf30, MTERFD1 and GRSF1 were found consistently in the eluate. The four groups of DDX28-interacting proteins described here form the core of the mitochondrial RNA granule and suggest that ribosome assembly could occur largely within, or in the proximity of, this compartment.
CONCLUSION
DEAD-box proteins are RNA-dependent ATPases that use ATP to rearrange the structures of RNA or ribonucleoprotein complexes. They are involved in every aspect of RNA metabolism, including nuclear transcription, pre-mRNA splicing, ribosome biogenesis, nucleo-cytoplasmic transport, translation, and RNA decay (Linder and Jankowsky, 2011). DEAD-box proteins also participate in organellar RNA metabolism, although their roles in mitochondrial gene expression remain poorly understood. This study demonstrates that DDX28 plays a role in biogenesis of the mt-LSU by interacting with it and with the 16S rRNA. We propose that DDX28 interacts early with 16S rRNA to catalyze the formation of a stable intermediate and remains associated with the growing pre-39S particle until its completion. DDX28 could be acting as an RNA chaperone to allow remodeling of early 16S rRNA-MRP interactions as shown for some bacterial DEAD box proteins (Linder and Jankowsky, 2011) and for yeast Mrh4 (De Silva et al., 2013). Future studies will focus on analyses of the specific 16S rRNA helices to which DDX28 binds.
Mitochondrial ribosome subunits and assembly factors have been found associated with, or in the vicinity of, nucleoids (Bogenhagen et al., 2014; He et al., 2012b; Hensen et al., 2013; Iborra et al., 2004; Lee et al., 2013) and RNA granules (Antonicka et al., 2013; Jourdain et al., 2013; Lee et al., 2013). These studies suggest a close association among nucleoids, RNA granules and the mitochondrial protein synthesis machinery. Recent data has shown that mitoribosome assembly could be initiated within or near the nucleoids (Bogenhagen et al., 2014; Dalla Rosa et al., 2014; He et al., 2012a), possibly in a co-transcriptional manner as it occurs in bacteria (Shajani et al., 2011). Our data on the localization of DDX28 within RNA granules, its participation in mt-LSU assembly and the composition of its interactome, all point toward the conclusion that most steps of mitoribosome biogenesis, at least for the mt-LSU, could occur in the RNA granules. We envision, however, the RNA granules as dynamic structures. Newly transcribed rRNAs and/or early mitoribosome assembly intermediates are transferred from nucleoids to the RNA granules, where mitoribosome assembly is completed. These mitochondrial matrix subcompartments are reminiscent of the nucleolus. Within the nucleus, the membrane-less nucleolus is organized around the chromosomal regions that contain the genes for the rRNAs, and is the site of rRNA transcription and processing and of ribosome assembly. Equivalent features pertain to the mitochondrial RNA granule, which we propose to term “mitochondriolus”.
In conclusion, we have identified DDX28 as a DEAD-box protein that plays an essential role in early stages of mt-LSU biogenesis, a process that takes place within the compartment defined by the RNA granule or mitochondriolus, the factory where human mitoribosome production is accomplished.
Methods
Human Cell Lines and Culture Conditions
HEK293T (obtained from the NIH AIDS Research and Reference Reagent Program), HeLa (obtained from ATCC), the osteosarcoma 143B.TK− and their rho0 derivative (143B206) (King and Attadi, 1996) cell lines were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate and 50 μg/ml uridine at 37°C in an atmosphere of 5% CO2.
siRNA Transfection
RNA interference was implemented for transient knockdown of DDX28 in HEK293T cells. We used three Stealth RNAi duplexes against human DDX28 (HSS125053, HSS125054 and HSS125055, Invitrogen, Life Technologies, Grand Island, NY) designed using Block-iT RNAi Express (http://rnaidesigner.invitrogen.com/rnaiexpress). Each stealth siRNA, and an ON-TARGETplus Non-targeting siRNA #1 control (Thermo Scientific, Pittsburgh PA) was transiently transfected into HEK293T cells at a final concentration of 20 nM using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s specifications. Transfection was repeated on days 3 and 6, and the cells were harvested on days 3, 6 or 9 for analysis.
Mitochondrial Preparation and DDX28 Localization Experiments
Mitochondria were isolated from HEK293T cells essentially as described (Clemente et al., 2013). Subcellular fractionation of HEK293T mitochondria was performed as described (Clemente et al., 2013) using sonication and proteinase K protection assays in mitochondria and mitoplasts. DDX28 localization in whole cells was determined by immunocytochemistry as explained in the supplemental material.
Antibodies
We used the services of Open Biosystems/Thermo Scientific (Huntsville, AL) to generate an affinity-purified rabbit polyclonal antibody against a DDX28 peptide. The peptide, RRRSLPGLASSVKEPLPQAT, comprises amino acids 520 to 540 on DDX28. The commercial antibodies used in this study are listed in the supplemental experimental procedures.
Pulse Labeling of Mitochondrial Translation Products
Cells were labeled for 15, 30, 45 or 60 minutes at 37°C in methionine-free DMEM medium containing either 150 μCi/ml [35S] methionine (PerkinElmer Life Sciences, Boston, MA) (for cytoplasmic protein labeling) or 150 μCi/ml [35S] methionine and 100 μg/ml emetine (for mitochondrial protein labeling) as described (Leary and Sasarman, 2009).
Sucrose Gradient Sedimentation
The sedimentation properties of DDX28 and the ribosomal proteins in sucrose gradients were analyzed essentially as described (Barrientos et al., 2004) using extracts from 2 mg of protein prepared from wild-type HEK293T cells or 400 μg of mitochondria isolated from non-targeting siRNA- and siDDX28-treated cells. Experimental details are described in the supplemental material. All gradients were performed at least in triplicate using independent mitochondrial preparations. The gradients reported are representative of each cell line or extraction condition because the patterns observed were reproducible.
RNA analysis
Methods for RNA isolation, quantitative RT-PCR, Northern blot analyses and primer extension analyses of the 16S rRNA are described in the supplemental experimental procedures.
Statistical Analysis
All experiments were done at least in triplicate. All data are presented as means ± SD of absolute values or percentage of control. Values were analyzed for statistical significance by Student’s t-test. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Dr. Flavia Fontanesi, Dr. Brant Watson and Dasmanthie De Silva for critical reading of the manuscript. This research was supported by grants from the National Institutes of Health (NIH) RO1 GM071775-06, GM105781-01 and GM112179-01 (to AB) and an American Heart Association (AHA) predoctoral fellowship (to YT).
Footnotes
Supplemental information includes four figures (S1 to S4) and two spreadsheets.
CONFLICTS OF INTEREST
The authors declare that they do not have any conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonicka H, Sasarman F, Nishimura T, Paupe V, Shoubridge EA. The Mitochondrial RNA-Binding Protein GRSF1 Localizes to RNA Granules and Is Required for Posttranscriptional Mitochondrial Gene Expression. Cell Metab. 2013;17:386–398. doi: 10.1016/j.cmet.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Barrientos A, Korr D, Barwell KJ, Sjulsen C, Gajewski CD, Manfredi G, Ackerman S, Tzagoloff A. MTG1 codes for a conserved protein required for mitochondrial translation. Mol Biol Cell. 2003;14:2292–2302. doi: 10.1091/mbc.E02-10-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF, Martin DW, Koller A. Initial steps in RNA processing and ribosome assembly occur at mitochondrial DNA nucleoids. Cell Metab. 2014;19:618–629. doi: 10.1016/j.cmet.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, Szczesny RJ. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 2013;41:1223–1240. doi: 10.1093/nar/gks1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Christian BE, Spremulli LL. Evidence for an active role of IF3mt in the initiation of translation in mammalian mitochondria. Biochemistry. 2009;48:3269–3278. doi: 10.1021/bi8023493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Ohira T, Sakaguchi Y, Goshima N, Nomura N, Nagao A, Suzuki T. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 2012;40:8033–8047. doi: 10.1093/nar/gks506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente P, Peralta S, Cruz-Bermudez A, Echevarria L, Fontanesi F, Barrientos A, Fernandez-Moreno MA, Garesse R. hCOA3 stabilizes cytochrome c oxidase 1 (COX1) and promotes cytochrome c oxidase assembly in human mitochondria. J Biol Chem. 2013;288:8321–8331. doi: 10.1074/jbc.M112.422220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Rosa I, Durigon R, Pearce SF, Rorbach J, Hirst EM, Vidoni S, Reyes A, Brea-Calvo G, Minczuk M, Woellhaf MW, et al. MPV17L2 is required for ribosome assembly in mitochondria. Nucleic Acids Res. 2014;19 doi: 10.1093/nar/gku513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SM, Rackham O, Shearwood AM, Hamilton KL, Narsai R, Whelan J, Filipovska A. Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett. 2009;583:1853–1858. doi: 10.1016/j.febslet.2009.04.048. [DOI] [PubMed] [Google Scholar]
- De Silva D, Fontanesi F, Barrientos A. The DEAD-Box protein Mrh4 functions in the assembly of the mitochondrial large ribosomal subunit. Cell Metab. 2013;18:712–725. doi: 10.1016/j.cmet.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung S, Nishimura T, Sasarman F, Shoubridge EA. The conserved interaction of C7orf30 with MRPL14 promotes biogenesis of the mitochondrial large ribosomal subunit and mitochondrial translation. Mol Biol Cell. 2013;24:184–193. doi: 10.1091/mbc.E12-09-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gomez JJ, Lebaron S, Froment C, Monsarrat B, Henry Y, de la Cruz J. Dynamics of the putative RNA helicase Spb4 during ribosome assembly in Saccharomyces cerevisiae. Mol Cell Biol. 2011;31:4156–4164. doi: 10.1128/MCB.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leitner A, Bieri P, Voigts-Hoffmann F, Erzberger JP, Leibundgut M, Aebersold R, Ban N. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;505:515–519. doi: 10.1038/nature12890. [DOI] [PubMed] [Google Scholar]
- He J, Cooper HM, Reyes A, Di Re M, Kazak L, Wood SR, Mao CC, Fearnley IM, Walker JE, Holt IJ. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 2012a;40:6097–6108. doi: 10.1093/nar/gks257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Cooper HM, Reyes A, Di Re M, Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A, et al. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 2012b;40:6109–6121. doi: 10.1093/nar/gks266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensen F, Cansiz S, Gerhold JM, Spelbrink JN. To be or not to be a nucleoid protein: a comparison of mass-spectrometry based approaches in the identification of potential mtDNA-nucleoid associated proteins. Biochimie. 2013;24:00331–00333. doi: 10.1016/j.biochi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Holt IJ, He J, Mao CC, Boyd-Kirkup JD, Martinsson P, Sembongi H, Reyes A, Spelbrink JN. Mammalian mitochondrial nucleoids: organizing an independently minded genome. Mitochondrion. 2007;7:311–321. doi: 10.1016/j.mito.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Kimura H, Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain AA, Koppen M, Wydro M, Rodley CD, Lightowlers RN, Chrzanowska-Lightowlers ZM, Martinou JC. GRSF1 Regulates RNA Processing in Mitochondrial RNA Granules. Cell Metab. 2013;17:399–410. doi: 10.1016/j.cmet.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Stuart RA. Truncation of the Mrp20 protein reveals new ribosome-assembly subcomplex in mitochondria. EMBO Rep. 2011;12:950–955. doi: 10.1038/embor.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal PS, Sharma MR, Booth TM, Haque EM, Tung CS, Sanbonmatsu KY, Spremulli LL, Agrawal RK. Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome. Proc Natl Acad Sci U S A. 2014;111:7284–7289. doi: 10.1073/pnas.1401657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Attadi G. Mitochondria-mediated transformation of human rho (0) cells. Methods Enzymol. 1996;264:313. doi: 10.1016/s0076-6879(96)64030-0. [DOI] [PubMed] [Google Scholar]
- Kotani T, Akabane S, Takeyasu K, Ueda T, Takeuchi N. Human G-proteins, ObgH1 and Mtg1, associate with the large mitochondrial ribosome subunit and are involved in translation and assembly of respiratory complexes. Nucleic Acids Res. 2013;41:3713–3722. doi: 10.1093/nar/gkt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary SC, Sasarman F. Oxidative phosphorylation: synthesis of mitochondrially encoded proteins and assembly of individual structural subunits into functional holoenzyme complexes. Methods Mol Biol. 2009;554:143–162. doi: 10.1007/978-1-59745-521-3_10. [DOI] [PubMed] [Google Scholar]
- Lee KW, Bogenhagen DF. Assignment of 2′-O-methyltransferases to Modification Sites on the Mammalian Mitochondrial Large Subunit 16S rRNA. J Biol Chem. 2014;29 doi: 10.1074/jbc.C114.581868. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Okot-Kotber C, Lacomb JF, Bogenhagen DF. Mitochondrial rRNA Methyltransferase Family Members are Positioned to Modify Nascent rRNA in Foci Near the mtDNA Nucleoid. J Biol Chem. 2013;288:31386–31399. doi: 10.1074/jbc.M113.515692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Metodiev MD, Spahr H, Loguercio Polosa P, Meharg C, Becker C, Altmueller J, Habermann B, Larsson NG, Ruzzenente B. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014;10:e1004110. doi: 10.1371/journal.pgen.1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk M, He J, Duch AM, Ettema TJ, Chlebowski A, Dzionek K, Nijtmans LG, Huynen MA, Holt IJ. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39:4284–4299. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- O’Brien TW. The general occurrence of 55 S ribosomes in mammalian liver mitochondria. J Biol Chem. 1971;246:3409–3417. [PubMed] [Google Scholar]
- Rackham O, Davies SM, Shearwood AM, Hamilton KL, Whelan J, Filipovska A. Pentatricopeptide repeat domain protein 1 lowers the levels of mitochondrial leucine tRNAs in cells. Nucleic Acids Res. 2009;37:5859–5867. doi: 10.1093/nar/gkp627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorbach J, Boesch P, Gammage PA, Nicholls TJ, Pearce SF, Patel D, Hauser A, Perocchi F, Minczuk M. MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol Biol Cell. 2014;9:01–0014. doi: 10.1091/mbc.E14-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorbach J, Gammage PA, Minczuk M. C7orf30 is necessary for biogenesis of the large subunit of the mitochondrial ribosome. Nucleic Acids Res. 2012;40:4097–4109. doi: 10.1093/nar/gkr1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzenente B, Metodiev MD, Wredenberg A, Bratic A, Park CB, Camara Y, Milenkovic D, Zickermann V, Wibom R, Hultenby K, et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- Sharpe Elles LM, Sykes MT, Williamson JR, Uhlenbeck OC. A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 2009;37:6503–6514. doi: 10.1093/nar/gkp711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Schlessinger D. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc Natl Acad Sci U S A. 1988;85:7144–7148. doi: 10.1073/pnas.85.19.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgardsdottir R, Brede G, Eide LG, Frengen E, Prydz H. Cloning and characterization of MDDX28, a putative dead-box helicase with mitochondrial and nuclear localization. J Biol Chem. 2001;276:32056–32063. doi: 10.1074/jbc.M011629200. [DOI] [PubMed] [Google Scholar]
- Valgardsdottir R, Ottersen OP, Prydz H. Regulated compartmentalization of the putative DEAD-box helicase MDDX28 within the mitochondria in COS-1 cells. Exp Cell Res. 2004;299:294–302. doi: 10.1016/j.yexcr.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Wredenberg A, Lagouge M, Bratic A, Metodiev MD, Spahr H, Mourier A, Freyer C, Ruzzenente B, Tain L, Gronke S, et al. MTERF3 regulates mitochondrial ribosome biogenesis in invertebrates and mammals. PLoS Genet. 2013;9:e1003178. doi: 10.1371/journal.pgen.1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.