Abstract
The nucleolus is responsible for the production of ribosomes, essential machines which synthesize all proteins needed by the cell. The structure of human nucleoli is highly dynamic and is directly related to its functions in ribosome biogenesis. Despite the importance of this organelle, the intricate relationship between nucleolar structure and function remains largely unexplored. How do cells control nucleolar formation and function? What are the minimal requirements for making a functional nucleolus? Here we review what is currently known regarding mammalian nucleolar formation at nucleolar organizer regions (NORs), which can be studied by observing the dissolution and reformation of the nucleolus during each cell division. Additionally, the nucleolus can be examined by analyzing how alterations in nucleolar function manifest in differences in nucleolar architecture. Furthermore, changes in nucleolar structure and function are correlated with cancer, highlighting the importance of studying the determinants of nucleolar formation.
Keywords: Nucleolus, Cancer, Ribosomopathies, Ribosome Biogenesis
Introduction
The nucleolus is a non-membrane bound nuclear organelle where ribosomes, the powerhouses of translation, are synthesized and assembled. The energetically expensive process of ribosome biogenesis requires over 200 assembly factors, many small nucleolar ribonucleoproteins (snoRNPs), and four different ribosomal RNAs (rRNAs). Ribosomal assembly begins with the transcription of the 47S polycistronic precursor rRNA (pre-rRNA) by RNA polymerase I (Pol I) in the nucleolus. The 47S rRNA is then chemically modified and processed into the mature 5.8S, 18S, and 28S rRNAs which are assembled into the small (18S) and large (5.8S and 28S) subunits of the functional ribosome. An additional rRNA, the 5S rRNA, is transcribed outside of the nucleolus by RNA polymerase III. Ribosome assembly begins in the nucleolus, moves to the nucleoplasm, and concludes in the cytoplasm. While this complex process has traditionally been studied in the yeast Saccharomyces cerevisiae (reviewed in Henras et al. 2008; Woolford Jr. and Baserga 2013), efforts have recently been made to understand the process in mammalian cells (O'Donohue et al. 2010; Sloan et al. 2013; Tafforeau et al. 2013; Wang et al. 2014; Wild et al. 2010). In addition to ribosome assembly, the nucleolus may play several other roles. Such roles include ribonucleoprotein biogenesis, p53 management as part of the stress response, and cell cycle regulation (reviewed in Pederson and Tsai 2009; Warner and McIntosh 2009; Golomb et al. 2014). The nucleolus is therefore a dynamic organelle which performs multiple essential cell functions.
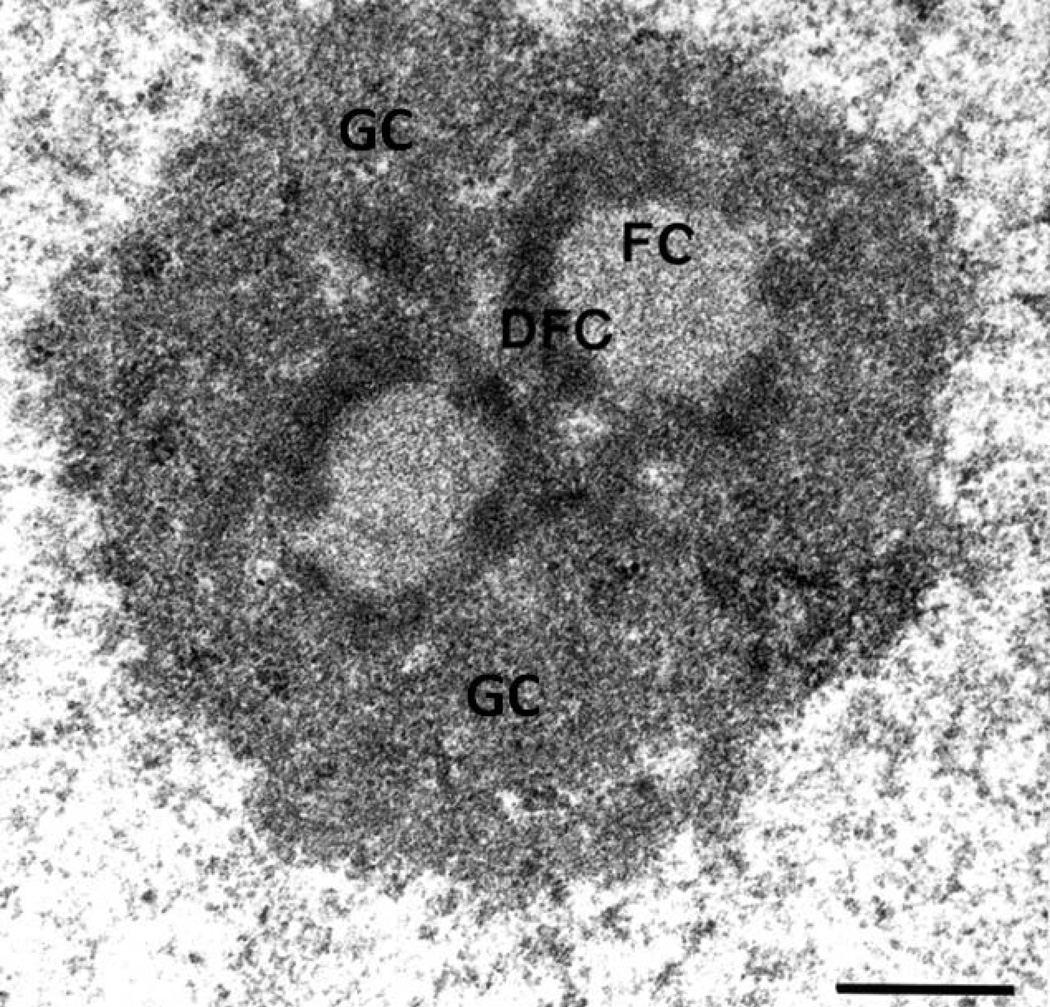
When viewed by light microscopy, nucleoli feature prominently in the cell nucleus. Additionally, silver nitrate preferentially stains a group of argyrophilic proteins which localize at transcriptionally active nucleolar organizing regions (NORs), allowing for visualization of nucleoli in cyto-histopathological samples (Goodpasture and Bloom 1975). During metaphase, these NORs which had been active in the preceding interphase appear as achromatic gaps, termed secondary constrictions, when stained with DAPI (4’,6-diamidino-2-phenylindole; Sumner 1982). The nucleoli of higher eukaryotes are comprised of three distinct subcompartments: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC), which can be seen by electron microscopy (Fig. 1; Scheer and Weisenberger 1994). Most proteins remain in the nucleolus for only tens of seconds (Phair and Misteli 2000), making precise assignment of subcompartment constituents and processes difficult.
Fig. 1.
An electron micrograph of a HeLa cell demonstrates that the nucleolus is comprised of three subcompartments: the fibrillar center (FC), dense fibrillar component (DFC) and granular component (GC). Bar, 0.5 µm. Reprinted with permission from (Sirri et al. 2002)
The nucleolar ultrastructure is likely a product of the functions it performs: ribosome biogenesis. rDNA transcription starts, most likely, at the interface of the FC and the DFC (Cheutin et al. 2002; Koberna et al. 2002). Pre-rRNA processing and pre-ribosome assembly proceed away from the FC-DFC interface, until the ribosomal subunits are exported into the cytoplasm for the final steps of maturation (Fig. 1). Current evidence therefore suggests that ribosome biogenesis occurs directionally away from the fibrillar center (Raska et al. 2006), and the nucleolar subcompartments are formed from the process of building a ribosome.
In contrast with current views that the vectorial nature of rRNA processing determines nucleolar structure (Thiry and Lafontaine 2005; Olson and Dundr 2005), it has also been suggested that the fluid, liquid-like behavior of the nucleolus is adequate to determine its size and shape. Recently, Brangwynne et al. used the germinal vesicles (GV); nucleus of amphibian oocytes to demonstrate that the nucleolus behaves like a liquid droplet on a timescale of tens of seconds (Brangwynne et al. 2011). The surface tension of the droplet is thus responsible for the spherical shape of the nucleolus. Interestingly, the authors were able to visualize such properties as nucleoli came into contact with each other and fused to form one larger nucleolus (Brangwynne et al. 2011). The authors utilized this fusion process to demonstrate that the volumes of nucleoli followed a power-law distribution characteristic of aggregation processes (Brangwynne et al. 2011). Additionally, Handwerger et al. used Xenopus GV nucleoli to suggest a “sponge model” in which the size of the molecule and the density of the subcompartment determine the movement of proteins in and out of the subcompartment (Handwerger et al. 2005). Whether or not these findings translate from Xenopus GV nucleoli to human nucleoli remains to be seen, however, as Xenopus GV nucleoli are extrachromosomal and are more numerous than human nucleoli (Wu and Gall 1997). It should also be noted that the above Xenopus GV experiments were conducted using isolated nucleoli in mineral oil. The influence of such preparations on the behavior of nucleoli has not yet been determined. Nevertheless, the nucleolus is comprised of a specific ultrastructure which directly relates to its function in ribosome biogenesis.
NORs dictate nucleolar formation
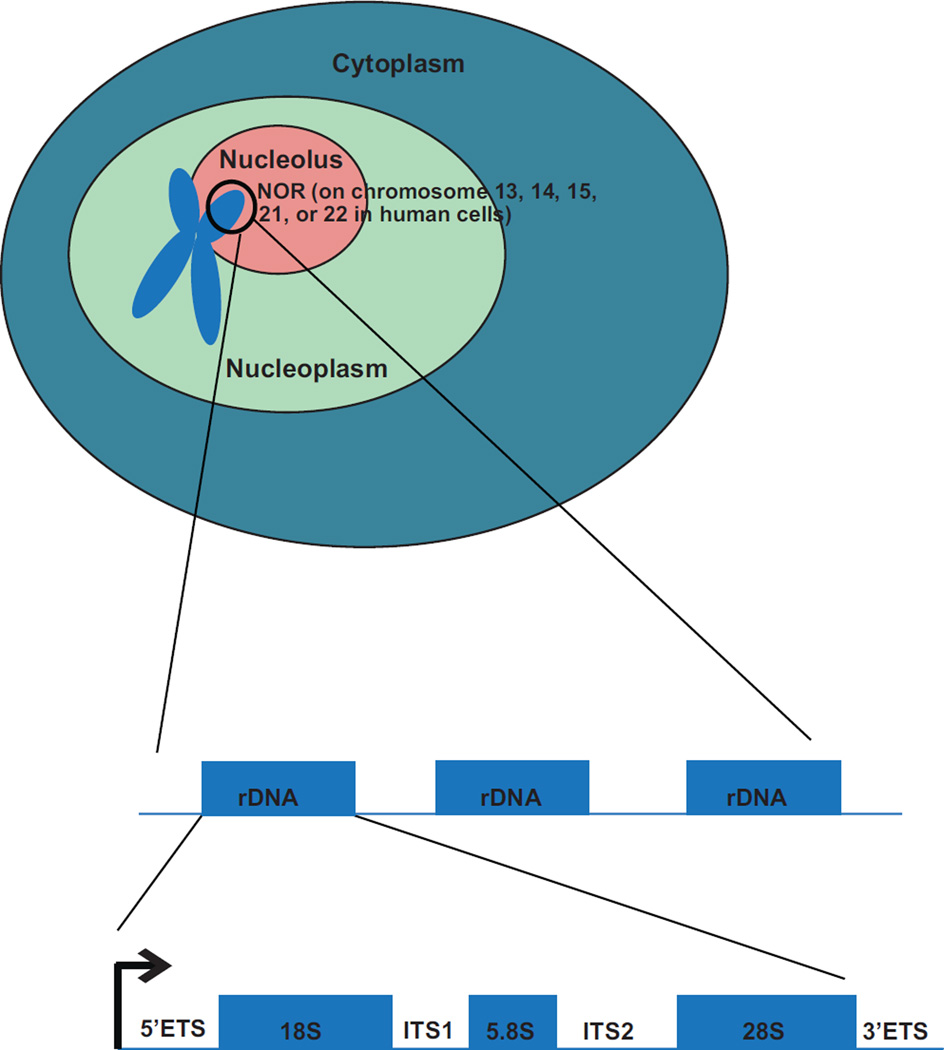
The first comprehensive description of the nucleolus is currently attributed to both Wagner (1835) and Valentin (1836). However, Fontana reported “un corps oviforme” existing in eel epidermal cells as early as 1781 (Fontana and Nyon 1781). It was not for another one hundred years that progress was made on the function of the nucleolus. In 1934, Barbara McClintock accurately characterized the NORs through studying a chromosomal translocation in Zea mays, stating that the nucleolus originates from “an organized body in the chromosome directly adjacent to the stalk of the satellite” (McClintock 1934). Each NOR is comprised of a cluster of ribosomal DNA (rDNA) repeats which contain the 47S pre-rRNA (Fig. 2). NORs are located on the short arms of the acrocentric chromosomes. The number of potential nucleoli therefore depends on the number of acrocentric chromosomes, which ranges between species. In humans, the acrocentric chromosomes are chromosomes 13, 14, 15, 21, and 22 (Henderson et al. 1972). Therefore, there are 10 NORs in diploid human cells and 10 possible nucleoli per cell.
Fig. 2.
The nucleolus forms around nucleolar organizer regions (NORs), located on the short arms of the acrocentric chromosomes. NORs are made of rDNA repeats. One rDNA repeat contains the transcript for the pre-rRNA. In human cells, the 47S pre-rRNA consists of the 18S, 5.8S, and 28S rRNAs flanked by external transcribed spacers (5’ETS and 3’ETS) and internal transcribed spacers (ITS1 and ITS2). The 18S, 5.8S, and 28S rRNAs are included into the small and large subunits of the forming ribosome
Not all NORs are transcriptionally active. Because transcriptional activity is required for the formation of functional nucleoli (Grob et al. 2014), not every NOR forms a nucleolus. These inactive NORs are not associated with the Pol I transcription machinery and so do not stain positively in silver nitrate staining (AgNOR) methods (McStay and Grummt 2008). In HeLa cells, it has been shown that only six of ten NORs are transcriptionally active at a time (Roussel et al. 1996). In addition, not all rDNA repeats within a NOR are transcriptionally active. The precise mechanisms that determine which rDNA repeats are active and which are inactive remain unknown. Thus far, researchers have found important roles for methylation, nucleosome position, and chromatin remodeling complexes (such as the activating Cockayne Syndrome protein B and NoRC repressive remodeling complex) in the maintenance of active and silent rDNA repeats (reviewed in McStay and Grummt 2008; Guetg and Santoro 2012). Furthermore, Haaf, et al. found that rDNA transcriptional activity varies according to cell type and may change with stages of development (Haaf et al. 1991). Given the intricate relationship between rDNA transcription and nucleolar formation, knowledge of the mechanisms governing transcriptional activity is essential.
Nucleolar formation observed through the cell cycle
The intricacy of the relationship between nucleolar formation and nucleolar function is exemplified in cell division. During mitosis in human cells, the nuclear envelope breaks down, and nucleolar components are dispersed throughout the dividing cell. The nucleolus then reforms after mitosis. Throughout this process, components of the Pol I transcription machinery (i.e. the Pol I subunits, the promoter selectivity factor SL1, and upstream binding factor UBF) remain associated with the rDNA while the remaining nucleolar components diffuse throughout the cell (Roussel et al. 1996). The process of dissolution begins in prophase, when ribosome production is halted (Gebrane-Younes et al. 1997). It is then that rRNA processing components, found in the GC and DFC of active nucleoli, relocate to form the perichromosomal compartment (Gautier et al. 1992). Later in prophase, cyclin B-cyclin dependent kinase 1 (CDK1) phosphorylates SL1 (Heix et al. 1998). When phosphorylated, SL1 is unable to associate with UBF, inhibiting the formation of the Pol I pre-initiation complex and thus halting rDNA transcription in mitosis (Heix et al. 1998). During or shortly after this, the nuclear envelope breaks down, and the nucleolus is no longer visible (Hernandez-Verdun 2004).
Beginning in telophase, the nucleoli reassemble. In HeLa cells, it has been shown that the nucleolar assembly process takes on average 1.5 hours to complete from the start of anaphase (Savino et al. 2001). At the end of mitosis, NORs which remained associated with the Pol I transcription machinery (previously active NORs) resume transcription through the dephosphorylation of several key proteins, including cyclin B-CDK1, by the phosphatases PP1 (Wu et al. 2009) and PP2A (Mochida et al. 2009). The remaining components necessary for nucleolar function are organized into prenucleolar bodies (PNBs). These extranucleolar PNBs contain processing proteins, snoRNAs, ribosomal proteins, and unprocessed pre-rRNAs (Azum-Gelade et al. 1994; Jimenez-Garcia et al. 1994). PNBs, however, are distinct from nucleoli as they do not contain Pol I transcriptional machinery or rDNA (Jimenez-Garcia et al. 1989). For reassembly of the functional nucleolus, the PNBs must therefore redistribute their components back to each active NOR, forming multiple nucleoli. This dispersal process is completed within the 2 hours following telophase in HeLa cells (Muro et al. 2010). Nucleolar fusion then occurs (Savino et al. 2001) to form mature nucleoli through processes that remain largely unknown. Questions remain regarding how many NORs fuse to form one nucleolus and what properties govern the fusion process. Notably, Floutsakou et al. have paved the way to answering those questions by characterizing the DNA sequences surrounding the NORs (Floutsakou et al. 2013). This new information will allow for the development of hybridization-based approaches to specifically visualize each NOR as nucleoli are formed.
Requirements for nucleolar assembly and function
The nucleolus is remarkable in its ability to disassemble and reassemble into a functioning entity after each mitosis, but which components of the nucleolus are required for function? Formation of functional nucleoli requires rDNA which can be transcribed by Pol I. To begin active transcription of the rDNA by Pol I, the pre-initiation complex must be formed, requiring the association of UBF and SL1 to recruit initiation-competent Pol I (Learned et al. 1985; Learned et al. 1986). While the rDNA to be transcribed exists as repeats in eukaryotic NORs, Karpen et al. demonstrated that insertion of only one rRNA gene in the polytene chromosomes of Drosophila melanogaster caused the formation of “mini-nucleoli” which produce pre-rRNA and recruit a nucleolar antigen (Karpen et al. 1988). It should be noted, however, that the amplification process of polytenization may have increased the number of juxtaposed rRNA genes, enhancing Pol I recruitment and pre-rRNA production (Oakes et al. 2006). Therefore, the results from Karpen et al. raised questions regarding whether a single rDNA repeat in somatic mammalian cells is sufficient to induce nucleolar formation.
Recently, efforts to determine the minimal requirements for nucleolar function have focused on the formation of synthetic nucleoli. Mais et al. were able to create “pseudo-NORs” through the insertion of UBF binding sequences called Xenopus Enhancer elements (XEns), normally present in the intergenic spacers of Xenopus rDNA, into the DNA of a human fibrosarcoma cell line (Mais et al. 2005). Visually, the “pseudo-NORs” appear the same as active NORs because they form secondary constrictions which silver stain (Mais et al. 2005). Despite having the correct NOR structure, these “pseudo-NORs” lack the promoter sequence for the production of rRNA. Thus, “pseudo-NORs” are not transcriptionally active and do not form functional nucleoli which produce ribosomes (Mais et al. 2005). Therefore, the next step in the construction of synthetic nucleoli was to include Pol I transcription. Active transcription was achieved by Grob et al. who made “neo-NORs” (Grob et al. 2014). These “neo-NORs” intersperse the XEns included in “pseudo-NORs” with human rDNA promoters, mouse pre-rRNA coding sequences, and mouse transcriptional terminators (Grob et al. 2014). The “neo-NORs” were transcriptionally active, processed pre-rRNA, produced ribosomes, and coalesced into endogenous NORs in HT1080 cells to form larger nucleoli (Grob et al. 2014). The addition of rDNA transcription units allowed “neo-NORs” to form functionally compartmentalized nucleoli, while “pseudo-NORs” only formed the FC seen at the bookmarking stage of UBF binding (Grob and McStay 2014). Therefore, formation of functional nucleoli requires at least one rRNA gene, recruitment of the Pol I transcription machinery including UBF and SL1, and active transcription of the rDNA.
Variations in nucleolar number and size
There are therefore only a few absolute requirements for the formation of functional nucleoli, resulting in much variation in their number, shape, and size across different species and cell types. Nucleolar size/area is one parameter known to fluctuate greatly in human cells. This parameter has been examined mainly in the context of cancer. Studies using various cancer cell lines have shown that increases in nucleolar area per nucleus are directly related to increased Pol I activity of the cell as well as to increased UBF, DNA topoisomerase I, and fibrillarin expression (Derenzini et al. 1998). Additionally, depletion of proteins responsible for controlling proliferation, such as p53 and pRb, causes an increase in nucleolar area (Treré et al. 2004). This finding is logical as increased proliferation is linked to increased production of ribosomes and therefore linked to increased rRNA transcription. However, proliferation alone does not fully explain differences in nucleolar size. For example, rapidly proliferating small-cell anaplastic lung cancer cells have a small nucleolar area per nucleus, while slower growing large-cell lung carcinoma cells have a much larger nucleolar area per nucleus (Zink et al. 2004). While these abnormal, malignant cells provide elegant model systems for examining nucleolar size/area, more studies are needed to understand the mechanisms governing these parameters in normal human cells.
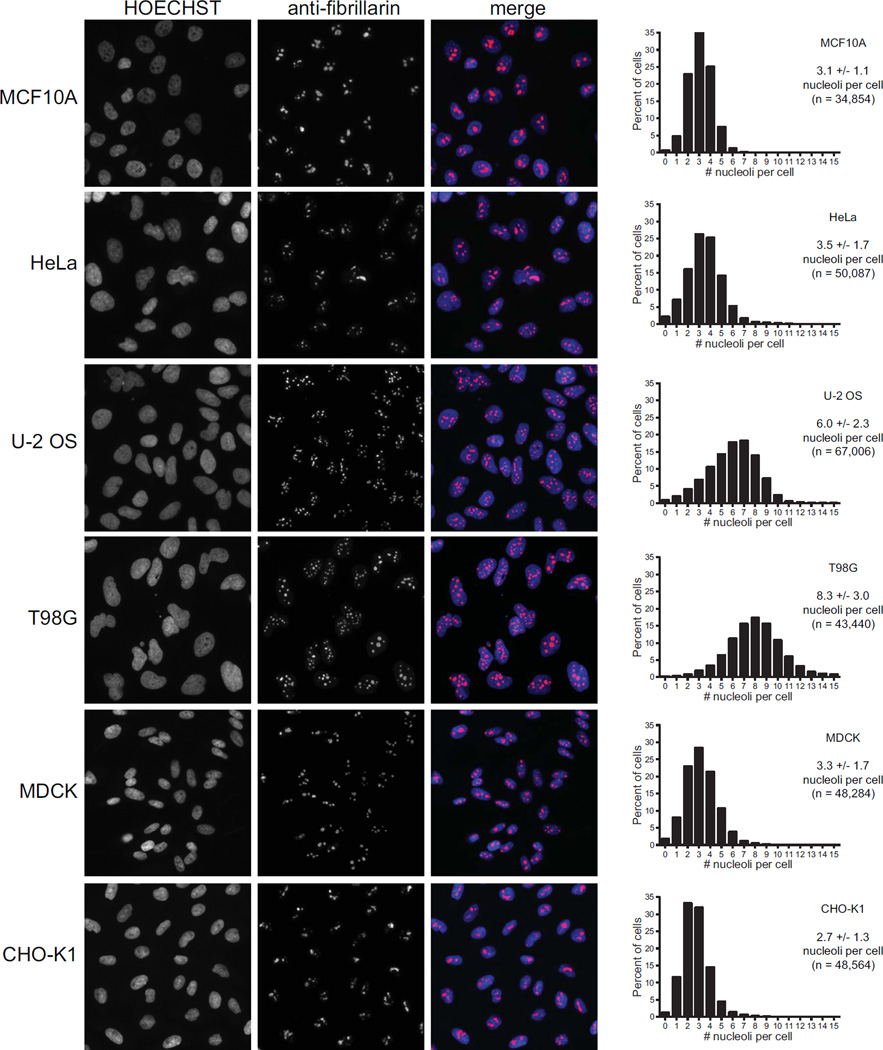
Nucleolar number also varies for unknown reasons. Because there are 10 NORs located on the acrocentric chromosomes in humans, a maximum of 10 nucleoli in human cells is possible. However, most human cells have far fewer active nucleoli, and many questions remain as to the mechanisms controlling nucleolar number. To examine nucleolar number variation in mammalian cells, we visualized nucleoli in multiple cell lines using an antibody to the nucleolar protein fibrillarin. The number of nucleoli per nucleus was then determined via a pipeline in CellProfiler (Carpenter et al. 2006). From this analysis, we found that the number of nucleoli per cell fluctuates greatly within a given population of the same cell line. Examining a frequency distribution of the number of nucleoli per cell shows a normal curve with mean and variance that differ by cell line (Fig. 3). Multiple mammalian cell lines, including HeLa human cervical carcinoma cells, MCF-10A human mammary epithelial cells, MDCK dog kidney cells, and CHO Chinese hamster ovary cells, contained an average of roughly 3 nucleoli per cell (Fig. 3). Interestingly, nucleolar number is not directly related to the ploidy of the cell type, indicating the presence of mechanisms which control for nucleolar number. For example, both MCF10A and HeLa cells have the same average number of nucleoli per nucleus, but MCF10A cells have a normal diploid karyotype while HeLa cells are aneuploid. Also, the average number of nucleoli per cell is unchanged between multiple mammalian species such as dog, hamster, and human. However, the mean number of nucleoli per nucleus is different for other cell lines. For example, U20S human bone osteosarcoma cells have an average of 6 nucleoli per nucleus, while T98G human glioblastoma multiforme cells have 8 nucleoli per nucleus (Fig. 3). Other studies have shown that approximately 91% of HT1080 human fibrosarcoma cells have greater than 3 nucleoli per cell (Krystosek 1998). In addition, as early as the 1960s, Shea and Leblond demonstrated that nucleolar number in different mouse tissue sections ranges between one and six nucleoli per cell with an average of 2–3 nucleoli per cell (Shea and Leblond 1966). As with nucleolar size/area, information regarding nucleolar number in normal, non-transformed mammalian tissues is lacking. Nucleolar number therefore varies greatly among mammalian cells and between tissue types. In order to better understand the mechanisms governing nucleolar number determination, a comprehensive account of the average nucleolar number for all tissues and cell lines is needed.
Fig. 3.
Nucleolar number varies greatly among tissue culture cells. The indicated cell lines were fixed with paraformaldehyde and stained with an antibody to the nucleolar protein fibrillarin (72B9; Reimer et al. 1987). Cells were also stained with HOECHST for visualization of the nucleus. Images were analyzed using a CellProfiler pipeline which counts the number of nucleoli per cell (Carpenter et al. 2006). Representative images for each cell line are depicted with a frequency distribution of the number of nucleoli per nucleolus shown to the right
While nucleolar number varies, changes in the number of nucleoli can occur through multiple mechanisms. In a human fibrosarcoma cell line treated with 8-chloro-cAMP, a protein kinase A agonist, the nucleoli of nondividing cells condensed from multiple nucleoli into one large nucleolus (Krystosek 1998). This nucleolar coalescence involves a movement of the acrocentric chromosomes from being dispersed throughout the nucleus to a single central location (Krystosek 1998). More recent findings show that there may also be a genetic factor in nucleolar number determination. Freed et al. showed that depletion of the ribosome biogenesis factors Utp4/Cirhin and NOL11 in MCF-10A cells caused a significant shift in the number of nucleoli from the average 2–3 nucleoli per cell to one nucleolus per cell (Freed et al. 2012). This is likely due to the essential functions of hUTP4/Cirhin and NOL11 in pre-rRNA transcription and/or processing. Another theory holds that the coalescence of rRNA genes is dependent on protein-protein interactions between the heterochromatin regions of the different chromosomes (Carmo-Fonseca et al. 2000). Finally, the role of transcription in maintenance of nucleolar structure cannot be overlooked. Depletion of UBF, which is necessary for Pol I transcription in vivo and may also play a role in maintaining the active chromatin state, causes a coalescence of nucleoli to form one large body in mouse embryonic fibroblasts (Hamdane et al. 2014). With the many factors that regulate nucleolar number in human cells, it is clear that nucleolar number determination is a non-stochastic process. More studies are needed to describe all of the components and molecular mechanisms involved in regulating this process.
Cancer and the nucleolus
Cancer cells present another example of variation in the structure and function of nucleoli. The importance of the nucleolus in cancer was realized as early as 1896, when it was noted that malignant cells had large, irregular nucleoli (Pianese 1896). With the advent of silver-staining, Ploton et al. examined human prostatic cancer cells and discovered that the malignant cells had larger nucleoli with more silver stained dots than benign hyperplastic glands and normal lymphocytes (Ploton et al. 1986). After this study, nucleolar size was found to be an accurate prognostic indicator of clinical outcome in a number of other cancers (reviewed in Derenzini et al. 2009; Pich et al. 2000). The change in nucleolar size is thought to reflect the rate of cell proliferation (Derenzini et al. 1998). While nucleolar size does function as an accurate prognostic indicator of malignant vs benign lesions, the nucleolar size parameter is not an accurate diagnostic tool (Derenzini et al. 2009). This is because not all tumors proliferate rapidly, so many of the cells within the tumor may contain small nucleoli while the tumor is still classified as malignant (Derenzini et al. 1998). Nucleolar alterations in cancer have therefore long been observed, but questions remain as to the role of the nucleolus in cancer.
Malignant cells must upregulate rRNA synthesis and the production of ribosomes in order to proliferate. Therefore, the nucleolus has historically been a desirable target for cancer therapeutics. Multiple drugs have been approved which affect the essential nucleolar process of Pol I transcription (reviewed in Drygin et al. 2010; Hannan et al. 2013). Current chemotherapies target rRNA transcription as well as early and late rRNA processing (Burger et al. 2010). However, low selectivity of the approved therapeutics makes it difficult to determine whether their therapeutic effect is due solely to transcription inhibition of Pol I. Enhancing selectivity could improve efficacy and decrease toxicity. Current therapeutics therefore hope to improve selectivity to target only Pol I transcription in malignant cells. For example, a Phase I clinical trial is underway in patients with advanced hematological malignancies for the selective Pol I inhibitor, CX-5461. Preliminary studies have shown that CX-5461 inhibits PIC formation by preventing the binding of SL1 to the rDNA (Drygin et al. 2011). Treatment with CX-5461 induces the p53 stress response pathway and causes apoptosis in wild-type p53 human lymphoma and leukemia cell lines (Bywater et al. 2012). A second new and promising compound is BMH-21. BMH-21 is a small molecule which binds to GC rich portions of the rDNA, resulting in reduced rRNA transcription and the degradation of the large catalytic subunit of Pol I, RPA194 (Peltonen et al. 2014). These new compounds with increased specificity may usher in a new era in Pol I-targeted anticancer therapeutics.
Previously, changes in nucleolar structure were thought of as a byproduct of cell transformation, but could changes in the structure and function of the nucleolus drive transformation as well? Recent insights suggest that changes in proteins which affect nucleolar size/number and function can drive cancer. For example, increased ribosome biogenesis caused by depletion of the cell-cycle control protein ADP ribosylation factor like 2 (Arl-2) also showed increases in nucleolar number, nucleolar area, and aggressivity of the tumor (Belin et al. 2009). In addition, bystin-like (BYSL), a protein involved in pre-18S rRNA processing, may play a role in driving tumor formation as its inhibition has been shown to prevent tumor formation in nude mice (Wang et al. 2009). Another way of examining how the nucleolus drives cancer is through ribosomopathies. These disorders, caused by altered ribosome biogenesis and function, often impair development of certain tissues. In addition, an increased susceptibility to cancer exists in patients with many ribosomopathies, such as Diamond Blackfan anemia (Vlachos et al. 2012). The precise mechanisms describing how the proteins mutated in ribosomopathies, which cause dysregulations in nucleolar function, cause cancer remain to be defined. It is also possible, that defects in these ribosome biogenesis proteins drive cancer in an indirect manner. For example, the ribosome biogenesis proteins involved may have extraribosomal functions which contribute to cancer development, the overall decrease in available ribosomes could alter translation of genes involved in transformation, or byproducts of ribosome biogenesis defects may cause transformation (Montanaro et al. 2008). Regardless, more information must be provided to elucidate the crucial role of the nucleolus in cancer.
Conclusions and perspectives
The nucleolus is a highly dynamic organelle with a complex structure which is intricately related to its primary function of ribosome biogenesis. The variations in number, size, and shape of nucleoli demonstrate the complexity of processes governing nucleolar development. Therefore, further studies are needed to elucidate the molecular mechanisms which guide the nucleolar structure/function relationship. Recently, Neumϋller et al. conducted a study to uncover genetic determinants of nucleolar size in both Drosophila melanogaster and Saccharomyces cerevisiae (Neumuller et al. 2013). While the authors found a number of interesting candidates, no studies have yet been conducted to identify proteins which regulate nucleolar size in human cells. In addition, determinants of nucleolar number have not been examined. Identification of the proteins involved in regulating nucleolar size and number would be the first step in elucidating the non-stochastic molecular mechanisms used by human cells to control nucleolar functions. Additionally, understanding such mechanisms could shed light on the driving role of the nucleolus in cancer progression, leading to the production of new selective therapeutics.
Acknowledgements
We thank Kathleen L. McCann for her critical reading of this manuscript.
Funding: This work was supported by the National Institutes of Health (CMB TG T32GM007223-40 to Katherine Farley and NIH R01 GM52581 and a pilot grant from the Yale Cancer Center to Susan Baserga).
Footnotes
Compliance with Ethical Standards
Ethical Approval: This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of Interest: Katherine I. Farley declares that she has no conflict of interest. Yulia Surovtseva declares that she has no conflict of interest. Janie Merkel declares that she has no conflict of interest. Susan J. Baserga declares that she has no conflict of interest.
References
- Azum-Gelade MC, Noaillac-Depeyre J, Caizergues-Ferrer M, Gas N. Cell cycle redistribution of U3 snRNA and fibrillarin. Presence in the cytoplasmic nucleolus remnant and in the prenucleolar bodies at telophase. Journal of cell science. 1994;107(Pt 2):463–475. doi: 10.1242/jcs.107.2.463. [DOI] [PubMed] [Google Scholar]
- Belin S, et al. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PloS one. 2009;4:e7147. doi: 10.1371/journal.pone.0007147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proceedings of the National Academy of Sciences. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger K, et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem. 2010;285:12416–12425. doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. doi:S1535-6108(12)00216-4 [pii] 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nature cell biology. 2000;2:E107–E112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, et al. Three-dimensional organization of active rRNA genes within the nucleolus. Journal of cell science. 2002;115:3297–3307. doi: 10.1242/jcs.115.16.3297. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Montanaro L, Trere D. What the nucleolus says to a tumour pathologist. Histopathology. 2009;54:753–762. doi: 10.1111/j.1365-2559.2008.03168.x. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs RL. Nucleolar function and size in cancer cells. The American journal of pathology. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- Drygin D, et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer research. 2011;71:1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annual review of pharmacology and toxicology. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- Floutsakou I, Agrawal S, Nguyen TT, Seoighe C, Ganley AR, McStay B. The shared genomic architecture of human nucleolar organizer regions. Genome research. 2013;23:2003–2012. doi: 10.1101/gr.157941.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana F, Nyon JL. Traité sur le vénin de la vipere sur les poisons americains sur le laurier-cerise et sur quelques autres poisons végetaux: on y a joint des observations sur la structure primitive du corps animal : Différentes expériences sur la reproduction des nerfs et la description d'un nouveau canal de l'oeil. chez Nyon l'Ainé. 1781 [Google Scholar]
- Freed EF, Prieto JL, McCann KL, McStay B, Baserga SJ. NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS genetics. 2012;8:e1002892. doi: 10.1371/journal.pgen.1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier T, Robert-Nicoud M, Guilly MN, Hernandez-Verdun D. Relocation of nucleolar proteins around chromosomes at mitosis. A study by confocal laser scanning microscopy. Journal of cell science. 1992;102:729–737. doi: 10.1242/jcs.102.4.729. [DOI] [PubMed] [Google Scholar]
- Gebrane-Younes J, Fomproix N, Hernandez-Verdun D. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. Journal of cell science. 1997;110:2429–2440. doi: 10.1242/jcs.110.19.2429. [DOI] [PubMed] [Google Scholar]
- Golomb L, Volarevic S, Oren M. p53 and ribosome biogenesis stress: the essentials. FEBS letters. 2014;588:2571–2579. doi: 10.1016/j.febslet.2014.04.014. [DOI] [PubMed] [Google Scholar]
- Goodpasture C, Bloom SE. Visualization of nucleolar organizer regions im mammalian chromosomes using silver staining. Chromosoma. 1975;53:37–50. doi: 10.1007/BF00329389. [DOI] [PubMed] [Google Scholar]
- Grob A, Colleran C, McStay B. Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes & development. 2014;28:220–230. doi: 10.1101/gad.234591.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob A, McStay B. Construction of synthetic nucleoli and what it tells us about propagation of sub-nuclear domains through cell division. Cell cycle. 2014;13:2501–2508. doi: 10.4161/15384101.2014.949124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C, Santoro R. Formation of nuclear heterochromatin: the nucleolar point of view. Epigenetics. 2012;7:811–814. doi: 10.4161/epi.21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Hayman DL, Schmid M. Quantitative determination of rDNA transcription units in vertebrate cells. Experimental cell research. 1991;193:78–86. doi: 10.1016/0014-4827(91)90540-b. doi: http://dx.doi.org/10.1016/0014-4827(91)90540-B. [DOI] [PubMed] [Google Scholar]
- Hamdane N, et al. Conditional inactivation of upstream binding factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body. PLoS genetics. 2014;10:e1004505. doi: 10.1371/journal.pgen.1004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger KE, Cordero JA, Gall JG. Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Molecular biology of the cell. 2005;16:202–211. doi: 10.1091/mbc.E04-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan RD, Drygin D, Pearson RB. Targeting RNA polymerase I transcription and the nucleolus for cancer therapy. Expert Opinion on Therapeutic Targets. 2013;17:873–878. doi: 10.1517/14728222.2013.818658. [DOI] [PubMed] [Google Scholar]
- Heix J, Vente A, Voit R, Budde A, Michaelidis TM, Grummt I. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. The EMBO journal. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, Atwood KC. Location of ribosomal DNA in the human chromosome complement. Proceedings of the National Academy of Sciences of the United States of America. 1972;69:3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cellular and molecular life sciences : CMLS. 2008 doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D. Behavior of the Nucleolus During Mitosis. In: Olson MOJ, editor. The Nucleolus. Intelligence Unit. Landes Bioscience. 2004. [Google Scholar]
- Jimenez-Garcia LF, Rothblum LI, Busch H, Ochs RL. Nucleologenesis: use of non-isotopic in situ hybridization and immunocytochemistry to compare the localization of rDNA and nucleolar proteins during mitosis. Biology of the cell / under the auspices of the European Cell Biology Organization. 1989;65:239–246. doi: 10.1111/j.1768-322x.1989.tb00795.x. [DOI] [PubMed] [Google Scholar]
- Jimenez-Garcia LF, Segura-Valdez ML, Ochs RL, Rothblum LI, Hannan R, Spector DL. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Molecular biology of the cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Schaefer JE, Laird CD. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes & development. 1988;2:1745–1763. doi: 10.1101/gad.2.12b.1745. [DOI] [PubMed] [Google Scholar]
- Koberna K, et al. Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of "Christmas trees" in situ. The Journal of cell biology. 2002;157:743–748. doi: 10.1083/jcb.200202007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystosek A. Repositioning of human interphase chromosomes by nucleolar dynamics in the reverse transformation of HT1080 fibrosarcoma cells. Experimental cell research. 1998;241:202–209. doi: 10.1006/excr.1998.4046. [DOI] [PubMed] [Google Scholar]
- Learned RM, Cordes S, Tjian R. Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Molecular and cellular biology. 1985;5:1358–1369. doi: 10.1128/mcb.5.6.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned RM, Learned TK, Haltiner MM, Tjian RT. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell. 1986;45:847–857. doi: 10.1016/0092-8674(86)90559-3. [DOI] [PubMed] [Google Scholar]
- Mais C, Wright JE, Prieto JL, Raggett SL, McStay B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes & development. 2005;19:50–64. doi: 10.1101/gad.310705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The relation of a particular chromosomal element to the development of the nucleoli in Zea mays. ZZellforsch. 1934;21:294–326. [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annual review of cell and developmental biology. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Mochida S, Ikeo S, Gannon J, Hunt T. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. The EMBO journal. 2009;28:2777–2785. doi: 10.1038/emboj.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro L, Trere D, Derenzini M. Nucleolus, ribosomes, and cancer. The American journal of pathology. 2008;173:301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro E, Gebrane-Younis J, Jobart-Malfait A, Louvet E, Roussel P, Hernandez-Verdun D. The traffic of proteins between nucleolar organizer regions and prenucleolar bodies governs the assembly of the nucleolus at exit of mitosis. Nucleus. 2010;1:202–211. doi: 10.4161/nucl.1.2.11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, et al. Conserved regulators of nucleolar size revealed by global phenotypic analyses. Science signaling. 2013;6:ra70. doi: 10.1126/scisignal.2004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donohue MF, Choesmel V, Faubladier M, Fichant G, Gleizes PE. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. The Journal of cell biology. 2010;190:853–866. doi: 10.1083/jcb.201005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes ML, Johzuka K, Vu L, Eliason K, Nomura M. Expression of rRNA Genes and Nucleolus Formation at Ectopic Chromosomal Sites in the Yeast Saccharomyces cerevisiae. Molecular and cellular biology. 2006;26:6223–6238. doi: 10.1128/MCB.02324-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MO, Dundr M. The moving parts of the nucleolus. Histochemistry and cell biology. 2005;123:203–216. doi: 10.1007/s00418-005-0754-9. [DOI] [PubMed] [Google Scholar]
- Pederson T, Tsai RYL. In search of nonribosomal nucleolar protein function and regulation. The Journal of cell biology. 2009;184:771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen K, et al. A Targeting Modality for Destruction of RNA Polymerase I that Possesses Anticancer Activity. Cancer cell. 2014;25:77–90. doi: 10.1016/j.ccr.2013.12.009. doi: http://dx.doi.org/10.1016/j.ccr.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. doi: http://www.nature.com/nature/journal/v404/n6778/suppinfo/404604a0_S1.html. [DOI] [PubMed] [Google Scholar]
- Pianese G. Beitrag zur Histologie und Aetiologie der Carcinoma. Histologische und experimentelle Untersuchungen. Beitr Pathol Anat Allg Pathol. 1896;142:1–193. [Google Scholar]
- Pich A, Chiusa L, Margaria E. Prognostic relevance of AgNORs in tumor pathology. Micron. 2000;31:133–141. doi: 10.1016/s0968-4328(99)00070-0. [DOI] [PubMed] [Google Scholar]
- Ploton D, Menager M, Jeannesson P, Himber G, Pigeon F, Adnet JJ. Improvement in the staining and in the visualization of the argyrophilic proteins of the nucleolar organizer region at the optical level. The Histochemical journal. 1986;18:5–14. doi: 10.1007/BF01676192. [DOI] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Current opinion in cell biology. 2006;18:325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Reimer G, Pollard KM, Penning CA, Ochs RL, Lischwe MA, Busch H, Tan EM. Monoclonal autoantibody from a (New Zealand black x New Zealand white)F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis and rheumatism. 1987;30:793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- Roussel P, Andre C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. The Journal of cell biology. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino TM, Gébrane-Younès J, De Mey J, Sibarita J-B, Hernandez-Verdun D. Nucleolar Assembly of the Rrna Processing Machinery in Living Cells. The Journal of cell biology. 2001;153:1097–1110. doi: 10.1083/jcb.153.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Weisenberger D. The nucleolus. Current opinion in cell biology. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Shea JR, Leblond CP. Number of nucleoli in various cell types of the mouse. Journal of Morphology. 1966;119:425–433. doi: 10.1002/jmor.1051190404. [DOI] [PubMed] [Google Scholar]
- Sirri V, Hernandez-Verdun D, Roussel P. Cyclin-dependent kinases govern formation and maintenance of the nucleolus. The Journal of cell biology. 2002;156:969–981. doi: 10.1083/jcb.200201024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Mattijssen S, Lebaron S, Tollervey D, Pruijn GJ, Watkins NJ. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. The Journal of cell biology. 2013;200:577–588. doi: 10.1083/jcb.201207131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner AT. The nature and mechanisms of chromosome banding. Cancer Genetics and Cytogenetics. 1982;6:59–87. doi: 10.1016/0165-4608(82)90022-x. doi: http://dx.doi.org/10.1016/0165-4608(82)90022-X. [DOI] [PubMed] [Google Scholar]
- Tafforeau L, et al. The Complexity of Human Ribosome Biogenesis Revealed by Systematic Nucleolar Screening of Pre-rRNA Processing Factors. Molecular cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Thiry M, Lafontaine DL. Birth of a nucleolus: the evolution of nucleolar compartments. Trends in cell biology. 2005;15:194–199. doi: 10.1016/j.tcb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Treré D, Ceccarelli C, Montanaro L, Tosti E, Derenzini M. Nucleolar Size and Activity Are Related to pRb and p53 Status in Human Breast Cancer. Journal of Histochemistry & Cytochemistry. 2004;52:1601–1607. doi: 10.1369/jhc.4A6454.2004. [DOI] [PubMed] [Google Scholar]
- Valentin G. Repertorium für Anatomie und Physiologie kritische Darstellung fremder und Ergebnisse eigener Forschung. 1836 [Google Scholar]
- Vlachos A, Rosenberg PS, Atsidaftos E, Alter BP, Lipton JM. Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry vol 119. 2012;16 doi: 10.1182/blood-2011-08-375972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. Einige Bemerkungen und Fragen über das Keimbläschen (vesicular germinativa) Muller's Arch Anat Physiol U Wiss Med. 1835;268:373–377. [Google Scholar]
- Wang H, et al. Bystin-like protein is upregulated in hepatocellular carcinoma and required for nucleologenesis in cancer cell proliferation. Cell Res. 2009;19:1150–1164. doi: 10.1038/cr.2009.99. [DOI] [PubMed] [Google Scholar]
- Wang M, Anikin L, Pestov DG. Two orthogonal cleavages separate subunit RNAs in mouse ribosome biogenesis. Nucleic acids research. 2014;42:11180–11191. doi: 10.1093/nar/gku787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Molecular cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild T, et al. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS biology. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL, Jr, Baserga SJ. Ribosome Biogenesis in the Yeast Saccaromyces cerevisiae. Genetics. 2013;195:1–39. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, et al. PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nature cell biology. 2009;11:644–651. doi: 10.1038/ncb1871. doi: http://www.nature.com/ncb/journal/v11/n5/suppinfo/ncb1871_S1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Gall JG. "Micronucleoli" in the Xenopus germinal vesicle. Chromosoma. 1997;105:438–443. doi: 10.1007/BF02510480. [DOI] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]