Abstract
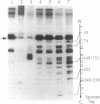
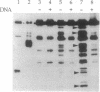
A two-step lysine-modification procedure has been devised to chemically footprint protein surfaces involved in macromolecular interactions. A protein tagged at one particular end, in the free state or in a complex, is first treated lightly with a reversible lysine-modifying reagent. The protein is then unfolded and treated extensively with an irreversible lysine reagent to block those lysines that did not react previously; next, the first lysine modification is reversed, and a lysine-specific endoproteinase is used to cleave the tagged polypeptide at the deblocked lysines. Separation of the proteolytic products by size and identification of the tagged fragments map the positions of these lysines. In this procedure, the reversible lysine reagent serves as the chemical footprinting agent, as cleavage of the polypeptide ensues only at the sites of reaction with this reagent. Lysines involved in macromolecular contacts are identified from differences in proteolytic patterns of the tagged protein when the first lysine modification is done with the protein in the free form and in a complex. Application of the method to vaccinia virus topoisomerase identifies a number of lysines that are involved in its binding to DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker M. M., Wang J. C. Use of light for footprinting DNA in vivo. Nature. 1984 Jun 21;309(5970):682–687. doi: 10.1038/309682a0. [DOI] [PubMed] [Google Scholar]
- Becker P. B., Ruppert S., Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987 Nov 6;51(3):435–443. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- Biemann K. Mass spectrometry of peptides and proteins. Annu Rev Biochem. 1992;61:977–1010. doi: 10.1146/annurev.bi.61.070192.004553. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson G. R., Schaffer M. H., Stark G. R., Vanaman T. C. Specific chemical cleavage in high yield at the amino peptide bonds of cysteine and cystine residues. J Biol Chem. 1973 Oct 10;248(19):6583–6591. [PubMed] [Google Scholar]
- Jay D. G. A general procedure for the end labeling of proteins and positioning of amino acids in the sequence. J Biol Chem. 1984 Dec 25;259(24):15572–15578. [PubMed] [Google Scholar]
- Johnson A., Meyer B. J., Ptashne M. Mechanism of action of the cro protein of bacteriophage lambda. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1783–1787. doi: 10.1073/pnas.75.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juillerat M., Parr G. R., Taniuchi H. A biologically active, three-fragment complex of horse heart cytochrome c. J Biol Chem. 1980 Feb 10;255(3):845–853. [PubMed] [Google Scholar]
- Kolodziej P. A., Young R. A. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- Lindsay D. G., Shall S. The acetylation of insulin. Biochem J. 1971 Mar;121(5):737–745. doi: 10.1042/bj1210737a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley J. E., Wang J. C. Proteolysis patterns of epitopically labeled yeast DNA topoisomerase II suggest an allosteric transition in the enzyme induced by ATP binding. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10485–10489. doi: 10.1073/pnas.88.23.10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn R. M., Bjornsti M. A., Caron P. R., Wang J. C. Peptide sequencing and site-directed mutagenesis identify tyrosine-727 as the active site tyrosine of Saccharomyces cerevisiae DNA topoisomerase I. Proc Natl Acad Sci U S A. 1989 May;86(10):3559–3563. doi: 10.1073/pnas.86.10.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morham S. G., Shuman S. Covalent and noncovalent DNA binding by mutants of vaccinia DNA topoisomerase I. J Biol Chem. 1992 Aug 5;267(22):15984–15992. [PubMed] [Google Scholar]
- Shaffer R., Traktman P. Vaccinia virus encapsidates a novel topoisomerase with the properties of a eucaryotic type I enzyme. J Biol Chem. 1987 Jul 5;262(19):9309–9315. [PubMed] [Google Scholar]
- Sharma A., Hanai R., Mondragón A. Crystal structure of the amino-terminal fragment of vaccinia virus DNA topoisomerase I at 1.6 A resolution. Structure. 1994 Aug 15;2(8):767–777. doi: 10.1016/s0969-2126(94)00077-8. [DOI] [PubMed] [Google Scholar]
- Shuman S., Golder M., Moss B. Characterization of vaccinia virus DNA topoisomerase I expressed in Escherichia coli. J Biol Chem. 1988 Nov 5;263(31):16401–16407. [PubMed] [Google Scholar]
- Shuman S., Kane E. M., Morham S. G. Mapping the active-site tyrosine of vaccinia virus DNA topoisomerase I. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9793–9797. doi: 10.1073/pnas.86.24.9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S., Moss B. Identification of a vaccinia virus gene encoding a type I DNA topoisomerase. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7478–7482. doi: 10.1073/pnas.84.21.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S., Prescott J. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J Biol Chem. 1990 Oct 15;265(29):17826–17836. [PubMed] [Google Scholar]
- Shuman S. Site-specific DNA cleavage by vaccinia virus DNA topoisomerase I. Role of nucleotide sequence and DNA secondary structure. J Biol Chem. 1991 Jan 25;266(3):1796–1803. [PubMed] [Google Scholar]
- Shuman S. Site-specific interaction of vaccinia virus topoisomerase I with duplex DNA. Minimal DNA substrate for strand cleavage in vitro. J Biol Chem. 1991 Jun 15;266(17):11372–11379. [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tullius T. D. Physical studies of protein-DNA complexes by footprinting. Annu Rev Biophys Biophys Chem. 1989;18:213–237. doi: 10.1146/annurev.bb.18.060189.001241. [DOI] [PubMed] [Google Scholar]
- Wu L. C., Laub P. B., Elöve G. A., Carey J., Roder H. A noncovalent peptide complex as a model for an early folding intermediate of cytochrome c. Biochemistry. 1993 Sep 28;32(38):10271–10276. doi: 10.1021/bi00089a050. [DOI] [PubMed] [Google Scholar]
- Zinn K., Maniatis T. Detection of factors that interact with the human beta-interferon regulatory region in vivo by DNAase I footprinting. Cell. 1986 May 23;45(4):611–618. doi: 10.1016/0092-8674(86)90293-x. [DOI] [PubMed] [Google Scholar]
- de Boer H. A., Comstock L. J., Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]