Abstract
Heterotopic ossification (HO) associated with traumatic neurological or musculoskeletal injuries remains a major clinical challenge. One approach to understanding better and potentially treating this condition is to silence one or more genes believed to be responsible for osteogenesis by small interfering RNA (siRNA) post-injury. Improved methods of delivering siRNA to myoprogenitor cells as well as relevant cell culture models of HO are needed to advance this approach. We utilize a model of HO featuring C2C12 myoprogenitor cells stimulated to the osteogenic phenotype by addition of BMP-2. For siRNA delivery, we utilize a nanocomposite consisting of DOTAP- based cationic liposomes coated with a graft copolymer of poly(propylacrylic acid) grafted with polyetheramine (Jeffamine), as this system has been shown previously to deliver antisense oligonucleotides safely into cells and out of endosomes for gene silencing in vitro and in vivo. Delivery of siRNA targeting Runx2, a transcription factor downstream of BMP-2, to stimulated C2C12 cells produced greater than 60% down-regulation of the Runx2 gene. This level of gene silencing was sufficient to inhibit alkaline phosphatase activity over the course of several days and calcium phosphate deposition over the course of 2 weeks. These results show the utility of the BMP-2/C2C12 model for capturing the cellular cell-fate decision in HO. Further, they suggest DOTAP/PPAA-g-Jeffamine as a promising delivery system for siRNA– based therapy for HO.
Introduction
Heterotopic ossification (HO), which was first identified by Dejerne and Ceillier in 1918 in the patients of World War II, describes the progression of unwanted bone formation in the soft tissues following injury or trauma.1 HO can arise following severe musculoskeletal trauma2 or injuries to the spinal cord or central nervous system3 that stimulate multipotent progenitor cells to start growing bone. Differentiation and proliferation of these cells in the soft tissues of the body cause the calcified bone formation that defines HO. During the acute phase of HO, the condition may become painful, may cause internal injuries to the surrounding tissues and eventually may restrict the patient's movement. Treatments for HO include surgical excision of the calcified soft tissue, radiation therapy, and chemical treatment with etidronate, which is intended to block calcification without inhibiting bone matrix formation.4 As none of these treatments provides satisfactory outcomes, there is considerable interest in understanding better the pathophysiology of HO and developing improved treatments for it.
While the molecular pathways responsible for ossification in soft tissues remains unclear, several studies have implicated various growth factors, including bone morphogenetic proteins (BMPs), transforming growth factor-beta (TGF-β), basic fibroblast growth factor (bFGF), insulin-like growth factor (IGF) and others.5 One study indicated that depending upon the type of injury, the cells in the CNS release several bone forming mediators that activate the expression of BMP2/4 at the trauma site.6 Furthermore, the local presence of BMP elicits early neurogenic inflammation, which might induce the migration or release of pre-osteoblastic cells or other multipotent cells that trigger HO from the nervous system.7 Given the prominent role of BMPs in osteogenesis, its downstream mediator, runt-related transcription factor 2 (Runx2)/CBFA1, has emerged as an important player in the differentiation of osteoblast. 8
RNA interference (RNAi) is an attractive approach to the study of heterotopic ossification, as it can both be used as a tool to dissect the molecular mechanisms governing the syndrome and also as a therapeutic modality. Silencing of Runx2 has been achieved in vivo using adenovirus vectors and plasmid-encoded short interfering RNA (siRNA).9 However, the poor safety record of viral delivery and inefficiency of naked plasmid delivery motivate the development of safe and efficient non-viral approaches to exploit the full potential of RNA interference technique.
We have developed a multifunctional, non-viral poly(nucleic acid) delivery system that is designed to overcome delivery barriers at the systemic, cellular and intracellular levels. Its core consists of the cationic lipid N-[1-(2, 3-Dioleoyloxy) propyl]-N, N, N-trimethylammonium methyl-sulfate (DOTAP), which electrostatically binds and protects the poly(nucleic acid). Onto this lipoplex between DOTAP and the poly(nucleic acid) is adsorbed an anionic polymer based on the endosomolytic polyelectrolyte poly(propylacylic acid) (PPAA).10 Optionally grafted onto the PPAA are poly(alklyene oxide) chains, which can be either hydrophilic poly(ethylene oxide) (PEO) or amphiphilic poly(oxyalkylene amines) (JeffamineTM), thus allowing controlled modification of the hydrophilic/lipophilic balance of the resulting ternary liposome/poly(nucleic acid)/graft copolymer complex. We have recently utilized this system to deliver antisense oligonucleotides (AONs) and silence genes efficiently in several cell lines10c, d as well as in vivo (Peddada et al., in preparation). In the present work, we have employed the modified delivery vector to introduce siRNA, targeting Runx2 mRNA, in a myoprogenitor (C2C12) cell-based pathological in vitro model of HO. The effect of Runx2 silencing on the osteogenic differentiation of C2C12 cells grown in the pathophysiological environment was investigated.
Materials and Methods
Materials
Azobisisobutyronitrile (AIBN), diethyl ether, dimethylformamide (DMF), methanol, and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) were purchased from Aldrich Chemical company (Milwaukee, WI). 2-propylacrylic acid was purchased from Polymer Source (Quebec, Canada). 1-hydroxybenzotriazole hydrate was purchased from AK scientific (Union City, CA). JeffamineTM M-2070 was donated by Huntsman (Chocolate Bayou, TX). All chemicals were used as received.
Synthesis and Characterization of Poly(propylacrylic acid)
Poly (propylacrylic acid) (PPAA) was prepared by standard bulk free radical polymerization of 2-propylacrylic acid monomer under nitrogen using AIBN as the catalyst/initiator 11. Briefly, 16.7 mg (1.02 x 10−1 mmole) of AIBN was dissolved in 5.99g (52.5 mmole) 2-propylacrylic acid in a 30mL scintillation vial. The mixture was purged with nitrogen, placed in the Polyblock-4 reaction block (HEL INC, Lawrenceville, NJ), and stirred at 250 rpm at 30°C for 1 hour to ensure thorough mixing of initiator and monomer. The temperature of the reaction mixture was then increased to 61°C for 50 hours, maintaining a stir rate of 250 rpm. The resulting PPAA polymer in the scintillation vial was then dissolved in 6 mL of methanol and precipitated in 300 mL of diethyl ether. The precipitation process was performed twice to remove any unreacted monomer and initiator. Polymer was then filtered with a Buchner funnel with fritted disk and was further dried under vacuum to remove solvent. Poly (propylacrylic acid) was characterized via gel permeation chromatography (GPC) (Malvern Viscotek, Houston, TX) and proton nuclear magnetic resonance (1H NMR) (500 MHz-Varian Inova, Santa Clara, CA). 1H NMR (DMSO-d6) δ 0.80 (s, CH3), 1.0–2.1 (br, CH2), 12.0 (s, COOH). The weight-average molecular weight, Mw, and number-average molecular weight, Mn, of the PPAA polymer backbone were determined to be 71 kDa and 27 kDa, respectively.
Synthesis and Characterization of Poly(propylacrylic acid)-graft-Jeffamine
The graft copolymers were prepared by carbodiimide coupling of Jeffamine M-2070 pendent chains to the acrylate backbone chain (PPAA) as previously described.10d Briefly, for the 1% theoretical Jeffamine graft density copolymer, 100 mg of PPAA (1.41 x 10−3 mmole), 17.5 mg of Jeffamine M-2070 (8.76 x 10−3 mmole), 11.8 mg of HOBt (8.76 x 10−2 mmole) and 2.5 mL of DMF were mixed in a 4 mL glass vial until the solids were completely dissolved. Then 16.8 mg of EDC (8.76 x 10−2 mmole) were added, and the reaction vial was placed in the Polyblock-4 reaction block and stirred at 250 rpm at room temperature for 50 hours. This procedure was repeated for the 10% theoretical grafting by increasing the amounts of Jeffamine and HOBt accordingly. The graft copolymers were purified by equilibrium dialysis using a 12–14 kDa MWCO regenerated cellulose dialysis tube (Cellu-Sep, Seguin, TX). The dialysis tube containing 10 mL of the reaction mixture was placed in 500 mL of DMF, and the external dialysate solution was exchanged at 48 hr with fresh DMF. After 96 hr dialysis with DMF, the dialysis was repeated for another 96 hr against methanol. The contents of the dialysis bag were mixed with 10 mL of deionized water. The methanol was allowed to evaporate overnight and the resultant graft copolymer mixture in water was lyophilized. 1H NMR analysis was used to determine the experimental graft densities by the ratio of the areas of the CH2CH2 peak of Jeffamine and the CH2 peak of the backbone chain, and the molecular weights were determined by GPC. 1H NMR (DMSO-d6) δ 0.85 (s, CH3 of PPAA), 1.0–2.1 (br, CH2 of PPAA), 1.1 (s, CH3 of PPO), 3.20 (s, OCH3), 3.20–3.45 (m, CHCH2 of PPO), 3.5 (s, OCH2CH2O), 7.6 (s, C (O)NH), 12.0 (s, COOH). The Mw/Mn of PPAA-g-1%Jeffamine and PPAA-g-10%Jeffamine were 67 kDa / 28 kDa and 109 kDa / 52 kDa, respectively. The experimental graft densities based upon 1H NMR analyses were 0.1% and 5.9% for the theoretical PPAA-g-1%Jeffamine and PPAA-g- 10%Jeffamine, respectively.
Cell Culture
Mouse myoprogenitor cells (C2C12; ATCC: CRL-1772™) were cultured in Dulbecco's Modified Eagle's Medium (DMEM; ATCC: 30-2002) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA), 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA) at 37°C in a humidified atmosphere of 5% CO2 in air. For maintenance and passaging, cells were detached from tissue culture surface using trypsin-EDTA (Invitrogen, Carlsbad, CA) at 60–70% confluence and re-cultured at 1:6 split ratio. The cells were cultured at 4000/cm2 seeding density in standard tissue culture flasks (BD Falcon™, Franklin Lakes, NJ).
Screening siRNA Candidates
Small interfering RNA (siRNA) sequences targeting mouse Runx2 were designed on the basis of thermodynamic considerations, using the RNAstructure algorithms 12, followed by heuristic pruning to restrict selection to sequences containing no more than three of the same nucleotide in succession and to those containing all four nucleotide bases. The shortlisted siRNA sequences shown in Table 1 were custom synthesized and purchased from Dharmacon (Chicago, IL). Nonspecific control siRNA labeled with fluorescent Cy5 dye at the 3’ end (AllStars Negative Control siRNA) was purchased from Qiagen Sciences (Germantown, MD). The control siRNA was validated by the manufacturer to have no homology to any known mammalian gene.
Table 1.
Candidate siRNAs based on RNAStructure Analysis
| siRNA | Sequence | |
|---|---|---|
| S-1 | Sense | 5’-CAGACAAGUGAAGAGGUUUU-3’ |
| Antisense | 5’-AACCUCUUCACUUGUCUGUU-3’ | |
| S-2 | Sense | 5’-GCAUAAAGGGAGAAGCAGUU-3’ |
| Antisense | 5’-CUGCUUCUCCCUUUAUGCUU-3’ | |
| S-3 | Sense | 5’-AUUGAAGAAGAAGCACACUU-3’ |
| Antisense | 5’-GUGUGCUUCUUCUUCAAUUU-3’ |
C2C12 cells, seeded at 4000 cells/cm2 in 12-well plates, were transfected with each of the three candidate Runx2 specific siRNAs or scrambled siRNA in Opti-MEM reduced serum media (Invitrogen, Carlsbad, CA) at a final siRNA concentration of 160 nM using Lipofectamine 2000 (Invitrogen) for 4 hours. Cells were cultured in 100 ng/ml human recombinant bone morphogenic-2 protein (hrBMP2) supplemented complete growth media after transfection to induce osteoblastic differentiation, or they were cultured in normal growth media post transfection in case of control. Three siRNAs were tested against the target gene Runx2, and the siRNA with the most potent and specific gene silencing effect was selected for further studies. The cells were assayed for the activity of alkaline phosphatase enzyme on day 3 post-BMP-2 treatment.
SiRNA Treatment of C2C12 Cells with Nanocomplexes
Appropriate volumes of the cationic liposome DOTAP (Roche Applied Science, Indianapolis, IN) and siRNA to achieve a weight ratio of 10:1 (charge ratio (+/−) of 3.8) were first diluted in HBS and PBS buffers respectively as per the manufacturer’s guidelines and then complexed by vortex mixing, followed by incubation for 30 minutes at room temperature. The anionic polyelectrolyte (PPAA, PPAA-g-1%Jeffamine or PPAA-g-10%Jeffamine) was added to the DOTAP-siRNA complex to produce a theoretical net charge ratio (+/−) of 1.0, which we have found to be optimal for antisense oligonucleotide delivery10d, and the ternary complexes were incubated for an additional 30 minutes at room temperature. Theoretical charge ratios on nano-complexes were calculated by assuming 100% ionization for DOTAP and siRNA and 33% protonation of carboxylic acids in PPAA at physiological pH.10b Lipofectamine 2000, used as the control delivery agent, was complexed with siRNA at the optimal weight ratio of 2:1 per manufacturer’s instructions.
For transfection, the C2C12 cells were cultured in 12 well plates, and siRNA was used at a concentration of 160 nM siRNA. Twenty four hours post-seeding, the conditioned media was removed, and the cells were washed with the fresh media. Next the cells were incubated with the DOTAP/siRNA/polyelectrolye complexes in the growth media containing 10% FBS for 4 hours. Subsequently, the transfection mixture was replaced by growth media supplemented with 100 ng/ml BMP-2. The cells were grown for 3–5 days in BMP2 supplemented media, which was changed every 48 hours.
Physical characterization of nanocomplexes
The complexes were self-assembled in PBS as described above. Hydrodynamic sizes and zeta potentials of the complexes were measured immediately after the formation of the DOTAP/siRNA/polyelectrolyte complexes using a Malvern Instruments Zetasizer Nano ZS-90 instrument (Southboro, MA). Dynamic light scattering (DLS) measurements were performed at a 90° scattering angle at 37°C. Each recorded data value was the average of three (for hydrodynamic diameter) and ten (for zeta potential) scans per measurement.
Cellular uptake of nanocomplexes by C2C12 Cells
C2C12 cells were treated with nanocomplexes as described above, using Cy5 labeled negative control siRNA. Cells were analyzed for Cy5- siRNA uptake 24 hours following transfection using fluorescence activated cell sorting (FACS) and laser-scanning confocal microscopy. Geometric mean Cy5 fluorescence (FL4 channel) intensity for 10,000 cells was recorded on the FACS Calibur three-laser flow cytometer (BD Biosciences, San Jose, CA). CellQuest software equipped with the flow cytometer was used for the data analysis.
For confocal microscopy, cells were seeded and transfected in the Lab-tekTM Chamber SlideTM system (Nunc Brand Products, Rochester, NY) at 24 hours post seeding. After four hours of incubation, the transfection mixture was removed and the cells were fixed using 4% paraformaldihyde. DAPI (Invitrogen, Carlsbad, CA) and CellMaskTM Deep Red (Invitrogen) stains were used to stain nucleus and plasma membrane respectively as per the manufacturer’s guidelines. The cell samples were then analyzed using a Leica TCS SP2 laser-scanning confocal microscope (Leica Microsystems Inc, Buffalo Grove, IL).
Runx2 Gene Expression Measurement
The expression of Runx2 gene was measured by quantitative real-time PCR (rtPCR). Total RNA was extracted from the cell samples using an RNeasy Mini kit (Qiagen, Valencia, CA), and the cDNA templates were transcribed using RetroScript reverse transcription kit (Ambion, Austin, TX) according to the manufacturer’s instructions. NanoDropTM (Thermo Scientific, Wilmington, DE) was used to determine the concentration and purity of the RNA preparations. SYBR Green master-mix (Qiagen) was used in RT-PCR reactions according to the manufacturer's instructions, and the reactions were performed in a Lightcycler 3.5 Real-Time PCR System (Roche Diagnostics, Indianapolis, IN). All primers were designed in our lab and synthesized by Integrated DNA Technologies, Inc. (IDT, Coralville, IA). The Comparative Ct (ΔΔCt) method was used to calculate the relative gene expression levels, where ΔΔCt corresponds to the cycle threshold difference between the reactions containing target gene primer pair and the 18S rRNA housekeeping gene primer pair in each sample. Each experiment was repeated at least two times in triplicates. Data were expressed as means ± S.D. of the relative gene expression levels. The primers were designed using the Primer3 web utility13 to obtain the following sequences (all murine): 18S (forward) 5’-CCCTGCCCTTTGTACACACC-3’; 18S (reverse) 5’-CGATCCGAGGGCCTCACTA-3’; Runx2 (forward) 5’-CCGCACGACAACCG ACCAT-3’; Runx2 (reverse) 5’-AGCCACCAAGGCTGGAGTCTT-3’.
Alkaline Phosphatase Assay
For the histochemical detection of alkaline phosphatase expression, cells were fixed with 4% paraformaldehyde for 15 minutes at room temperature. After fixation, the cells were stained by incubation for 30 min in a mixture of 0.1 mg/ml Naphthol AS-MX phosphate (Sigma-Aldrich, St. Louis, MO), 2% N,N-dimethylformamide (Sigma-Aldrich, St. Louis, MO) and 1 mg/ml solution of fast blue BB salt (Sigma-Aldrich, St. Louis, MO) in 0.1 M Tris-HCl, pH 8.2, at room temperature, and were observed under bright field microscope.
For quantitative analysis of alkaline phosphatase activities, cells were lysed by incubation in CelLytic™ (Sigma-Aldrich) for 30 minutes on ice. Cells were collected in Eppendorf tubes by scraping off the wells and then sonicated for 1 minute at 4°C. Twenty microliters of cell lysate were reacted for 30 minutes with 100 μl p- nitrophenyl phosphatase (pNPP) (Sigma-Aldrich) in 96 well plate in triplicates. The colorimetric reaction was stopped after 30 minutes by adding 20 μl 2N NaOH, and the absorbance was measured at 420 nm. Alkaline phosphatase activity was normalized by total protein content, which was determined using Thermo Scientific Pierce BCA Protein Assay Kit.
Bone mineralization assays
Calcium deposition in osteogenically differentiated C2C12 cells was evaluated using an Alizarin Red S (ARS) staining method.14 The transfected and untransfected cells were cultured in 100 ng/ml BMP-2 supplemented media in 24 well plates for two weeks. Media were changed every 48 hours. On day 14, cells were fixed with 4% paraformaldehyde, then stained with a 2% solution of ARS dye at pH 4.2. After 20 minutes incubation followed by washing, the samples were analyzed under a bright field microscope.
Statistical Analysis
Each experiment was repeated two to three times independently. Measurements in each experiment were made in triplicate. All results are expressed as mean ± standard deviation (S.D.). The statistical significance was analyzed by Student's t test or ANOVA. A p value < 0.05 was considered to be statistically significant.
Results
Runx2 SiRNA Screening
C2C12 cells are undifferentiated multipotent cells that have a potential to differentiate into musculoskeletal lineages, including bone and skeletal muscle tissues. It has been established previously that bone morphogenic protein-2 (BMP-2) treatment of C2C12 cell cultures down-regulates the expression of myogenic transcription factors, inhibits the formation of multinucleated myotubes, and promotes differentiation towards the osteoblastic lineage.15 Here, we utilize induction of C2C12 cells by BMP-2 (100 ng/ml) as a cell culture model that recapitulates the pathogenesis of HO. Among the regulatory factors governing the differentiation and the lineage commitment of undifferentiated progenitor cells, Runx2 has been identified as an essential transcription factor and initial target for gene silencing in these studies.9b, 16
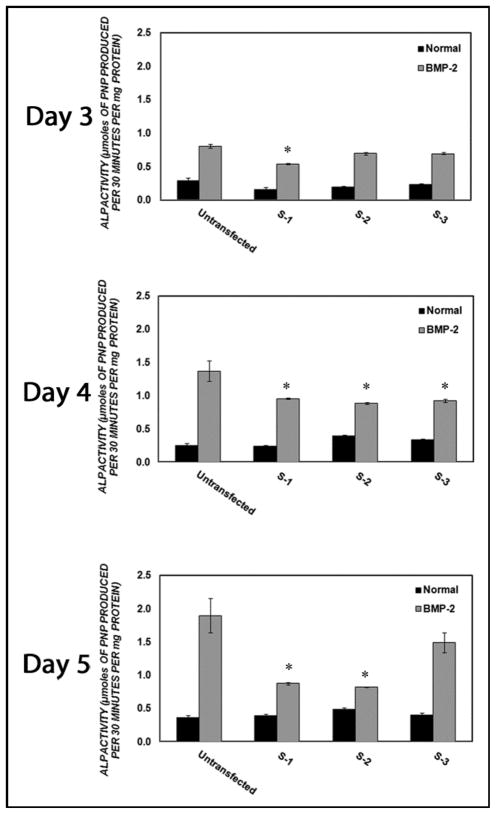
We selected three candidate siRNA sequences against Runx2 mRNA using sequence and structural considerations (Table 1). Each of these was transfected into C2C12 cells using Lipofectamine2000 followed by osteogenic stimulation with 100 ng/ml BMP-2 supplemented media. The endogenous alkaline phosphatase activity of the C2C12 cells was measured on parallel samples grown in the absence of BMP-2. The endogenous alkaline phosphatase activity among cell cultures transfected with the three different siRNA’s was not significantly different (p > 0.05) from the alkaline phosphatase activity of untransfected controls on the respective days. Untransfected control cell cultures displayed statistically significantly enhanced alkaline phosphatase activities (185%, 448% and 440%) in BMP2 supplemented media on days 3, 4 and 5 post BMP2 treatment respectively as compared to the cells grown in the absence of BMP-2 (Figure 1). All three siRNA sequences inhibited alkaline phosphatase (ALP) activity in C2C12 cells stimulated with BMP-2 on at least one day of measurement. ALP activity was decreased significantly by S-3 treatment at day 4, by S-2 treatment on days 3 and 4, and by S-1 treatment on days 3, 4 and 5 (Figure 1). As sequence S-1 exerted the most efficient and durable knockdown of Runx2 in the C2C12 cell culture, it was selected as the siRNA sequence to target Runx2 in all subsequent experiments.
Figure 1. Screening of candidate siRNA sequences.
Cells were transfected with 160 nM Runx2- targeting siRNA sequences S-1, S-2 and S-3 for 4 hours using Lipofectamine 2000 in OptiMEM. Cells were cultured in normal or 100 ng/ml BMP-2 supplemented growth media after transfection. Alkaline phosphatase activity was measured on days 3, 4 and 5 post-treatment. The asterisk (*) represents statistically significant difference (p<0.05) in alkaline phosphatase activity from that of untransfected control when cells were treated in BMP-2 supplemented media.
Cytotoxicity
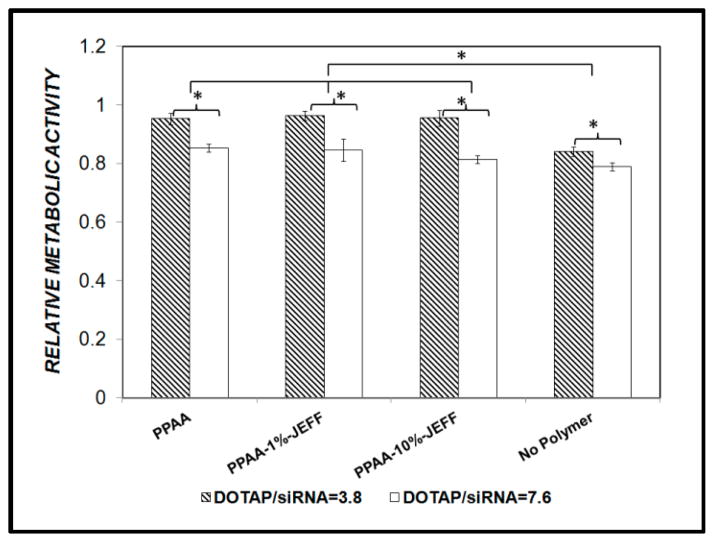
The use of polymer and liposome based carriers is often limited by their inherent cytotoxicities. The cytotoxicity caused by the DOTAP/siRNA/ anionic polyelectrolyte nanocomplexes was determined by monitoring the metabolic activity of C2C12 cells transfected with the nanocomplexes 24 hours post transfection. When mixed at a 10:1 weight ratio to give a net charge ratio 3.8, the DOTAP/siRNA lipoplexes reduced the metabolic activity of the C2C12 cells to 86% of the untreated cells (Figure 2, ‘No Polymer’). The ternary complexes of the DOTAP/siRNA with PPAA or PPAA-g-Jeffamine provided a cytoprotective effect, with 95% basal metabolic activity observed at an overall charge ratio of 1.0 irrespective of the copolymer graft density. When the DOTAP:siRNA ratio was increased to 20:1 to give the initial lipoplex charge ratio (+/−) of 7.6, the metabolic activity was further reduced in all samples but again the anionic polyelectrolytes provided a cytoprotective effect Nonetheless, an initial DOTAP/siRNA charge ratio of 3.8 was used for subsequent studies.
Figure 2. Effect of DOTAP/siRNA/polyelectrolyte complexes on the metabolic activity of C2C12 cells.
Metabolic activity, measured by the MTS assay, was compared among the cells transfected with initial DOTAP/siRNA charge ratio 3.8 and 7.6 alone (No Polymer) or upon complex formation with PPAA, PPAA-g-1% Jeffamine or PPAA-g-10% Jeffamine to a final net charge ratio of 1.0. Cells were treated with 160 nM siRNA for 4 hr, in 10% FBS-containing cell culture media, and assayed 24 h post-treatment.
Size and zeta-potential of DOTAP/ siRNA/ polyelectrolyte complexes
We have previously utilized the pH-sensitive polymer poly(propylacrylic acid) (PPAA) and the graft copolymer PPAA-g-Jeffamine to improve DOTAP: lipolex delivery of antisense oligonucleotides (AONs) into tumor cells in serum-free and serum-containing conditions, respectively.10d, 17 In the present work, we have adopted this system for siRNA delivery. Given the greater size and charge on siRNA molecules relative to AONs, we first determined the hydrodynamic sizes and zeta potentials of the nanocomplexes formed in the presence of siRNA as a probe of self-assembled complex formation.
The cationic liposomes comprised of DOTAP alone have a positive zeta potential of 23 mV (Figure 3A). Upon addition of siRNA to form the lipoplex, a decrease in zeta potential to 15 mV was observed, which was consistent with partial charge neutralization of the liposome’s cationic charge. Addition of anionic polymer PPAA to the DOTAP/siRNA complexes reduced the zeta potential of the resultant ternary complex to 3.9 mV, which was consistent with electrostatic neutralization of the residual positive DOTAP charge by the anionic copolymer. The siRNA complexes formed with the PPAA-g-1% Jeffamine and PPAA-g-10% Jeffamine had slightly negative zeta potentials, which was consistent with more extended chain conformations of the bound polyelectrolytes at the DOTAP/siRNA lipoplex surface, as has been observed with dendrimer condensation of DNA.18.
Figure 3. Physical characterization of nanocomplexes. (A).
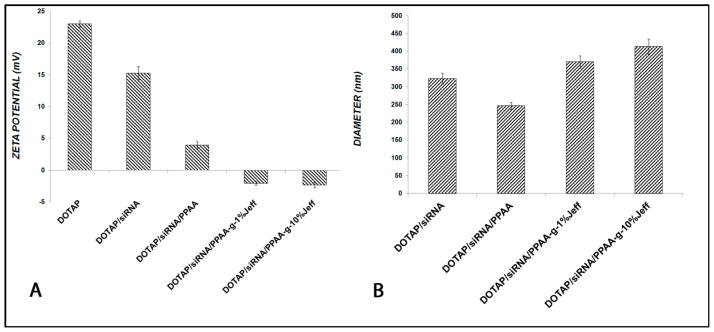
Zeta potential of the self-assembled complexes formed in the absence of (DOTAP/siRNA) or in the presence of the indicated polyelectrolyte. The amounts of DOTAP/siRNA correspond to a theoretical charge ratio of 3.8. Upon addition of polyelectrolyte to DOTAP/siRNA lipoplexes, the resultant theoretical charge ratio on the ternary complexes was 1.0. (B) Hydrodynamic diameters of the DOTAP/siRNA complexes, formed as in (A), were measured using dynamic light scattering.
The hydrodynamic diameter of the DOTAP/siRNA/PPAA complexes decreased to 245 ± 10 nm compared with 323 ± 15 nm for DOTAP/siRNA alone. This compaction effect was likely driven by the reduction of electrostatic repulsion forces between adjacent DOTAP quaternary amine functional groups caused by binding of the anionic polyelectrolyte, consistent with Manning’s counterion condensation theory.19 However, in the presence of the graft copolymers, the nanoparticle sizes were found to increase to 369 ± 16 nm for the 1% graft density copolymer and further to 412 ± 22 nm for the 10% graft density copolymer. These increases paralleled the zeta potential measurements and were consistent with more extended polyelectrolyte chain conformations, which would increase with increased graft densities and resultant molecular weights.
Cellular Uptake of Nanocomplexes by C2C12 Cells
Our recent results in delivery of AONs suggest that the hydrophilic/lipophilic balance (HLB) of the copolymer governs its interaction with lipid bilayers and that this property can be correlated directly to cellular uptake and gene silencing.10c Here, the effect of Jeffamine graft density on the intracellular delivery of siRNA by the DOTAP/siRNA/PPAA-g-Jeffamine ternary nanocomplexes was investigated. Confocal fluorescence microscopy images revealed that the nanocomplexes formed in the presence of PPAA-g-1%Jeffamine resulted in the greatest extent of intracellular siRNA uptake (Figure 4). The diffuse red fluorescent regions in each of the images were indicative of successful escape from the endosomal vesicles by the nanocomplexes with PPAA and the two graft copolymers. This trend was confirmed by flow cytometry, which provided the quantitative comparison that the cellular uptake of the nanocomplexes in C2C12 cells was enhanced by 13-fold and 7-fold when they were formed using PPAA-g-1%-Jeffamine and PPAA-g-10% Jeffamine, respectively, as compared to the DOTAP/siRNA/PPAA complexes and by 82-fold and 42-fold respectively when compared to DOTAP/siRNA (Figure 4D). These results are similar to those we have observed for AON delivery to tumor cells.10c
Figure 4. Cellular uptake of nanocomplexes containing Cy5-labeled siRNA in C2C12 cells.
Imaging of uptake using confocal microscopy. Nucleus and cell membrane were stained with DAPI (blue) and Cell mask (faint red) stains. Bright red fluorescent spots represent internalized Cy5 labeled siRNA. Transfection was carried out using A) DOTAP/siRNA/PPAA, B) DOTAP/siRNA/PPAA-g-1%Jeffamine and C) DOTAP/siRNA/PPAA-g-10%Jeffamine. (D) The uptake was also measured quantitatively using flow cytometry. “No polymer” refers to DOTAP/siRNA and the other samples to the indicated polymer added to DOTAP/siRNA.
Silencing of Runx2
To determine whether the cellular uptake observed is sufficient to produce gene silencing by RNA interference, siRNA sequence S-1, specific for mouse Runx2, was delivered to C2C12 cells using each of the nanocomplexes. Real-time PCR analysis confirmed the down regulation of Runx2 by the siRNA, both under basal (BMP-free) conditions as well as when transfected cells were cultured subsequently in BMP-2 (100 ng/ml) supplemented media. The transfection of C2C12 cells carried out using complexes formed in the presence of PPAA-g-1% Jeffamine showed maximum down regulation (>69% in the absence of BMP-2) of Runx2 gene that remained substantially (more than 60%) silenced even in osteogenic conditions when Runx2 is normally induced strongly (Figure 5).
Figure 5. Silencing of Runx2 expression in mouse mesenchymal progenitor cells (C2C12).
Cells were transfected with the indicated formulations in the same way as in Figures 3 and 4. The mRNA levels of Runx2 were determined by real-time PCR. Gene expression in each sample is normalized to expression of the 18S ribosomal RNA, and the expression is then normalized to the Runx2 expression of untransfected cells untreated with BMP-2. Single asterisk (*) represents statistical significance (p < 0.05) relative to No Polymer condition in the absence of BMP-2, and double asterisk (**) represents statistical significance (p < 0.05) relative to No Polymer condition in the presence of BMP-2.
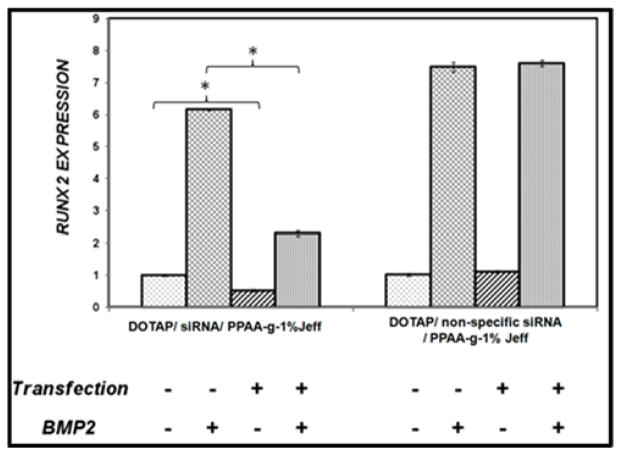
To ensure that the silencing of Runx2 was due to RNA interference rather than to the delivery system, we also transfected non-specific siRNA sequence separately into C2C12 cells carried by the ternary nanocomplexes formed in the presence of PPAA-g-1%-Jeffamine, as 1% grafting density led to the greatest suppression of the Runx2 as shown in Figure 5. Real-time PCR analysis revealed that the Runx2 specific siRNA yielded more than 50% silencing in the endogenous Runx2 level and approximately 63% in osteogenic conditions forty-eight hours following transfection, while no change in Runx2 expression was observed upon transfection with non-specific siRNA (Figure 6).
Figure 6. Silencing using DOTAP/siRNA/PPAA-g-1% Jeffamine nanocomplexes is specific.
Expression of Runx2 was determined following transfection of either Runx2 specific siRNA or nonspecific control.
Effect of Runx2 Silencing on Osteogenesis
Having established the ability of DOTAP/siRNA/PPAA-g-1% Jeffamine complexes to mediate gene silencing of Runx2 in the C2C12 cell line, we next examined whether knockdown of Runx2 is sufficient to affect osteoblastic differentiation in C2C12 cells stimulated with BMP-2. The post transfection alkaline phosphatase (ALP) activity was measured by quantitative assay as well as histochemically in normal and osteogenic conditions. The histochemical analysis qualitatively showed that silencing of Runx2 using the DOTAP/siRNA /PPAA-g-1%-Jeffamine complexes effectively down-regulated the activity of ALP. Furthermore, the staining results indicated that the complexes formed in the presence of 1% grafted PPAA were more effective than Lipofectamine 2000 (Figure 7). A quantitative assay showed that the ALP activity of the C2C12 cells, following silencing with Runx2-specific siRNA complexed with DOTAP/PPAA-g-1% Jeffamine, was reduced by ~60% on day 4 and over 45% on day 5 post BMP-2 treatment (Figure 7E).
Figure 7. Down-regulation of Runx2 inhibits the alkaline phosphatase activity in differentiating mouse mesenchymal progenitors cells (C2C12).

Transfection with Runx2 siRNA and BMP stimulation were as described in Figure 5. Cells were not transfected (A) and B), or transfected with 160 nM Runx2 siRNA sequence S-1 using C) Lipofectamine 2000 or D) DOTAP/siRNA/PPAA-g-1%Jeffamine and were induced to differentiate into osteblastic cell lineage with BMP-2 (B, C and D) after transfection. The histochemical analysis was performed on day 5 following treatemnt. E) Quantitative assay of alkaline phosphatase activity was performed using p-nitrophenyl phosphatase as a substrate. The asterisks represent statistical significant differences (p<0.05) in the alkaline phosphatase activity after transfection from that of untransfected control on days 3 and 4 post BMP-2 treatment respectively.
Discussion
Several investigations to understand the etiology of acquired HO in animal models, simulating traumatic events like fracture, total joint arthroplasty, traumatic muscle or CNS injuries along with signifying enhanced BMP signaling pathways, confirmed that the dysregulation of BMP signaling and post trauma tissue microenvironment coordinate to produce ectopic bone growth.20 The growing molecular understanding of HO motivates the use of RNA interference as an approach that can be used both to further our understanding of the mechanisms of pathological bone formation as well as to provide a specific means of halting this condition without untoward side effects. In the present work, to determine the effectiveness of our ternary liposome/siRNA/copolymer nanocomplexes in inhibiting the post-transcriptional encoding of Runx2 mRNA, we have developed an in vitro cell culture model to simulate the pathology of heterotopic ossification. Experiments were conducted on progenitor cells (C2C12) that normally form myogenic bundles and tubes when they reach a critical density in culture. However, they can be induced to differentiate into an osteoblastic cell lineage by exogenously administering human recombinant BMP-2. The plasticity of the C2C12 cells in response to an external cue (BMP-2) provides a valuable system for determining how to inhibit this process that mimics the pathophysiology of HO.
The efficient delivery of siRNA cargo, and consequently the silencing of the targeted gene, is the cumulative outcome of several steps, including survival of the complexes in physiological environment, efficient internalization of the complexes by the cells, and their escape from the endosomolytic pathway.21 Construction of a delivery system that is effective in each of these ways remains a formidable challenge to the field. The polyelectrolyte PPAA (ungrafted) demonstrates outstanding membrane disruption ability at endosomal pH and forms nanocomplexes with DOTAP/siRNA of reasonable size and charge, but the transfection carried out by DOTAP/siRNA/PPAA resulted in inefficient silencing of the target gene, similar to previous results we have seen for this system for antisense oligonucleotide delivery.10d Accumulation of serum proteins on the surface of nanocomplexes may disrupt their interaction with the plasma membrane and thereby affect nucleic acid delivery.22
To address this limitation, we developed copolymers of PPAA grafted with Jeffamine M2070, a copolymer of ethylene oxide and propylene oxide with a terminal amine. Grafting of the Jeffamine onto PPAA abates the low pH membrane disruption activity of the copolymer but augments significantly the resistance to serum attack.10c, d Furthermore, grafting of Jeffamine chains onto the PPAA imparts non-ionic amphiphilic character to the polyelectrolyte, enhancing its tendency to interact with lipophilic cell membranes. Among the graft copolymers that we have synthesized to date, PPAA-g-1% Jeffamine exhibits the greatest extent of membrane interaction. Our results with antisense oligonucleotides have suggested that a minimum level of grafting is necessary for serum stability, but once this is attained, a lower hydrophilic-lipophilic balance is desirable, as is the case for the graft copolymers with low extent of grafting and incorporation of the more lipophilic propylene oxide moieties (i.e., Jeffamine). In this light, it is reasonable that the nanocomplexes formed in the presence of PPAA-g-1% Jeffamine exhibit maximum efficiency to knock down Runx2 expression and inhibit the early and late phenotypic markers of osteoblastic differentiation in C2C12 cells. Transfection of C2C12 cells by specific siRNA for 4 hours efficiently knocked down expression of Runx2 in myoprogenitor cells by 24 hours post transfection. Notably, the biological effect of single-dose silencing was maintained at a phenotypic level 2 weeks post transfection.
In the present work, we have established a relatively simple cell culture model that recapitulates the defining feature of heterotopic ossification, namely a switch from myoblastic to osteoblastic phenotype. In the experiments presented here, the osteoblastic differentiation was driven by BMP-2, which is understood to be a major molecular mediator of HO. The C2C12 cell culture system can be also driven to an osteoblastic phenotype via an osteogenic medium containing ascorbic acid, beta-glycerophosphate and dexamethasone.23,24 Thus, it serves as an attractive model in which to investigate the molecular pathways driving this cell fate decision. Here, we showed that silencing of Runx2, mediated by potent delivery using the serum-stable DOTAP/siRNA/PPAA-g-1%Jeffamine formulation, is sufficient to halt the osteogenic switch driven by BMP-2. The fact that silencing was substantial and robust in serum-containing media bodes well for use of this delivery formulation in vivo, as we have found in applying it to AON delivery in tumors (Peddada et al., manuscript in preparation). Based on this experience, we expect the ternary nanocomplex approach utilized here to have impact both in validation of osteogenic genes as well as in evaluation of siRNA or siRNA/drug treatment therapies for HO and other diseases for which a molecular pathogenesis can be defined.
Figure 8. Down-regulation of Runx2 decreased the deposition of calcium phosphate in differentiating mouse myoblastic progenitors cells (C2C12).
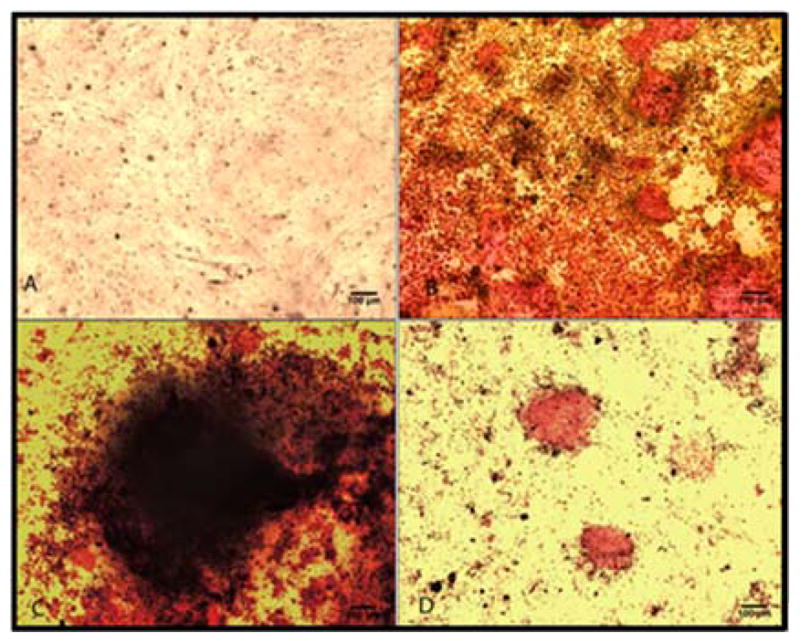
A and B represent the untransfected control. Cells were transfected with C) Runx2 siRNA (sequence S-1) using Lipofectamine 2000, D) Runx2 siRNA sequence using DOTAP/siRNA/ PPAA-g-1% Jeffamine complexes and were induced to differentiate into osteoblastic cell lineage in BMP-2 supplemented media (B, C and D). Histochemical analysis was performed on day 14 of BMP-2 treatment using Alizarin Red S.
Acknowledgments
Support for this work was provided by a grant from the Department of Defense (W81-WXH-10-2-0139). The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of the Army or the Department of Defense. We thank Dr. Prabhas Moghe for access to the confocal microscopy collaborative resources under the auspices of NIH grant P41 EB001046 for the biomedical technology resource RESBIO and George Dorfman for his help in bio-imaging.
Contributor Information
Swati Mishra, Email: swmishra@rutgers.edu.
Asa D. Vaughn, Email: asa.d.vaughn@us.army.mil.
David I. Devore, Email: david.devore@us.army.mil.
Charles M. Roth, Email: cmroth@rutgers.edu.
References
- 1.Shehab D, Elgazzar AH, Collier BD. Heterotopic ossification. J Nucl Med. 2002;43:346–53. [PubMed] [Google Scholar]
- 2.(a) Garland DE. A clinical perspective on common forms of acquired heterotopic ossification. Clin Orthop Relat Res. 1991:13–29. [PubMed] [Google Scholar]; (b) Singer BR. Heterotopic ossification. Br J Hosp Med. 1993;49:247–51. 254–5. [PubMed] [Google Scholar]
- 3.Donald GN, Resnick L. 2. Diagnosis of bone and joint disorders. Vol. 5 W.B. Saunders; Philadelphia, PA: 1988. [Google Scholar]
- 4.(a) Chao ST, Joyce MJ, Suh JH. Treatment of heterotopic ossification. Orthopedics. 2007;30:457–64. doi: 10.3928/01477447-20070601-18. quiz 465–6. [DOI] [PubMed] [Google Scholar]; (b) Stover SL, Niemann KM, Miller JM., 3rd Disodium etidronate in the prevention of postoperative recurrence of heterotopic ossification in spinal-cord injury patients. J Bone Joint Surg Am. 1976;58:683–8. [PubMed] [Google Scholar]; (b) Cipriano C, Pill SG, Rosenstock J, Keenan MA. Radiation therapy for preventing recurrence of neurogenic heterotopic ossification. Orthopedics. 2009;32 doi: 10.3928/01477447-20090728-33. [DOI] [PubMed] [Google Scholar]
- 5.Liu K, Tripp S, Layfield LJ. Heterotopic ossification: review of histologic findings and tissue distribution in a 10-year experience. Pathol Res Pract. 2007;203:633–40. doi: 10.1016/j.prp.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Uwe Scherbel PR, Khurana Jasvir, Born Christopher, DeLong William. Expression of Bone Morphogenic Proteins in Rats with and without Brain Injury and a Tibia Fracture. The University of Pennsylvania Orthopaedic Journal. 2001;14:85–89. [Google Scholar]
- 7.Salisbury E, Rodenberg E, Sonnet C, Hipp J, Gannon FH, Vadakkan TJ, Dickinson ME, Olmsted-Davis EA, Davis AR. Sensory nerve induced inflammation contributes to heterotopic ossification. J Cell Biochem. 2011;112:2748–58. doi: 10.1002/jcb.23225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, Satake M, Takada K, Komori T. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32:633–8. doi: 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]
- 9.(a) Lin L, Shen Q, Leng H, Duan X, Fu X, Yu C. Synergistic inhibition of endochondral bone formation by silencing Hif1alpha and Runx2 in trauma-induced heterotopic ossification. Mol Ther. 2011;19:1426–32. doi: 10.1038/mt.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xue T, Mao Z, Lin L, Hou Y, Wei X, Fu X, Zhang J, Yu C. Non-virus-mediated transfer of siRNAs against Runx2 and Smad4 inhibit heterotopic ossification in rats. Gene Ther. 2010;17:370–9. doi: 10.1038/gt.2009.154. [DOI] [PubMed] [Google Scholar]
- 10.(a) Jones RA, Cheung CY, Black FE, Zia JK, Stayton PS, Hoffman AS, Wilson MR. Poly(2-alkylacrylic acid) polymers deliver molecules to the cytosol by pH-sensitive disruption of endosomal vesicles. Biochem J. 2003;372:65–75. doi: 10.1042/BJ20021945. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kyriakides TR, Cheung CY, Murthy N, Bornstein P, Stayton PS, Hoffman AS. pH-sensitive polymers that enhance intracellular drug delivery in vivo. J Control Release. 2002;78:295–303. doi: 10.1016/s0168-3659(01)00504-1. [DOI] [PubMed] [Google Scholar]; (c) Mishra S, Peddada LY, Devore DI, Roth CM. Poly(alkylene oxide) Copolymers for Nucleic Acid Delivery. Acc Chem Res. 2012 doi: 10.1021/ar200232n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Peddada LY, Harris NK, Devore DI, Roth CM. Novel graft copolymers enhance in vitro delivery of antisense oligonucleotides in the presence of serum. J Control Release. 2009;140:134–40. doi: 10.1016/j.jconrel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun D, Cherdron H, Rehahn M, Ritter H, Voit B. Polymer Synthesis: Theory and Practice. 4. Springer; Berlin: 2005. [Google Scholar]
- 12.(a) Lu ZJ, Mathews DH. OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 2008;36:W104–8. doi: 10.1093/nar/gkn250. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lu ZJ, Mathews DH. Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Res. 2008;36:640–7. doi: 10.1093/nar/gkm920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steve Rozen HJS. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz MSS, editor. Humana Press; Totowa: 2000. pp. 365–386. [Google Scholar]
- 14.Kochanowska I, Chaberek S, Wojtowicz A, Marczynski B, Wlodarski K, Dytko M, Ostrowski K. Expression of genes for bone morphogenetic proteins BMP-2, BMP-4 and BMP-6 in various parts of the human skeleton. BMC Musculoskelet Disord. 2007;8:128. doi: 10.1186/1471-2474-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 1994;127:1755–66. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin L, Chen LX, Wei XL, Fu X, Zhang JY, Ma KT, Zhou CY, Yu CL. Adenovirus-mediated transfer of siRNA against Runx2/Cbfa1 inhibits the formation of heterotopic ossification in animal model. Biochem Bioph Res Co. 2006;349:564–572. doi: 10.1016/J.Bbrc.2006.08.089. [DOI] [PubMed] [Google Scholar]
- 17.Lee LK, Williams CL, Devore D, Roth CM. Poly(propylacrylic acid) enhances cationic lipid-mediated delivery of antisense oligonucleotides. Biomacromolecules. 2006;7:1502–8. doi: 10.1021/bm060114o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merkel OM, Mintzer MA, Sitterberg J, Bakowsky U, Simanek EE, Kissel T. Triazine dendrimers as nonviral gene delivery systems: effects of molecular structure on biological activity. Bioconjug Chem. 2009;20:1799–806. doi: 10.1021/bc900243r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning GS. Simple Model for the Binding of a Polyelectrolyte to an Oppositely Charged Curved Surface. J Phys Chem B. 2003;107:11485–11490. [Google Scholar]
- 20.Lounev VY, Ramachandran R, Wosczyna MN, Yamamoto M, Maidment ADA, Shore EM, Glaser DL, Goldhamer DJ, Kaplan FS. Identification of Progenitor Cells That Contribute to Heterotopic Skeletogenesis. Journal of Bone and Joint Surgery-American Volume. 2009;91A:652–663. doi: 10.2106/Jbjs.H.01177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery (vol 8, pg 129, 2009) Nat Rev Drug Discov. 2010;9:412–412. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nie S. Understanding and overcoming major barriers in cancer nanomedicine. Nanomedicine (Lond) 2010;5:523–8. doi: 10.2217/nnm.10.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maniatopoulos C, Sodek J, Melcher AH. Bone formation in vitro by stromal cells obtained from bone marrow of young adult rats. Cell Tissue Res. 1988;254:317–30. doi: 10.1007/BF00225804. [DOI] [PubMed] [Google Scholar]
- 24.Park JB. The Effects of Dexamethasone, Ascorbic Acid, and beta- Glycerophosphate on Osteoblastic Differentiation by Regulating Estrogen Receptor and Osteopontin Expression. J Surg Res. 2012;173:99–104. doi: 10.1016/J.Jss.2010.09.010. [DOI] [PubMed] [Google Scholar]