Abstract
Salmonella enterica utilizes flagellar motility to swim through liquid environments and on surfaces. The biosynthesis of the flagellum is regulated on various levels, including transcriptional and posttranscriptional mechanisms. Here, we investigated the motility phenotype of 24 selected single gene deletions that were previously described to display swimming and swarming motility effects. Mutations in flgE, fliH, ydiV, rfaG, yjcC, STM1267 and STM3363 showed an altered motility phenotype. Deletions of flgE and fliH displayed a non-motile phenotype in both swimming and swarming motility assays as expected. The deletions of STM1267, STM3363, ydiV, rfaG and yjcC were further analyzed in detail for flagellar and fimbrial gene expression and filament formation. A ΔydiV mutant showed increased swimming motility, but a decrease in swarming motility, which coincided with derepression of curli fimbriae. A deletion of yjcC, encoding for an EAL domain-containing protein, increased swimming motility independent on flagellar gene expression. A ΔSTM1267 mutant displayed a hypermotile phenotype on swarm agar plates and was found to have increased numbers of flagella. In contrast, a knockout of STM3363 did also display an increase in swarming motility, but did not alter flagella numbers. Finally, a deletion of the LPS biosynthesis-related protein RfaG reduced swimming and swarming motility, associated with a decrease in transcription from flagellar class II and class III promoters and a lack of flagellar filaments.
Introduction
The gram-negative enteropathogen Salmonella enterica is the causative agent of salmonellosis. Typical symptoms include gastroenteritis, including diarrhea, abdominal cramps and in rare cases enteric fever, which can be life threatening, especially for immune-compromised people [1]. After ingestion of contaminated food or water Salmonella employs a battery of virulence factors, such as toxin injection devices (injectisomes) [2,3], fimbriae or flagella to successfully colonize and persist within the host. The flagellum, a rotating, rigid, helical motility organelle, enables bacteria to chemotactically swim towards nutrients or away from harmful substances [4]. Furthermore, flagella facilitate biofilm formation and adherence to the host epithelium. By enabling bacteria-host interactions, flagella therefore enhance bacterial pathogenesis [5].
The flagellum consists of three main structures: the basal body, the hook and the filament. The basal body includes rotor and stator protein complexes necessary for motor-force generation and thereby flagellar rotation [6,7]. Additionally, a rod spans the periplasmic space from the inner membrane through the cell wall to the outer membrane. The hook extends from the cell surface to a length of approximately 55 nm and functions as a flexible linking structure between the membrane-embedded basal body and the rigid filament. The 10–15 μm long filament is built up of thousands of subunits of a single protein, flagellin (FljB or FliC in Salmonella enterica). Every flagellar subunit has to be exported to enable a proper assembly at the tip of the growing structure. For this purpose, Salmonella uses a flagellar-specific type-III secretion system (fT3SS) that is structurally and functionally related to the virulence-associated T3SS (vT3SS) of injectisome devices [8]. The secretion of flagellar components is proton-motive force-dependent and coupled to ATP hydrolysis and has to proceed in a timely and ordered fashion [9–11]. The flagellar regulon of Salmonella enterica serovar Typhimurium comprises more than 60 genes [12]. The biosynthesis of the flagellar apparatus relies on a hierarchical regulatory system, consisting of three flagellar promoter classes. On top of this hierarchy and under the control of a σ70-dependent class I promoter is the flagellar master operon flhDC. FlhD and FlhC form a heteromultimeric complex (FlhD4C2) that ultimately controls initiation of flagellar assembly [13,14]. Together with σ70/RNA polymerase, the FlhD4C2 complex activates transcription of genes that are under the control of class II promoters, resulting in the expression of proteins that make up the basal body and hook (HBB). The regulatory protein FlgM is expressed from a class II promoter and interacts with the flagellar-specific σ28-factor to prevent association of σ28 with RNA polymerase, which is needed for transcription from class III promoters. Upon completion of the hook-basal-body complex, the secretion mode of the fT3SS switches to export of late substrates. FlgM as a late secretion substrate is exported, which releases σ28 and results in expression of filament and chemosensory proteins, as well as motor force generators [15–18].
Regulation of flagellar biosynthesis occurs on various levels. A variety of environmental stimuli are integrated at the level of the class I promoter that ultimately determine expression of the master regulatory operon, flhDC. On a transcriptional level the cyclic AMP-catabolite activator protein (CAP) controls bacterial motility by activating the flhDC operon [19]. The histone-like nucleoid-structuring protein (H-NS) indirectly activates flhDC transcription by repressing a negative regulator, HdfR [20]. Transcription of flhDC is also activated by the iron-regulatory protein Fur [21] and the master regulator of the Salmonella pathogenicity island-1 encoded injectisome (Spi-1), HilD. HilD positively regulates flagellar gene expression through a direct binding to the P5 promoter of flhDC [22]. Furthermore, there are various negative regulators that bind to the flhDC promoter region, such as Spi-1 encoded RtsB, LrhA and SlyA [23–26]. On a posttranslational level, FlhD4C2 concentration is altered through FliZ, a flagellar protein that is under the control of class II and III promoters [27]. FliZ represses transcription of ydiV that encodes for a posttranscriptional anti-FlhD4C2 factor [28]. YdiV binds to FlhD, inhibits the DNA-binding properties of FlhD4C2 to class II promoters and targets the FlhD4C2 complex to ClpXP-mediated degradation [29,30].
Recently, Bogomolnaya et al. screened a library of 1023 single gene deletions in the virulent Salmonella enterica serovar Typhimurium background ATCC14028s for mutants that affected swimming and/or swarming motility [31,32]. Mutations in 130 genes that were not previously known to be involved in flagellar motility displayed an altered motility phenotype. Since various genes were linked to virulence in Salmonella, the flagellar system or were predicted to have a regulatory function, we were interested in the mechanisms of the altered motility phenotypes and investigated 24 presumed motility mutants in detail. We found that seven mutations (flgE, fliH, ydiV, rfaG, yjcC, STM1267, STM3363) affected swimming or swarming motility in our assays. A detailed analysis of flagella biosynthesis and flagellar gene expression for the mutant strains is described herein.
Results
Verification of novel factors modulating motility
We constructed single gene deletions of putative factors modulating motility [31] using the λ-Red recombination system as described by Datsenko and Wanner [33]. We were particularly interested in genes that were linked to virulence of Salmonella, the flagellar system or were predicted to have a regulatory function. To verify the previously published motility phenotypes of selected putative motility regulators, we performed swimming and swarming motility assays. Of the 24 tested mutants, only seven (flgE, fliH, ydiV, rfaG, yjcC, STM1267, STM3363) displayed altered motility compared to the wildtype (WT) control (Figs 1 and 2 and summarized in S4 Table). This was independent of growth rates and doubling times (S1 Fig, S3 Table), the incubation time of the motility plates (S2 Fig) and the used Salmonella enterica strain (S3 Fig). A ΔfliF strain served as negative control.
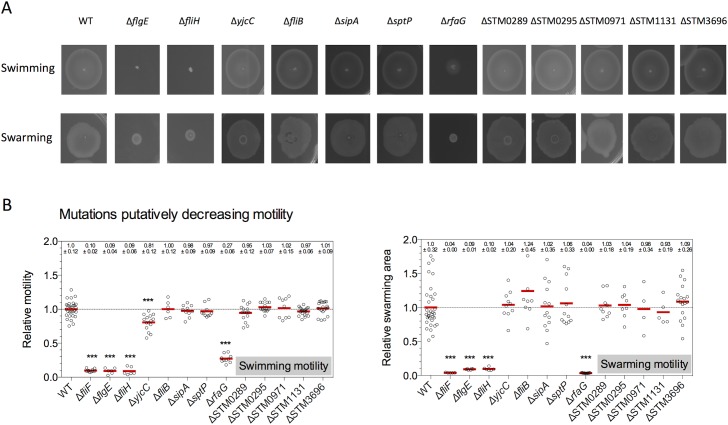
Fig 1. Swimming and swarming motility phenotypes of single gene deletion mutants putatively decreasing motility.
Motility phenotypes of mutants were analyzed on soft-agar plates (0.3% agar) after 4 hours incubation at 37°C for swimming motility. Swarming motility was analyzed on plates containing 0.6% agar after 9 hours incubation at 37°C. (A) Representative motility plates for each mutant tested (TH6622, EM824, EM1480, EM1508, EM1511, EM1512, EM1688, EM1690, EM2384, EM2385, EM2590, EM2591, and EM2605). The diameter of the motility swarm and the swarming area were measured and normalized to the wildtype. (B) Quantified relative motility. Biological replicates are shown as individual data points and analyzed by the Student’s t test. Asterisks indicate a significantly different motility phenotype (*** = P<0.001).
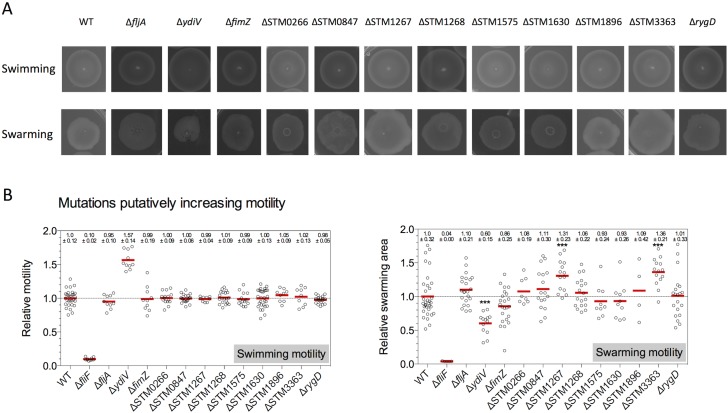
Fig 2. Swimming and swarming motility phenotypes of single gene deletion mutants putatively increasing motility.
(A) Representative motility plates for mutants potentially increasing motility (TH6622, EM824, EM2381, EM2382, EM2383, EM1507, EM1686, EM1481, EM1689, EM1509, EM1510, EM1482, EM1484, and EM1691). The diameter of the motility swarm and the swarming area were measured and normalized to the wildtype. (B) Quantified relative motility. Biological replicates are shown as individual data points and analyzed by the Student’s t test. Asterisks indicate a significantly different motility phenotype (*** = P<0.001).
In contrast to the swimming-only defect described in Bogomolnaya et al. [31], we found that flgE and fliH mutations resulted in a loss of both swimming and swarming motility (Fig 1). This was expected since FlgE, the flagellar hook protein, and FliH, a component of the fT3SS ATPase complex, are essential structural and functional components of the flagellum [34]. A deletion of ydiV showed increased swimming motility due to the posttranslational inhibition of flagellar class II gene expression via YdiV-mediated degradation of FlhD4C2 complex [29,30] (Fig 2). However, we found that swarming motility was decreased up to 50%. A deletion of rfaG showed a reduction in swimming (30% of WT) and a complete loss of swarming motility (Fig 1). Decreased swimming motility was also observed in a ΔyjcC mutant (80% of WT). Deletions of STM1267 and STM3363 –predicted genes with unknown functions—had no effect on swimming motility, but led to an increased swarming motility phenotype (Fig 2).
Characterization of mutants with an altered motility phenotype
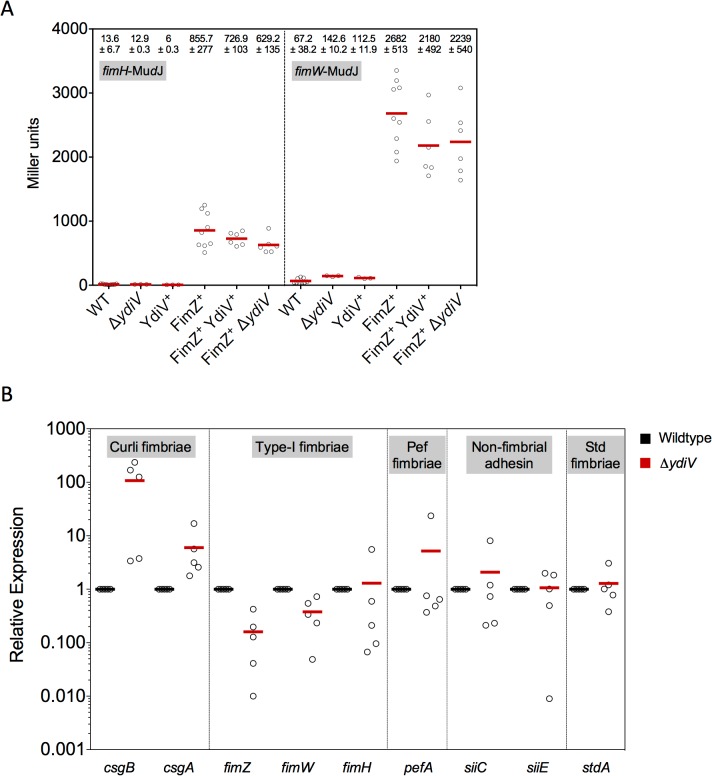
We next characterized selected mutants that displayed effects on motility by analyzing transcription of fimbrial or flagellar genes and filament formation. Since the ydiV mutation decreased swarming ability, we hypothesized that YdiV might be involved in expression of fimbriae as shown for Escherichia coli [35] and thereby resulting in a stickier phenotype of the ΔydiV deletion mutant in the swarm agar assay. Fimbrial gene transcription was analyzed by β-galactosidase activity assay using lac operon fusions to fimH, encoding for the fimbrial tip responsible for adhesion [36], and fimW, encoding the Type I fimbriae repressing protein FimW. A strain that overexpressed FimZ, a positive regulator of Type I fimbriae, was used as a control. Expression of FimZ led to higher transcription of fimH and fimW levels and these levels were neither influenced by a ydiV deletion, nor YdiV overexpression (Fig 3A), indicating that expression of Type I fimbriae was not affected by YdiV. Salmonella enterica serovar Typhimurium encodes for 13 fimbrial adhesins [37], and we next analyzed transcription of csgAB, fimZ, pefA, siiC, siiE, stdA adhesion genes by quantitative real-time PCR (Fig 3B). We found that in a ΔydiV mutant gene expression of csgA and csgB was 3- and 100-fold increased, respectively, whereas none of the other fimbrial genes displayed significantly altered transcript levels.
Fig 3. YdiV influences curli fimbriae gene transcription.
(A) Effects of a ΔydiV mutant and overproduced YdiV (YdiV+) and FimZ (FimZ+) on fimbrial gene expression were analyzed by β-galactosidase assay. Transcription of fimH that encodes a fimbrial structure component, and fimW that encodes for a repressor of fimbria, was examined in strains TH13438 (ΔaraBAD997::fimZ+ fimH56::MudJ), TH13439 (ΔaraBAD997::fimZ+ fimW57::MudJ), TH13498 (fimH56::MudJ), TH13502 (ΔaraBAD995::ydiV+ fimH56::MudJ), TH13505 (fimW57::MudJ), TH13509 (ΔaraBAD995::ydiV+ fimW57::MudJ), EM2527 (ΔydiV252 fimH56::MudJ), EM2528 (ΔydiV252 fimW57::MudJ), EM2724 (ΔaraBAD997::fimZ+ fimH56::MudJ ydiV240::Tn10dTc[del-25]), EM2725 (ΔaraBAD997::fimZ+ fimW57::MudJ ydiV240::Tn10dTc[del-25]), EM2726 (ΔaraBAD997::fimZ+ fimH56::MudJ ΔydiV251::tetRA) and EM2727 (ΔaraBAD997::fimZ+ fimW57::MudJ ΔydiV251::tetRA). At least three independent biological samples were analyzed. Error bars represent the standard errors of the means. Asterisks indicate gene expression levels that are significantly different to wildtype levels (*** = P< 0.001). (B) Relative csgA, csgB, fimZ, fimW, fimH, pefA, siiC, siiE and stdA gene expression of a ΔydiV::FKF mutant (EM2382) compared to the wildtype (TH6622) as analyzed using qRT-PCR. Experiments were performed with 5 biological replicates.
A deletion of yjcC (STM4264), a putative phosphodiesterase with EAL-motif, showed decreased swimming motility, but had no effect on swarming motility. To investigate this motility decrease in more detail, we analyzed β-galactosidase activity in strains that harbored lac-fusions to flagellar class I (flhC), class II (fliL) and class III (fliC) promoters. Transcription levels of all flagellar gene fusions tested were similar to WT levels, suggesting that YjcC does not play a role in flagellar gene expression (S4 Fig).
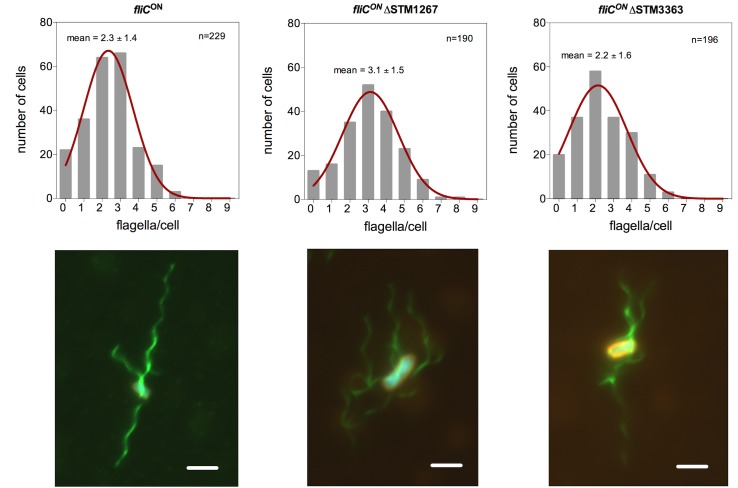
We next analyzed filament formation in two hypermotile mutants, ΔSTM1267 and ΔSTM3363, which showed an increased swarming motility phenotype. For that purpose, we analyzed flagellation levels by anti-FliC immunostaining in a strain locked in the FliC-phase. The WT strain assembled 2–3 flagella per cell (mean = 2.3 ± 1.4) under the experimental conditions (Fig 4). A ΔSTM3363 mutation did not affect filament formation (mean = 2.2 ± 1.6). A deletion in STM1267 slightly increased the number of flagella per cell (mean = 3.1 ± 1.5), which provided a possible explanation for the hypermotile phenotype.
Fig 4. Flagellar filament formation is increased in a STM1267 deletion mutant.
Top: Histogram of counted flagella per cell body of the wildtype, a STM1267::FRT and STM3363::FRT deletion mutant. The average filament numbers per cell based on a Gaussian non-linear regression analysis is indicated in the figure. Bottom: Flagellar filament formation was analyzed by flagellin immunostaining using α-FliC primary and α-rabbit conjugated Alexa Fluor 488 secondary antibodies (green) in a strain background locked for fliC expression. The membrane was stained with FM-64 (red) and DNA with DAPI (blue). Scale bar = 2 μm.
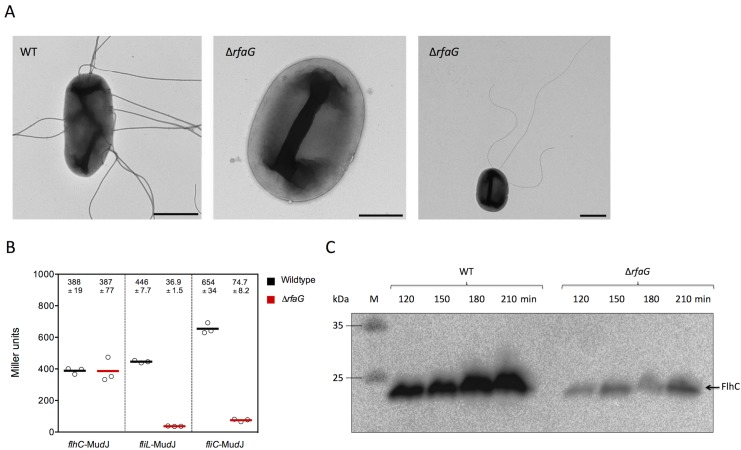
Upon deletion of rfaG, which encodes for a glucosyltransferase, swimming motility was reduced up to 80% and swarming motility was abolished completely. Thus, we investigated flagellation of a ΔrfaG mutant by transmission electron microscopy. In comparison to the highly flagellated WT, we observed a primarily deflagellated phenotype (Fig 5A left and middle panels). Only few bacteria possessed a low number of flagella (Fig 5A right panel), which was in agreement with the remaining 20% of swimming motility. We hypothesized that RfaG regulated flagellar gene expression on a transcriptional level, which would explain the non-flagellated phenotype. We performed β-galactosidase assays with lac-fusions to flagellar class I (flhC), class II (fliL) and class III (fliC) promoters in a ΔrfaG deletion mutant. Transcription of flhC was not altered, however, a ΔrfaG mutation reduced transcription of class II and class III promoters significantly (Fig 5B). FlhC protein levels were examined using a 3x FLAG tagged version of FlhC [29]. FlhC protein levels were reduced in a rfaG deletion mutant in contrast to the WT (Fig 5C), indicating that RfaG is involved in regulation of flagellar class II gene expression by posttranscriptionally affecting FlhDC stability or mRNA translation.
Fig 5. Flagella numbers and flagellar class II and class III gene expression are decreased in a ΔrfaG mutant.
(A) Transmission electron microscopy (TEM) of wildtype and ΔrfaG::FRT strains. With the exception of few individual cells the ΔrfaG::FRT mutant is non-flagellated. Scale bars: Left and right panel = 1 μm; Middle panel = 0.5 μm. (B) Effect of ΔrfaG::FRT mutation on flagellar gene transcription from class I (flhC-MudJ), class II (fliL-MudJ) and class III (fliC-MudJ) promoters. The β-galactosidase assay was performed in triplicate. Error bars represent the standard errors of the means. (C) FlhC-3xFLAG protein levels in the wildtype (EM1438) and ΔrfaG strain (EM2748). Bacteria were grown in LB medium and samples were taken 120, 150, 180 and 210 min after inoculation. Protein levels were monitored by SDS-PAGE and immunoblotting using monoclonal anti-FLAG antibodies.
Discussion
Flagellar motility in Salmonella enterica serovar Typhimurium is of great importance in respect to biofilm formation, adherence and virulence. Thus, motility is tightly regulated and assembly of flagella occurs in an ordered fashion. There are various levels on which regulation of motility takes place: transcription of the master operon flhDC is regulated by a myriad of factors. Furthermore, FlhDC complex is subject to several posttranscriptional regulation mechanisms. In a recent genome-wide screen for novel motility regulators by Bogomolnaya et al., 130 mutations in the Salmonella Typhimurium genome were implicated to influence motility [31]. Here, we sought to characterize the molecular mechanisms of regulation of selected putative novel motility regulators. Seven of the 24 tested mutants showed an altered motility phenotype in our motility assays, which differ slightly in media composition and inoculation methods from the methodology used in Bogomolnaya et al. as described in detail in Materials and Methods.
For flgE and fliH mutations, it was described that only swimming motility was affected [31]. However, we found that ΔflgE and ΔfliH mutants were non-motile in both swimming and swarming motility agar assays. This result was expected, since FlgE (encoding for the hook protein) and FliH (encoding for an accessory protein of the flagellum-associated ATPase complex) are essential components of the flagellar structure and type-III export apparatus, respectively.
YdiV was shown previously to repress motility posttranslationally via targeted degradation of FlhD [29]. Accordingly, a ydiV deletion resulted in increased motility due to upregulation of FlhDC activity. However, we additionally observed a 50% decrease in swarming motility in the ΔydiV mutant. For Escherichia coli it is known that YdiV represses Pap fimbriae [35]. Therefore, we hypothesized that in Salmonella YdiV might also affect fimbrial expression, leading to a stickier phenotype of the deletion mutant on swarming agar plates. However, type I fimbriae production was not upregulated in the ΔydiV mutant indicating that YdiV is not involved in repression of type I fimbriae in Salmonella. 12 other fimbrial loci are predicted for Salmonella enterica serovar Typhimurium strain LT2, including curli fimbriae, Pef and Std fimbriae [37]. Here, we showed that a ydiV deletion led to increased mRNA levels of csgA and csgB, the major and minor subunits of curli fimbriae. However, in a recent study by Anwar et al. [38] a ΔydiV deletion mutant showed reduced expression of CsgD, the master regulator of biofilm formation, and developed a reduced rdar (red, dry and rough) morphotype on congo red plates. This hints to a reduced expression of curli fimbriae. These contradictory results may be due to different media and growth conditions used. As described for Myxococcus xanthus or Pseudomonas aeruginosa, type IV pili play a role in twitching and gliding motility [39]. We thus suggest that the decreased swarming phenotype of the ΔydiV mutant is due to derepression of curli fimbriae.
We next validated the influence of YjcC on swimming motility as described by Bogomolnaya et al. [31] and observed that YjcC does not affect flagellar gene expression in Salmonella. YjcC is a putative phosphodiesterase containing an EAL-motif (Glu-Ala-Leu). EAL-domain containing proteins degrade of cyclic di(3’→5’)-guanylic acid (c-di-GMP) [40]. C-di-GMP levels are known to influence many mechanisms, e.g. virulence, biofilm formation and motility. In Salmonella Typhimurium and Escherichia coli, motility is regulated through c-di-GMP on a posttranslational level via binding to receptors, including the cellulose synthase BcsA and the c-di-GMP binding protein YcgR [41,42]. Upon binding of c-di-GMP to YcgR, the loose interaction of YcgR with flagellar motor proteins is tightened, resulting in a slow down of flagellar rotation [41,43]. It has been described that a knockout of another EAL-containing protein YhjH (STM3611) reduced swarming motility [44] and that a ΔyjcC mutant exhibits higher c-di-GMP levels [40]. A deletion mutant of yjcC was shown to increase expression of curli fimbriae and cellulose biosynthesis, leading to higher adhesion to glass surfaces, which would provide a possible mechanism for the decrease of motility in the ΔyjcC mutant [40].
The genes STM1267 and STM3363 are not yet characterized in detail, but are conserved among Salmonella species. We showed that both knockouts of STM1267 and STM3363 increased swarming, but not swimming motility. STM3363 encodes for a hypothetical protein that contains a conserved Barstar-like domain, which was shown to play a role in a toxin-antitoxin system in Bacillus subtilis [45]. STM1267 encodes for a hypothetical protein with a conserved domain of the YmgB superfamily. In Escherichia coli, YmgB, also called AriR, represses biofilm formation and decreases cellular motility [46]. We demonstrated that a deletion of STM3363 increased swarming motility independently on filament formation, whereas a ΔSTM1267 mutant showed a higher number of flagella per cell body, which could account for the increased swarming motility phenotype. We cannot exclude that a deletion of STM3363 affected the recently described PagM-mediated flagella-independent surface motility [47].
Finally, we showed a severe defect of a ΔrfaG mutant in both swimming and swarming motility. RfaG is involved in biosynthesis of lipopolysaccharides (LPS), connecting inner and outer core. In particular, the O-antigen of LPS has been reported to contribute to swarming motility for several gram-negative bacteria [48–51]. The core and O-antigen polysaccharide of LPS confers hydrophilicity to the cell surface and in addition, LPS reduces the surface tension between the agar and cell surface (reviewed in [52]). In Escherichia coli it was shown previously that genes involved in biosynthesis of LPS are required for swarming, but not for swimming motility [53]. It was described by Raetz et al. that knockouts of proteins responsible for the construction of the inner core also impaired swimming motility [54], suggesting that flagellar assembly and function may be influenced by altered LPS structures. Here, we show that a ΔrfaG deletion diminished flagellar assembly and significantly reduced transcription of flagellar class II and class III promoters, but not of the class I promoter. Moreover, FlhC protein levels were reduced in the ΔrfaG strain. We conclude that a defect in LPS biosynthesis regulates motility by affecting FlhDC stability or translation of its mRNA on a posttranscriptional level via an unknown mechanism.
In summary, we analyzed in this study the molecular mechanisms by which recently described novel factors modulate motility [31]. We were able to verify the published motility effects of seven of 24 selected putative motility regulators. We hypothesize that a combination of several factors might explain the lack of motility phenotype for several other putative novel motility factors. First, it appears possible that off-site effects of the λ-RED-mediated construction of the single deletion mutants might result in false-positive hits. To exclude secondary site mutations, it is advisable to re-transduce gene deletions constructed by λ-RED-mediated recombination into a clean background. Bogomolnaya et al. [31] did not perform a re-transduction of the deletion mutants of the high-throughput screen, but the authors advised such a procedure for follow-up studies. Second, in Bogomolnaya et al. an antibiotic resistance cassette was inserted in the location of the deleted gene, whereas in this study, the antibiotic resistance cassettes were removed for deletions in ydiV, fliB, STM0971, STM1267, STM1896, STM3363, STM0266, STM0295, STM1575, STM1630, yjcC, STM0289, STM0847, STM1131, STM1268, STM3696, rygD, flgE, fliH, and rfaG in order to minimize the possibility of polar effects. It appears possible that some of the motility effects observed by Bogomolnaya et al. were indirect due to polar effects of the inserted antibiotic resistance cassette. Third, the motility assays in Bogomolnaya et al. were performed using different media compositions for swimming motility plates (e.g. the motility plates of Bogomolnaya et al. contained yeast extract), inoculation methods and time points for quantification when compared to the methodology used here (see also Materials and Methods). It is possible that certain motility phenotypes are specific for the conditions and media composition used in Bogomolnaya et al. and it is also a possibility that—due to the high-throughput screening format of Bogomolnaya et al.–the quantification of small swimming and swarming motility areas might have lead to misinterpretation. Swarming motility is highly influenced by humidity and wetness that may contribute to alterations in motility phenotypes.
However, importantly, we were able to identify possible regulatory mechanisms of several novel factors that were originally described by Bogomolnaya et al. [31] to be involved in motility regulation, in particular of STM1267, STM3363, RfaG, YdiV and YjcC. Among the novel factors implicated by Bogomolnaya et al. [31] to be involved in swimming and swarming motility are many additional genes whose functions are currently unknown or poorly understood. We conclude that careful examination of the described motility defects is needed by independently constructed deletion mutants, but further studies on these genes have great potential to lead to a better understanding of the molecular mechanisms involved in motility of Salmonella.
Materials and Methods
Bacterial strains, plasmids and media
All bacterial strains used in this study are listed in S1 Table and were derived from ATCC14028s. Cells were grown in lysogeny broth (LB) at 37°C, whereas strains harboring a temperature-sensitive λ-RED plasmid were grown at 30°C [55]. Bacterial growth was measured via optical density at 600 nm in a Varioskan Flash plate reader (Thermo Scientific). Motility agar was prepared and swimming motility was examined as described before [56]. Swarming motility of overnight cultures was assayed on 0.6% Difco Bacto Agar (LB Miller base 25 g/l, 0.5% glucose) in closed containers with 100% humidity. The generalized transducing phage of Salmonella enterica serovar Typhimurium P22 HT105/1 int-201 was used in all transductional crosses [57]. Single gene deletions of genes ydiV, fliB, STM0266, STM0295, STM1575, STM1630, yjcC, STM1512, fljA, fimZ, sipA, sptP, flgE, and fliH were produced as previously described [33] and retained 10 codons of the respective coding region (15 bp each at either the 5’ and 3’ ends) in addition to the FRT scar after removal of the antibiotics resistance cassette. The complete coding region was deleted for genes STM3696, STM1131, STM0847, STM1268, STM0971, STM1267, STM1896 and STM3363. The rfaG mutation was inserted as described in [58]. The oligonucleotides used for strain construction are listed in S2 Table.
β-galactosidase assays
β-galactosidase activity was measured as described before using at least three independent biological replicates [59]. Cultures were supplemented with 0.2% arabinose if required for the induction of ParaB expression or 0.1 μg/ml anhydrotetracyclin to induce tetA-dependent expression. For fimbrial gene expression studies the cultures were grown under static growth conditions.
RNA isolation and quantitative real-time PCR
Strains were grown under static growth conditions in LB medium and total RNA isolation was performed using the RNeasy minikit (Qiagen). For removal of genomic DNA, RNA was treated with DNase using the TURBO DNA-free kit (ambion). Reverse transcription and quantitative real-time PCRs (qRT-PCR) were performed using the SensiFast SYBR No-ROX One Step kit (Bioline) in a Rotor-Gene Q lightcycler (Qiagen). Relative changes in mRNA levels were analyzed according to Pfaffl [60] and normalized against the transcription levels of multiple reference genes according to the method described by Vandesompele et al. [61]. The reference genes gyrB, gmk and rpoD were used as previously described [22, 59].
Fluorescent microscopy
Fluorescent microscopy analysis was performed as described before [62]. Logarithmically growing bacteria were fixed onto poly-L-lysine pre-coated coverslips by addition of 2% formaldehyde and 0.2% glutaraldehyde. Flagella were stained using polyclonal anti-FliC antibodies (rabbit) and anti-rabbit conjugated Alexa Fluor 488 secondary antibodies (Invitrogen). The cell membranes were stained using FM-64 (0.5 mg/ml, Invitrogen) and DNA staining was performed using DAPI (Sigma-Aldrich). Images were taken using an Axio Observer microscope with an Axiocam HR camera (Zeiss) at 1000x magnification. Images were analyzed and contrast and brightness were adjusted using ImageJ (http://rsbweb.nih.gov/ij/).
Electron microscopy
Strains were cultured in 5 ml LB or minimal media overnight. 650 μl glutaraldehyde (2%) was added to fixate the bacteria. The mixture was stored in the fridge at 4°C. For TEM observation bacteria were negatively stained with 2% aqueous uranyl acetate using a carbon film deposited on mica. Samples were examined in a Zeiss TEM 910 at an acceleration voltage of 80 kV with calibrated magnifications. Images were recorded digitally with a Slow-Scan CCD-Camera (ProScan, 1024 x 1024) with ITEM-Software (Olympus Soft Imaging Solutions). Contrast and brightness were adjusted with Adobe Photoshop CS3.
Analysis of protein levels
A single colony of the analyzed strains was cultured in LB medium overnight, diluted 1:100 into fresh LB medium and grown for 120–210 min. 2 ml samples of the culture were taken every 30 min and whole cell lysates were prepared by normalizing the samples to measured OD600 values using SDS sample buffer. Protein levels were analyzed by SDS polyacrylamide gel electrophoresis and immunoblotting using anti-FLAG antibodies (Sigma).
Supporting Information
Growth was measured via absorption at 600 nm every 15 minutes for 13 h with at least three biological replicates of each mutant strain (EM880, EM1480, EM1481, EM1482, EM1484, EM1507, EM1508, EM1509, EM1510, EM1511, EM1512, EM1686, EM1688, EM1689, EM1690, EM1691, EM2381, EM2382, EM2383, EM2384, EM2385, EM2590, EM2591 and EM2605). The dotted lines represent the standard error of the mean.
(TIF)
Swimming motility of single gene deletion mutants in STM1896 (EM1482 ΔSTM1896::FRT), STM1630 (EM1510 ΔSTM1630::FRT) and STM3696 (EM1690 ΔSTM3696::FRT) was monitored over time to exclude incubation time dependent swimming behaviors. The motility assay was performed with four biological replicates.
(TIF)
The Salmonella enterica serovar Typhimurium strains ATCC14028s, SL1344, LT2 and SJW1103 (TH6622, EM774, TH437 and TH8362) were examined on swimming motility agar. The diameter of the motility swarm was measured and normalized to ATCC14028s. Biological replicates are shown as individual data points.
(TIF)
Transcription of class I (flhC-MudJ), class II (fliL-MudJ) and class III (fliC-MudJ) promoters were monitored for wildtype strains (EM584, EM2586 and EM2585) and ΔyjcC::FRT mutants (EM2587, EM2588 and EM2589) by β-galactosidase assay. Three independent biological samples were analyzed. Error bars represent the standard errors of the means.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Significantly increased or decreased motility phenotypes are shown in bold.
(DOCX)
Acknowledgments
We are grateful to Kelly T. Hughes for the generous donation of strains and advice and we thank Michael Kolbe for sharing strain SL1344. We thank Nadine Körner and Ina Schleicher for expert technical assistance and Florian D. Fabiani for advice and helpful comments on the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Helmholtz Association Young Investigator grant number VH-NG-932 and the People Programme (Marie Curie Actions) of the European Unions’s Seventh Framework Programme grant number 334030 (to ME). JAD, IS and CK acknowledge support by the President’s Initiative and Networking Funds of the Helmholtz Association of German Research Centers (HGF) under contract number VH-GS-202. SF was funded in the Zoonosis PhD program via a Lichtenberg Fellowship from the Niedersächsiche Ministerium für Wissenschaft und Kultur (MWK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rubin RH, Einstein L. Salmonellosis: Microbiologic, Pathogenic, and Clinical Features. Stratton Intercontinental Medical Book Corp, New York: 1977. [Google Scholar]
- 2. Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000; 54:735–774. [DOI] [PubMed] [Google Scholar]
- 3. Galán J, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999; 284:1322–1328. [DOI] [PubMed] [Google Scholar]
- 4. Adler J, Templeton B. The effect of environmental conditions on the motility of Escherichia coli . J Gen Microbiol. 1967; 46:175–184. [DOI] [PubMed] [Google Scholar]
- 5. Duan Q, Zhou M, Zhu L, Zhu G. Flagella and bacterial pathogenicity. JBasic Microbiol. 2013; 53:1–8. [DOI] [PubMed] [Google Scholar]
- 6. Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003; 72:19–54. [DOI] [PubMed] [Google Scholar]
- 7. Blair DF, Berg H. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Annu Rev Biochem. 1990; 60:439–449. [DOI] [PubMed] [Google Scholar]
- 8. Blocker A, Komoriya K, Aizawa S. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc Natl Acad Sci USA. 2003; 100:3027–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Akeda Y, Galán JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005; 437:911–915. [DOI] [PubMed] [Google Scholar]
- 10. Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature. 2008; 451:485–488. 10.1038/nature06449 [DOI] [PubMed] [Google Scholar]
- 11. Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. Energy source of flagellar type III secretion. Nature. 2008; 451:489–492. 10.1038/nature06497 [DOI] [PubMed] [Google Scholar]
- 12. Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, et al. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006; 188:2233–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J Mol Biol. 2006; 355:798–808. [DOI] [PubMed] [Google Scholar]
- 14. Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994; 176:7345–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993; 262:1277–1280. [DOI] [PubMed] [Google Scholar]
- 16. Chadsey MS, Karlinsey JE, Hughes KT. The flagellar anti-sigma factor FlgM actively dissociates Salmonella typhimurium sigma28 RNA polymerase holoenzyme. Genes Dev. 1998; 12:3123–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chadsey MS, Hughes KT. A multipartite interaction between Salmonella transcription factor sigma28 and its anti-sigma factor FlgM: implications for sigma28 holoenzyme destabilization through stepwise binding. J Mol Biol. 2001; 306:915–929. [DOI] [PubMed] [Google Scholar]
- 18. Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993; 262:1277–1280. [DOI] [PubMed] [Google Scholar]
- 19. Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, et al. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol. 1999; 181:7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ko M, Park C. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J Bacteriol. 2000; 182:4670–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stojiljkovic I, Baumler AJ, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J Mol Biol. 1994; 236:531–545. [DOI] [PubMed] [Google Scholar]
- 22. Singer HM, Kühne C, Deditius JA, Hughes KT, Erhardt M. The Salmonella Spi1 virulence regulatory protein HilD directly activates transcription of the flagellar master operon flhDC . J Bacteriol. 2014; 196:1448–1457. 10.1128/JB.01438-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ellermeier CD, Slauch JM. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J Bacteriol. 2003; 185:5096–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli . Mol Microbiol. 2002; 45:521–532. [DOI] [PubMed] [Google Scholar]
- 25. Spory A, Bosserhoff A, von Rhein C, Goebel W, Ludwig A. Differential regulation of multiple proteins of Escherichia coli and Salmonella enterica serovar Typhimurium by the transcriptional regulator SlyA. J Bacteriol. 2002; 184:3549–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Libby SJ, Goebel W, Ludwig A, Buchmeier N, Bowe F, Fang FC, et al. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc Natl Acad Sci USA. 1994; 91:489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saini D, Brown JD, Aldridge PD, Rao CV. FliZ is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J Bacteriol. 2008; 190:4979–4988. 10.1128/JB.01996-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wada T, Tanabe Y, Kutsukake K. FliZ acts as a repressor of the ydiV gene, which encodes an anti-FlhD4C2 factor of the flagellar regulon in Salmonella enterica serovar Typhimurium. J Bacteriol. 2011; 193:5191–5198. 10.1128/JB.05441-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wada T, Morizane T, Abo T, Tominage A, Inoue-Tanaka K, Kutsukake K. EAL domain protein YdiV acts as an anti- FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J Bacteriol. 2011; 193:1600–1611. 10.1128/JB.01494-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takaya A, Erhardt M, Karata K, Winterberg K, Yamamoto T, Kughes KT. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol Microbiol. 2012; 83:1268–1284. 10.1111/j.1365-2958.2012.08007.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bogomolnaya LM, Aldrich L, Ragoza Y, Talamantes M, Andrews KD, McClelland M, et al. Identification of novel factors involved in modulating motility of Salmonella enterica serotype Typhimurium. PloS One. 2014; 9:e111513 10.1371/journal.pone.0111513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Santiviago CA, Reynolds MM, Porwollik S, Choi S-H, Long F, Andrews-Polymenis HL, et al. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PloS Path. 2009; 5:e1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Minamino T, González-Pedrajo B, Kihara M, Namba K, Macnab RM. The ATPase FliI can interact with the Type III flagellar protein export apparatus in the absence of its regulator, FliH. J Bacteriol. 2003; 185:3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spurbeck RR, Alteri CJ, Himpsl SD, Mobley HLT. The multifunctional protein YdiV represses P fimbria-mediated adherence in uropathogenic Escherichia coli . J Bacteriol. 2013; 195:3156–3164. 10.1128/JB.02254-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kisiela DI, Kramer JJ, Tchesnokova V, Aprikian P, Yarov-Yarovov V, Clegg S, et al. Allosteric catch bond properties of the FimH adhesion from Salmonella enterica serovar Typhimurium. J Biol Chem. 2011; 286:37136–38147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001; 413:852–856. [DOI] [PubMed] [Google Scholar]
- 38. Anwar N, Rouf SF, Römling U, Rhen M. Modulation of Biofilm-Formation in Salmonella enterica Serovar Typhimurium by the Periplasmic DsbA/DsbB Oxidoreductase System Requires the GGDEF-EAL Domain Protein STM3615. PLoS ONE 9(8): e106095 2014. 10.1371/journal.pone.0106095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999; 32:01–10. [DOI] [PubMed] [Google Scholar]
- 40. Simm R, Lusch A, Kader A, Andersson M, Römling U. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J Bacteriol. 2007; 189:3613–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, et al. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010; 141:107–116. 10.1016/j.cell.2010.01.018 [DOI] [PubMed] [Google Scholar]
- 42. Fang X, Gomelsky M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol. 2010; 76:1295–1305. 10.1111/j.1365-2958.2010.07179.x [DOI] [PubMed] [Google Scholar]
- 43. Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell. 2010; 38:128–139. 10.1016/j.molcel.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simm R, Morr M, Kader A, Nimtz M, Römling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004; 53:1123–1134. [DOI] [PubMed] [Google Scholar]
- 45. Hartley RW, Smeaton JR. On the reaction between the extracellular ribonuclease of Bacillus amyloliquefaciens (barnase) and its intracellular inhibitor (barstar). J Biol Chem. 1973; 248:5624–5626. [PubMed] [Google Scholar]
- 46. Lee J, Page R, Garcia-Contreras R, Palermino JM, Zhang XS, Doshi O, et al. Structure and function of the Escherichia coli protein YmgB: a protein critical for biofilm formation and acid-resistance. J Mol Biol. 2007; 373:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park SY, Pontes MH, Groisman EA. Flagella-independent surface motility in Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci USA. 2015; 112:1850–1855. 10.1073/pnas.1422938112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gygi D, Rahmann MM, Lai HC, Carlson R, Guard-Petter J, Hughes C. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis . Mol Microbiol. 1995; 17:1167–1175. [DOI] [PubMed] [Google Scholar]
- 49. Mireles JR, Toguchi A, Harshey RM. Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J Bacteriol. 2001; 183:5848–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Toguchi A, Siano M, Burkart M, Harshey RM. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J Bacteriol. 2000; 183:6308–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Q, Frye JG, McClelland M, Harshey RM. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol Microbiol. 2004; 52:169–187. [DOI] [PubMed] [Google Scholar]
- 52. Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol. 2003; 57:249–273. [DOI] [PubMed] [Google Scholar]
- 53. Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J Bacteriol. 2007; 189:950–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002; 71:635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bertani G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol. 2004; 186:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wozniak CE, Lee C, Hughes KT. T-POP array identifies EcnR and PefI-SrgD as novel regulators of flagellar gene expression. J Bacteriol. 2008; 191:1498–1508. 10.1128/JB.01177-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sanderson KE, Roth JR. Linkage map of Salmonella typhimurium, Edition VI. Microbiol Rev. 1983; 47:410–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss R. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar Typhimurium. Infect Immun. 2011; 79:4227–4239. 10.1128/IAI.05398-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Singer HM, Erhardt M, Hughes KT. RflM functions as transcriptional repressor in the autogenous control of the Salmonella flagellar master operon flhDC . J Bacteriol. 2013; 195:4274–4282. 10.1128/JB.00728-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001; 29:e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 2002, research 0034.1–0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica . The EMBO Journal. 2011; 30:2948–2961. 10.1038/emboj.2011.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth was measured via absorption at 600 nm every 15 minutes for 13 h with at least three biological replicates of each mutant strain (EM880, EM1480, EM1481, EM1482, EM1484, EM1507, EM1508, EM1509, EM1510, EM1511, EM1512, EM1686, EM1688, EM1689, EM1690, EM1691, EM2381, EM2382, EM2383, EM2384, EM2385, EM2590, EM2591 and EM2605). The dotted lines represent the standard error of the mean.
(TIF)
Swimming motility of single gene deletion mutants in STM1896 (EM1482 ΔSTM1896::FRT), STM1630 (EM1510 ΔSTM1630::FRT) and STM3696 (EM1690 ΔSTM3696::FRT) was monitored over time to exclude incubation time dependent swimming behaviors. The motility assay was performed with four biological replicates.
(TIF)
The Salmonella enterica serovar Typhimurium strains ATCC14028s, SL1344, LT2 and SJW1103 (TH6622, EM774, TH437 and TH8362) were examined on swimming motility agar. The diameter of the motility swarm was measured and normalized to ATCC14028s. Biological replicates are shown as individual data points.
(TIF)
Transcription of class I (flhC-MudJ), class II (fliL-MudJ) and class III (fliC-MudJ) promoters were monitored for wildtype strains (EM584, EM2586 and EM2585) and ΔyjcC::FRT mutants (EM2587, EM2588 and EM2589) by β-galactosidase assay. Three independent biological samples were analyzed. Error bars represent the standard errors of the means.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Significantly increased or decreased motility phenotypes are shown in bold.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.