Abstract
The human genome contains a variant form of the α7-nicotinic acetylcholine receptor (α7nAChR) gene that is uniquely human. This CHRFAM7A gene arose during human speciation and recent data suggests that its expression alters ligand tropism of the normally homopentameric human α7-AChR ligand-gated cell surface ion channel that is found on the surface of many different cell types. To understand its possible significance in regulating inflammation in humans, we investigated its expression in normal human leukocytes and leukocyte cell lines, compared CHRFAM7A expression to that of the CHRNA7 gene, mapped its promoter and characterized the effects of stable CHRFAM7A overexpression. We report here that CHRFAM7A is highly expressed in human leukocytes but that the levels of both CHRFAM7A and CHRNA7 mRNAs were independent and varied widely. To this end, mapping of the CHRFAM7A promoter in its 5′-untranslated region (UTR) identified a unique 1-kb sequence that independently regulates CHRFAM7A gene expression. Because overexpression of CHRFAM7A in THP1 cells altered the cell phenotype and modified the expression of genes associated with focal adhesion (for example, FAK, P13K, Akt, rho, GEF, Elk1, CycD), leukocyte transepithelial migration (Nox, ITG, MMPs, PKC) and cancer (kit, kitL, ras, cFos cyclinD1, Frizzled and GPCR), we conclude that CHRFAM7A is biologically active. Most surprisingly however, stable CHRFAM7A overexpression in THP1 cells upregulated CHRNA7, which, in turn, led to increased binding of the specific α7nAChR ligand, bungarotoxin, on the THP1 cell surface. Taken together, these data confirm the close association between CHRFAM7A and CHRNA7 expression, establish a biological consequence to CHRFAM7A expression in human leukocytes and support the possibility that this human-specific gene might contribute to, and/or gauge, a human-specific response to inflammation.
INTRODUCTION
In 1979, Richman described the presence of a functionally distinct nicotinic acetylcholine receptor (AChR) on human lymphocytes that appeared to have altered ligand binding (1). As recently reviewed by Costantini et al. (2) and Sinkus et al. (3), it was almost twenty years later that Gault et al. (4) sequenced the human α7nAChR gene on chromosome 15q13–14 and found it to be structurally similar to that of all other species. At the same time, however, they noted the presence of a second, human-specific partially duplicated α7nAChR-like gene that localized 1.6 Mb 5′ upstream from human CHRNA7 (4). With only 386 amino acids of the α7nAChR channel domain, this new human-specific gene was initially called dupα7nAChR and found to encode an amino terminus that originated from a kinase gene on chromosome 3 (5). The ultimate genetic rearrangement, which occurred after the divergence of humans from other primates (6,7), created a new, distinct and human-specific open reading frame (ORF) that produces an exclusively human α7nAChR now called CHRFAM7A (8). Many species, including human, great apes, mice and rats have orthologs of CHRNA7 that are generated by alternative splicing of their respective CHRNA7 mRNA. However, only the human genome has a distinct CHRFAM7A gene that can gauge the function and ligand tropism of the normal α7nAChR ligand-gated channel (3,9).
Since its discovery in 1998, CHRFAM7A has largely been the focus of neuroscience and mental health research because historically the α7nAChR was viewed as a neuron-specific, ligand-gated ion channel. More recently, however, its detection in normal human leukocytes (10,11) has gained particular attention because several in vitro studies have shown that CHRFAM7A modifies α7nAChR channel activity and changes ligand tropism (12–14). Because α7nAChR activation is closely tied to the inflammatory responses of peripheral tissues, these observations raise the possibility that CHRFAM7A may be particularly relevant to gauging inflammation in humans. The Tracey laboratories, for example, established that efferent signaling of the vagus nerve acts exclusively via α7nAChR activation in the spleen to regulate systemic cytokine responses to infection in mice (15–20). Similarly, Costantini and colleagues demonstrated the existence of a similar α7nAChR-dependent regulation of the local inflammatory response in tissues (21–27). With α7nAChR activation clearly essential to inflammation, not to mention vagus nerve responsiveness and leukocyte function (28,29), we reasoned that it was therefore critical to understand how a human-specific α7nAChR in human leukocytes might influence human leukocyte function, the regulation of its expression and the biological consequences of its expression. Because newly evolved genes such as CHRFAM7A disproportionately segregate with complex human disease (30,31), the results point to the possible existence of human-specific responses to inflammation that are not present in other species. If so, CHRFAM7A may gauge the α7nAChR-mediated effects of the human vagus nerve.
MATERIALS AND METHODS
Materials
The plasmid encoding the full-length CHRFAM7A (variant 1: NM_139320.1) was from OriGene (Rockville, MD, USA) and differs from a shorter CHRFAM7A (variant 2: NM_148911) in that it encodes a 90-amino-acid amino terminus containing the FAM7A sequence (NH2-MQKYCIYQHF QFQLLIQHLW IAANCDI) and a α7nAChR amino terminus peptide (ADE RFDATFHTNV LVNSSGHCQY LPPGIFKSSC YIDVRWFPFD VQHCKLKFGS WSYGGWSLDL). The plasmid encoding full-length CHRNA7 (variant 2: NM_001190455) was from GeneCopoeia (Rockville, MD, USA) and differs from the shorter CHRNA7 (variant 1: NM_000746) by codons that encode a GKATASPPSTPPWDPGHIPGASVRPAPGP peptide introduced at His18 of the α7nAChR subunit. The pGL4 promoterless expression plasmid encoding firefly luciferase was purchased from Promega (Madison, WI, USA). All other chemicals and reagents were the products of Sigma-Aldrich (St. Louis, MO, USA) unless specified otherwise.
Human Peripheral Leukocytes
Informed consent was obtained from healthy volunteers for the collection of peripheral blood. Volunteers were recruited and enrolled by the University of California San Diego (UCSD) Clinical Translational Research Institute (San Diego, CA, USA). Venous blood was collected by peripheral venipuncture in BD Vacutainer blood collection tubes containing ethylenediaminetetraacetic acid (EDTA) (BD Biosciences, San Jose, CA, USA) and placed on ice. Red blood cells were lysed using BD Pharm Lyse ammonium chloride solution (BD Biosciences) at room temperature for 15 min and leukocytes pelleted by centrifugation. Cell pellets were stored at −80°C until further analyses. The UCSD Institutional Review Board approved the enrollment of participants, consent forms and specimen collection protocols.
Cell Culture
All cell lines were originally purchased from ATCC (Manassas, VA, USA) and/or acquired through the UCSD Department of Surgery, Division of Trauma, Burns and Acute Care Surgery Cell Repository (San Diego, CA, USA). Thawed cells were washed in RPMI culture media containing 10% fetal calf serum (FCS), the pellet reconstituted in culture media and cells plated into six-well tissue culture plates. All cells were washed 48 h later and allowed to grow to 90% confluence and propagated with trypsin digestion as needed. For transduction studies, cells were seeded at 2 × 106 in 6-well tissue culture plates the day before the experiment. As indicated in each experiment, cells were harvested directly from the culture dishes for total RNA preparation, processed for stable or transient transfection or treated with 100 ng/mL LPS (L4391, Sigma-Aldrich) for 3 h. At the end of incubations, cells were harvested, total RNA isolated and the cDNA generated (see below) used for analyses of gene expression.
Lentivirus Constructs for CHRFAM7A Expression
The ORF of human CHRFAM7A (variant 1) was amplified by PCR from the CHRFAM7A-pCMV6-Entry plasmid (PS100001, OriGene, Rockville, MD, USA) using: forward primer: 5′-aGTcC TCGAGATGcaaaaatattgcatct-3′; reverse primer: 5′-attcGGATCCTTACGCAAAG TCTTTGGACACGGC-3′.
The PCR products were purified and cloned into the bicistronic pLVX-IRES-ZsGreen1 lentivirus plasmid as recommended by the manufacturer. The identity of the plasmid was confirmed by DNA sequencing (Retrogen, San Diego, CA, USA). Lentivirus was packaged using the Lenti-X HTX Packaging System (631247, Clontech, Mountain View, CA, USA) following vendor instructions. After 48 h, the supernatant containing virus was collected and used to transduce THP1 cells. Stable transfected green fluorescent protein (GFP)-positive THP1 cells were detected by fluorescence microscopy and purified by cell sorting. Cells were expanded without further selection and the stability of transduction monitored by weekly flow cytometry.
Flow Cytometry and Cell Sorting of THP1 Cells
For flow cytometry analyses, cells were washed and fixed with Cytofix according to the manufacturer’s recommendations (BD Biosciences) for 10 min on ice. Cells were then incubated with labeled bungarotoxin (BD Biosciences) in FACS buffer (1% bovine serum albumin [BSA] in phosphate buffered saline [PBS] containing 0.005% sodium azide) and washed in FACS buffer. Flow cytometry was performed with a Becton Dickinson FACSCalibur and data analysis performed with CellQuestPro software from Becton, Dickinson and Company (BD) (Franklin Lakes, NJ, USA), processed and analyzed using JFlow. To purify GFP-expressing THP1 cells, the transduced cells were sorted twice by FACS at the core facilities of the Center for AIDS Research at UCSD, selected for GFP expression and expanded as cell suspensions. Stable expression was monitored weekly for retention of > 85% cells expressing GFP as measured by flow cytometry. Cells were propagated in 10% RPMI1640.
RNAseq, Gene Expression and Pathway Analyses
Total RNA was prepared from transduced and sorted THP1 cells using RNeasy kit (Qiagen, San Diego, CA, USA) and was quantified using a NanoDrop Spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). One μg total RNA was used for RNAseq analyses and performed by contract with the Genomics Core, Cedars-Sinai Medical Center (Los Angeles, CA, USA). Bioinformatic analyses, differential gene expression and pathway analyses were performed by contract with Accura-Science (Johnston, IA, USA). For datasets and RNA-seq differential expression (DE) analysis, the BAM files for vector-and stable CHRFAM7A-transduced cells were generated by RNA-seq at the genome core facilities at Cedars-Sinai Genomics core at Cedars Sinai Medical Center and were used for differential gene expression analyses. Comparisons were made between CHRFAM7A and vector using two methods to define differentially expression genes. DESeq (32) is one of the few methods suitable with limited replicates (33) and controls for false positive signals (34,35). The Python package HTseq was used to produce the count table and a P value <0.05 was set as cutoff. In the second method, the results of DESeq were overlapped with Cuffdiff (36) and a P value <0.05 was chosen as cutoff. The differentially expressed gene groups defined by both analytical methods were for functional enrichment analysis and GOseq in bioconductor was used for gene ontology analysis (37) with up- and downregulated differentially expressed genes respectively. A P value cutoff of 0.05 was used to choose significant gene ontology (GO) terms. The Functional Class Scoring method implemented in GSEABase was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses (38) and a P value of <0.05 was used to define significant pathway categories.
Isolation of RNA from Cultured Cells and Preparation of cDNA for PCR and qPCR
Total RNA was prepared from cell lysates using the RNeasy kit and was quantified using a NanoDrop Spectrophotometer. One μg of the total RNA was reversed transcribed using iScript cDNA synthesis kit (Bio-Rad, San Diego, CA, USA) in a 20 μL reaction as described by the manufacturer and 1 μL was used for RT-PCR or real-time qPCR analyses.
RT-PCR and Quantitative RT-PCR for CHRFAM7A and CHRNA7
RT-PCR was performed in a 50 μL reaction containing 45 μL PCR blue mix (Invitrogen [Thermo Fisher Scientific]), 1 μL of each primer (10 μmol/L), 1 μL cDNA and 2 μL water. The cycling conditions were 94°C for 4 min followed by 35 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 60 s and a final extension at 72°C for 5 min. Ten μL of each PCR product were resolved on a 2% agarose gel and images were acquired using an Alpha Innotech imaging system (Fisher Scientific [Thermo Fisher Scientific]). Real-time qPCR was performed in a 25 μL reaction containing 12.5 μL 2× SYBR Green PCR Master Mix (Bio-Rad), 0.5 μL of each primer (10 μmol/L), 1 μL cDNA and 10.5 μL water. PCR cycling conditions were: 95°C for 10 min followed by 45 cycles of 94°C for 25 s, 60°C for 25 s and 72°C for 40 s. Primer efficiency for CHRFAM7A and CHRNA7 were 100% and 94% respectively.
Primers for CHRFAM7A were: sense, 5′-ATAGCTGCAAACTGCGATA-3′; antisense, 5′-cagcgtacatcgatgtagcag-3′. Primers for CHRNA7 were: sense, 5′-acATGcgctgctcgccggga-3′; antisense, 5′-gattgtagttcttgaccagct-3′. Primers for GAPDH were: sense, 5′-CATGAGAAGT ATGACAACAGCCT-3′; antisense, 5′-AGTCCTTCCACGATACCAAAGT-3′.
5′ RACE and Identification of CHRFAM7A Variant 1
5′ RACE was performed using SMARTer RACE cDNA Amplification Kit (Clontech) following vendor’s instructions. Briefly, total RNA was prepared from THP1 cell with RNeasy kit (Qiagen). Three μg total RNA was processed for mRNA using PolyA Spin mRNA Isolation Kit (New England Biolabs [NEB], Ipswich, MA, USA). One-fifth of the poly A mRNA was reverse transcribed and the resulting cDNA was amplified sequentially by PCR and nested PCR. Both gene-specific primers (GSP), GSP and nestGSP listed below, hybridize to the fifth exon of human CHRNA7/CHRFAM7A, with nestGSP 5′ to the GSP without overlapping. The nested PCR products were purified and cloned into pDrive (Qiagen). Colonies were sequenced to identify the 5′ initiation sites and the 5′ sequence upstream the fifth exon of CHRNA7/CHRFAM7A (GSP [5′-GCAGG TACTGGCAATGCCCAGAAG-3′], NestGSP [5′-TAGTGTGGAATGTGG CGTCAAAGCG-3′]).
Analyses of the CHRFAM7A Promoter
The putative CHRFAM7A promoter region spanning from −2363 to +22 relative to the open reading frame ATG start codon was amplified by PCR of genomic DNA isolated from HEK293 cells. The longest fragment was cloned into pGL4 promoterless luciferase reporter plasmid (Promega) according to the manufacturer’s specifications and the resulting plasmid, pGL4-CHRFAM7A (~2400 bp), was confirmed by DNA sequencing and thereafter referred to as f2400 to reflect its size.
The primers were: sense, 5′-ATCAG CTAGCTCTAGATAGACAGCATTTTA-3′ containing a NheI restriction site; antisense, 5′-GCATAGATCTGGTAGATGCA ATATTTTTGCAT-3′ containing a BglII restriction site.
Three serial 5′ deletion promoter constructs of 1800, 1000 and 500 bp were derived by PCR of the f2400 template using the same antisense primer described above, with one of three sense primers to obtain: f1800 (5′-ATCAGCTAGCAAGCC TTCATCAGTGGAAAT-3′); f1000 (5′-ATCAGCTAGCGTATGACTCAAGTCC TTGAC-3′); f500 (5′-ATCAGCTAGC CTTGCTGTATTCTCTAAACTA-3′).
The fragments generated were cloned into the pGL4 vector to create plasmids f1800, f1000 and f500, which were each sequenced to confirm their identity. These plasmids were then transiently transfected into THP1 cells as described below and luciferase activity was analyzed 30 h after transfection following the manufacturer’s instructions (Promega). Luciferase activity was normalized to protein concentration and the data presented as relative luciferase activity compared with the activity of promoterless pGL4 transfected cells.
Transfections of THP1 Cells for Promoter Analyses
THP1 cells, cultured in RPMI1640 supplemented with 1× GlutaMAX (Life Technologies [Thermo Fisher Scientific]) and 1× penicillin/streptomycin, were seeded at 5 × 105 per well in a 24-well plate 2 h before transfection. Transient transfection was performed using Lipofectamine 2000 (Invitrogen [Thermo Fisher Scientific]). Briefly, 2.5 μL of the Lipofectamine 2000 was added into 50μL OPTI-MEM (Invitrogen [Thermo Fisher Scientific]), vortexed for 5 s and then continued to incubate at room temperature for 5 min. One μg plasmid diluted into 50 μL OPTI-MEM was added into the above mixture, vortexed for 5 s and incubated at room temperature for 20 min. The DNA-complex was then added dropwise to cells and cells were incubated for 30 h. Cells were washed with PBS and lysed with 100 μL Passive Lysis buffer at room temperature for 30 min with shaking. The lysate was spun down and 10 μL of the supernatant was used for luciferase assay on POLARstar Omega plate reader (BMG Labtech, Cary, NC, USA). Luciferase activity normalized to protein concentration was expressed as fold changes over that of pGL4-transfected cells.
Statistical Analyses
CHRNA7 and CHRFAM7A gene expression were normalized to that of GAPDH and the fold changes calculated by ΔΔCt method using the relative expression software tool (REST) for group-wise comparisons and statistical analyses of relative expression results (39). The GraphPad (GraphPad Software Inc., La Jolla, CA, USA) program Prism 6 was used for other analyses including two-tailed unpaired t test, Wilcoxon signed rank test of unpaired t test and analysis of variance (ANOVA) as indicated in the text and in legends to the figures.
All supplementary materials are available online at www.molmed.org.
RESULTS
Detection of CHRFAM7A and CHRNA7 Expression in Human Leukocytes
During analysis of human-specific gene expression in leukocytes purified from the blood of normal volunteers, we examined whether we could specifically detect concomitant expression of mRNAs encoding CHRFAM7A and CHRNA7 (Figure 1). As described in Materials and Methods and illustrated in Figure 1A, the full-length CHRFAM7A (variant 1, NM_139320.1) differs from the shorter CHRFAM7A (variant 2, NM_148911) in that it encodes a unique FAM7A peptide sequence (NH2-MQKYCIYQHF QFQLLIQHLW IAANCDI). Accordingly, PCR primers could be designed to specifically hybridize with CHRFAM7A variant 1. Because the CHRFAM7A variant 2 mRNA does not contain unique sequences that allow it to be differentiated from either CHRFAM7A (variant 1) or CHRNA7 (see Figure 1A), no specific RT-PCR primers could be designed to detect CHRFAM7A (variant 2) so we used 5′ RACE (see below).
Figure 1.
Expression of CHRNA7 and CHRFAM7A in human leukocytes. In humans, there are two reported variants of both CHRFAM7A and CHRNA7 and the PCR primers can help distinguish the longer variant 2 from variant 1 of CHRNA7 based on size (A), they do detect or differentiate variant 2 of CHRFAM7A. RT-PCR of mRNA from human leukocytes cells isolated from 7 patients (lanes 1–7) was used to identify the presence of (B) the duplicate α7-nicotinic acetylcholine receptor (CHRFAM7A), (C) the human α7-nicotinic acetylcholine receptor (CHRNA7 ) or (D) GAPDH mRNA. Arrows show the expected size of the amplified sequence from the cognate plasmids. (E) This panel shows the results from quantitative RT-PCR for both CHRFAM7A and CHRNA7. Concentrations were determined against a plasmid standard curve generated for each gene and then expressed as copy number/μg of total mRNA amplified in the starting material. (NS = nonsignificant using two-tailed unpaired t test).
In contrast, the full-length CHRNA7 mRNA (variant 2: NM_001190455) differs from the shorter CHRNA7 mRNA (variant 1: NM_000746) by encoding a GKATASPPSTPPWDPGHIPGASVRP APGP peptide that allows both transcripts differentiated by size when analyzed by gel electrophoresis (see Figure 1A). They are also measured simultaneously by quantitative RT-PCR. As shown in Figure 1B, PCR of leukocyte cDNA prepared from the mRNA of seven volunteers established the presence of CHRFAM7A mRNA in all samples (see Figure 1B). CHRNA7 was readily detected in four of seven samples (Figure 1C) and in all samples with longer exposures (not shown). Increased PCR cycle numbers showed ubiquitous expression of CHRNA7. Interestingly, leukocytes from different individuals appeared to express different combinations of CHRNA7 variants 1 and 2. For example, three samples (lanes 3, 4 and 7) had the longer CHRNA7 variant 1 transcript while a fourth (lane 4) only had variant 2 (see Figure 1C). Because no difference in the signal was obtained in the analyses of GAPDH (Figure 1D), the results suggest significant individual variability in expression of both leukocyte CHRNA7 and CHRFAM7A. On average, however, the differences detected between the levels of both genes are not statistically different in normal human leukocytes (Figure 1E). Both CHRNA7 and CHRFAM7A gene expression is markedly variable, ranging from 100 to 20,000 copies/μg of starting mRNA. Any differences in the ratio CHRFAM7A and CHRNA7 gene expression, which ranges as much as 10,000-fold between different donors (N = 22), is most striking when paired expression of CHRFAM7A to CHRNA7 is assessed although the mean differences suggest that as much CHRFAM7A is expressed as CHRNA7, on average.
Identification of the CHRFAM7A Variant 1 mRNA Transcript in THP1 Cells
The 5′ RACE method was used to extend the CHRFAM7A cDNA clones from THP1 cells to detect CHRFAM7A variant 1 (see above) and to identify the 5′ UTR sequences of the corresponding CHRFAM7A mRNAs that might regulate gene expression. As shown in Figure 2A, we were able to identify several CHRFAM7A variant 1 transcripts expressed in THP1 cells but did not detect variant 2. We detected four transcription initiation sites at −94, −206, −356 and −446 bp 5′ to the CHRFAM7A ORF. This sequencing also allowed us to deduce the primary sequence of leukocyte CHRFAM7A protein (Figures 2B, D), which demonstrated that it is identical to the variant 1 that is published in public genomic databases. These data also established the difference with the protein encoded by the CHRNA7 gene (Figure 2C). Finally, the use of 5′ RACE allowed us to focus on 5′ untranslated region (5′ UTR) sequences that are responsible for the start of CHRFAM7A transcription and the potential promoter elements regulating CHRFAM7A gene expression.
Figure 2.
Identification of CHRFAM7A in THP1 cells. (A) 5′ RACE of human THP1 cells identified 11 of 14 CHRFAM7A transcripts sequenced as initiating at −446 bp from the CHRFAM7A open reading frame (A of the ATG = +1). (B) Translation of the DNA sequence established that the deduced CHRFAM7A protein has a unique 27 amino acid sequence that distinguishes CHRFAM7A from the amino terminus of CHRNA7. The CHRFAM7A exons and the amino acid sequence they encode are shown in blue and red. (C) The CHRNA7 gene encodes two protein variants that differ by the underlined sequence. (D) The common 386 amino acid sequence shared by both CHRFAM7A and CHRNA7 also shows the sequence encoded by the variant 2 of CHRFAM7A which is 100% identical between both genes but not detected in the RT-PCR experiments described here or observed after 5′RACE of THP1 cells.
CHRFAM7A and CHRNA7 Gene Expression in Leukocyte Cell Lines
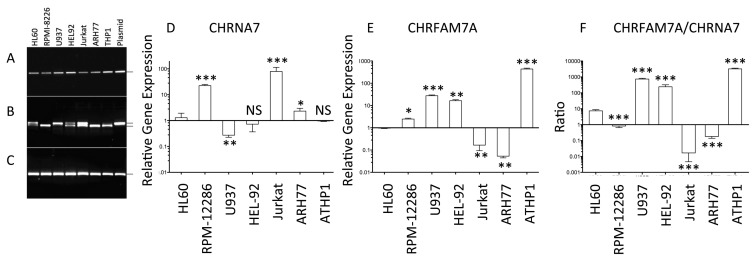
We used RT-PCR to survey the expression of both CHRFAM7A and CHRNA7 in human leukocyte lines (Figure 3). We found that HL60, RPMI-2286, U937, HEL92, Jurkat and ARH77 are like the premonocytic THP1 cell line and express both CHRFAM7A (Figure 3A) and CHRNA7 (Figure 3B) when compared with GAPDH (Figure 3C). As in normal leukocytes (see Figure 1), we detect both variant 1 and variant 2 mRNA transcripts of CHRNA7 (see Materials and Methods) depending on the cell evaluated. This raises the possibility that heteropentameric α7nAChRs exists on the leukocyte cell surface containing up to three distinct subunits: CHRNA7 variant 1; CHRNA7 variant 2; and CHRFAM7A. Three cell lines (HL60, HEL92 and Jurkat) express both transcripts 1 and 2 of CHRNA7 while U937 cells only express transcript 2 of CHRNA7. Three other cells (RPM-I2286, RH77 and THP1 cells) only express transcript 1 of CHRNA7. We conclude that significant heterogeneity of the α7nAChR subunits that compose the final cell surface pentameric channel may exist on the human leukocyte cell surface.
Figure 3.
Regulation of CHRNA7 and CHRFAM7A in leukocyte lines. mRNA was isolated from HL60, RPMI8226, U937, HEL92, Jurkat, ARH77 and THP1 cells and the cDNA prepared was probed for the presence of CHRFAM7A (A), CHRNA7 (B) and GAPDH (C) by PCR. The amplicons were compared with that generated with their respective plasmids encoding variant 1 of human CHRFAM7A, variant 1 of human CHRNA7 or human GAPDH. The differences in gene expression were quantified in cultures cells (N = 6) and are expressed as mean ± standard deviations after normalization to the levels of gene expression in HL60 cells (D and E). Significance of changes in gene expression was determined using the REST gene expression analysis program as described in Materials and Methods. The ratio of CHRFAM7A:CHRNA7 gene expression (F) varies over 10,000-fold between different cell types (for example, Jurkat versus THP1). Significance of changes in the ratio of CHRFAM7A:CHRNA7 gene expression was determined using Wilcoxon signed rank test of unpaired t test using the GraphPad PRISM 6.0. (*p < 0.05; **p < 0.01, ***p < 0.001)
There are no antibodies that can distinguish the different molecular forms of CHRNA7 and CHRFAM7A. Accordingly, we used qRT-PCR to quantify the expression of both CHRNA7 (Figure 3D) and CHRFAM7A (Figure 3E) in the different leukocyte cell lines. Because HL60 cells appear to express equal amounts of variants 1 and 2 of CHRNA7 (see Figure 3B) and intermediate amounts of CHRFAM7A (see Figure 3A), the expression levels of both genes were normalized to the levels detected in HL60 cells. As shown in Figure 3F, no consistent pattern was observed in the ratio of CHRFAM7A to CHRNA7 which varied 10-fold to 10,000-fold higher in some cells (for example, HL-60, U937, HEL92 and THP1 cells) but were either near equal in others (RPMI-2286 cells) or 10 to 100 lower, for example in Jurkat and ARH77 cells. If mRNA expression correlates with protein expression, then the data suggest the existence of significant heterogeneity in the CHRFAM7A–CHRNA7 pentamer that forms the human cell surface α7nAChR on human leukocytes.
CHRFAM7A Has a Unique and Distinct Promoter Regulating Gene Expression
Expression levels of CHRFAM7A and CHRNA7 in leukocytes suggest their independent regulation. Furthermore the 5′ UTR sequence and the translation initiation site of the CHRFAM7A gene from the 5′ RACE analyses (see Figure 2) allowed us to infer the existence of the CHRFAM7A variant 1 protein in human leukocytes that others have detected by immunoblotting with anti-CHRNA7 antibodies (10,11). With these sequences, we used a bioinformatic approach to identify potential transcription factor binding sites (40) and experimental promoter mapping techniques to assess the regulation of CHRFAM7A gene expression. As illustrated in Figure 4A, five 5′ UTR plasmid constructs were prepared with DNA sequences that ranged from +22 to −2400 bp of the CHRFAM7A variant 1 transcript using the adenosine in the ATG ORF as +1 (see Materials and Methods). Each 5′ UTR–containing plasmid was then tested for its ability to activate luciferase gene expression in transiently transfected THP1 cells (Figure 4B). Promoter activity was observed within 500 bp of the CHRFAM7A ORF. Interestingly, 5′ extensions of this fragment encode dominantly negative inhibitory sequences that decrease the luciferase activity observed within the first 500 bp of the 5′ UTR. These data point to the existence of both stimulatory and inhibitory transcriptional elements regulating CHRFAM7A gene expression, perhaps reflecting its reported downregulation by LPS treatment in leukocytes (10,11) and differential upregulation in human epithelial cells (41).
Figure 4.
Regulation of CHRFAM7A expression. A schematic representation of 5′ UTR CHRFAM7A (A) shows the potential transcription factor binding sites identified by consensus sequence analyses. The f2400-, f1800-, f1000- and f500-bp fragments of the 5′ UTR were prepared as described in the text and then tested for promoter activity by cloning sequences into the promoter reporter assay (see Materials and Methods). Transient transfection of THP1 cells (B) showed luciferase expression upregulated by the f500 fragment in THP1 cells but translation inhibitory elements are detected in further 5′ extensions of the 5′ UTR CHRFAM7A. Statistical differences were analyzed by ANOVA (**p < 0.01; *p < 0.05; N = 6).
Biological Consequence of CHRFAM7A Gene Expression in THP1 Cells
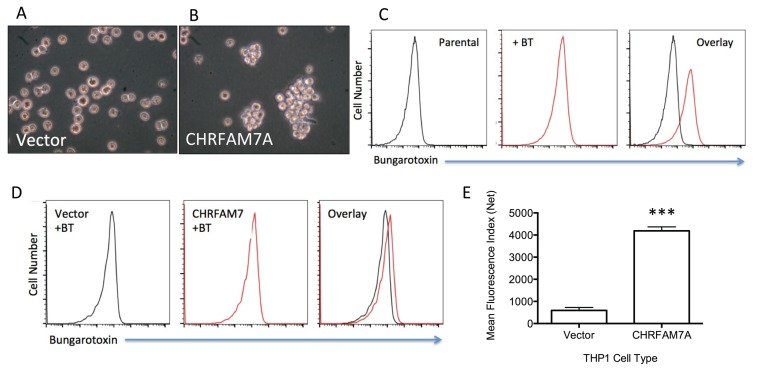
We used the transduction of THP1 cells with lentivirus to stably overexpress CHRFAM7A in THP1 cells and determine whether CHRFAM7A altered THP1 cell phenotype and/or gene expression. As shown in Figure 5, GFP-transduced THP1 cells (Figure 5A) can be differentiated from CHRFAM7A-transduced THP1 cells (Figure 5B) in that the latter tend to proliferate as loosely associated cell clusters that are reminiscent of expanding, transformed and presumably clonal cells. This is a marked contrast to the even distribution of both parental (not shown) and GFP-transduced THP1 cells, suggesting that CHRFAM7A overexpression affects cell–cell adhesion.
Figure 5.
Biological consequences of CHRFAM7A expression. THP1 cells appear as characteristic mononuclear cells growing in suspension (A) but with stable CHRFAM7A overexpression (B) the THP1 cells display an increased cell-cell adhesion phenotype demonstrating that CHRFAM7A is biologically active and alters THP1 cells. As shown in (C), parental THP1 cells bind the selective α7nAChR agonist bungarotoxin (BT) which can be measured by flow cytometry of THP1 cells incubated with labeled BT. Similar flow cytometry after stable transduction of THP1 cells with CHRFAM7A showed increased BT binding (D) when compared with THP1 cells transfected with only GFP. The difference in specific bungarotoxin binding can be quantified by measuring the increase in mean fluorescence (E). (***p < 0.001, N = 4)
To assess the functional consequence of stable CHRFAM7A overexpression, we used flow cytometry to first show that fluorescently labeled bungarotoxin (BT), a specific α7nAChR ligand, binds the leukocyte α7nAChR on THP1 cells (Figure 5C). BT is an irreversibly binding toxin that has been described extensively and shown to be a highly specific determinant of α7nAChR ligand binding. It also distinguishes the cell surface α7nAChR channel/receptor from other nicotinic receptors (42,43). As shown in Figure 5C, the addition of fluorescently labeled BT to THP1 cells produces a tenfold shift from baseline fluorescence labeling of THP1 cells that indicates the presence of the α7nAChR channel/receptor. This shift in BT binding was then compared between BT binding to GFP-vector and CHRFAM7A-transduced THP1 cells. As shown in Figure 5D, CHRFAM7A-transduced THP1 cells bind even more than labeled BT, suggesting that CHRFAM7A overexpression increases the α7nAChR channel and receptor at the THP1 cell surface. Quantitative analyses of the mean fluorescence intensity (MFI) in vector- and CHRFAM7A-transduced cells confirmed the significance of this shift (Figure 5E). These data suggest that stable CHRFAM7A overexpression either increased BT binding by altering ligand binding of a heteropentameric complex or by regulating CHRNA7 gene expression, thereby facilitating, not inhibiting, α7nAChR transport to the cell surface (3,9,12–14).
To further investigate the biological consequences of stable CHRFAM7A overexpression, its ability to increase α7nAChR dependent BT binding and induce a THP1 cell clumping phenotype, we isolated mRNA from both vector- and CHRFAM7A-transfected cells and analyzed their respective transcriptomes by RNA-seq. Clustering analyses of gene expression were performed using both DESeq (32) and CutDiff (36) analytical tools. In comparing the effects of CHRFAM7A and GFP-vector gene expression, DESeq identified 653 differentially expressed genes and 139 differentially expressed genes identified by Cuffdiff. The top 30 up-and downregulated differentially expressed genes are presented in Table 1 and sorted on the basis of the statistical significance of differential gene expression. As expected, the highest differentially expressed gene was CHRFAM7A (55.4-fold) in the CHRFAM7A-transduced cells. It is particularly noteworthy however that an increase in CHRNA7 expression (13.3-fold) was the second most significant difference in these stably transfected CHRFAM7A cells. This suggests that increased BT binding in CHRFAM7A cells (see Figure 5D) is the result of increased α7nAChR on the cell surface, rather then a reflection of an increase in a CHRFAM7A subunit in stably transfected cells. These data support the hypothesis that long-term and stable expression of CHRFAM7A modulates CHRNA7 availability to the surface. Alternatively, the CHRFAM7A protein may form a human-specific α7nAChR heteropentamer at the leukocyte cell surface with modified ligand specificity, tropism and binding kinetics, compared with the α7nAChR homopentamer found in all species (3,9,12–14).
Table 1.
Top differentially up- and downregulated genes, sorted by significance of change.
| Upregulated genes | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Ensmbl | Entrez | Gene | Gene name or description | Δ | p | |
| 1 | ENSG00000038427 | 1462 | VCAN | Versican | 4.5 | 7.40E-17 |
| 2 | ENSG00000175344 | 1139 | CHRNA7 | Cholinergic receptor, nicotinic, α7 (neuronal) | 13.3 | 2.50E-15 |
| 3 | ENSG00000189223 | 654433 | PAX8-AS1 | PAX8 antisense RNA 1 | 44.3 | 2.80E-12 |
| 4 | ENSG00000100181 | 387590 | TPTEP1 | Transmembrane phosphatase with tensin homology | 8.7 | 8.20E-12 |
| 5 | ENSG00000126709 | 2537 | IFI6 | Interferon, α-inducible protein 6 | 3.5 | 2.00E-11 |
| 6 | ENSG00000137965 | 10561 | IFI44 | Interferon-induced protein 44 | 6.7 | 1.50E-09 |
| 7 | ENSG00000088827 | 6614 | SIGLEC1 | Sialic acid binding Ig-like lectin 1, sialoadhesin | 3.6 | 3.80E-09 |
| 8 | ENSG00000166664 | 89832 | CHRFAM7A | Transfected gene | 55.4 | 7.80E-09 |
| 9 | ENSG00000166104 | 102466227 | MIR7162 | Micro RNA 7162 | 40.6 | 9.50E-07 |
| 10 | ENSG00000105655 | 51477 | ISYNA1 | Inositol-3-phosphate synthase 1 | 9.1 | 1.70E-06 |
| 11 | ENSG00000119922 | 3433 | IFIT2 | Interferon-induced protein tetratricopeptide repeats 2 | 3.6 | 2.20E-06 |
| 12 | ENSG00000206337 | 10866 | HCP5 | HLA complex P5 | 12 | 6.40E-06 |
| 13 | ENSG00000137959 | 10964 | IFI44L | Interferon-induced protein 44-like | 4.9 | 4.00E-05 |
| 14 | ENSG00000185745 | 3434 | IFIT1 | Interferon-induced protein tetratricopeptide repeats 1 | 3.9 | 5.20E-05 |
| 15 | ENSG00000183098 | 10082 | GPC6 | Glypican 6 | 5.1 | 6.30E-05 |
| 16 | ENSG00000170365 | 4086 | SMAD1 | SMAD family member 1 | 3.8 | 8.90E-05 |
| 17 | ENSG00000179796 | 116135 | LRRC3B | Leucine rich repeat containing 3B | 4.6 | 0.0002 |
| 18 | ENSG00000133106 | 94240 | EPSTI1 | Epithelial stromal interaction 1 | 3.3 | 0.0002 |
| 19 | ENSG00000165949 | 3429 | IFI27 | Interferon-induced protein 27 | 8.1 | 0.0006 |
| 20 | ENSG00000081923 | 5205 | ATP8B1 | ATPase, aminophospholipid transporter, type 8b member 1 | 0.31 | 0.0006 |
|
| ||||||
| Downregulated genes | ||||||
|
| ||||||
| Ensmbl | Entrez | Gene | Gene name or description | Δ | p | |
|
| ||||||
| 21 | ENSG00000174099 | 253827 | MSRB3 | Methionine sulfoxide reductase B3 | 0.37 | 1.80E-07 |
| 22 | ENSG00000156515 | 3098 | HK1 | Hexokinase 1 | 0.42 | 8.60E-07 |
| 23 | ENSG00000100060 | 4242 | MFNG | MFNG O-fucosyl 3-β-N-acetylglucosaminyl-transferase | 0.3 | 1.30E-06 |
| 24 | ENSG00000100234 | 7078 | TIMP3 | TIMP metallopeptidase inhibitor 3 | 0.51 | 1.40E-06 |
| 25 | ENSG00000182263 | 55137 | FIGN | Fidgetin | 0.08 | 3.30E-06 |
| 26 | ENSG00000165659 | 1602 | DACH1 | Dachshund family transcription factor 1 | 0.46 | 1.50E-05 |
| 27 | ENSG00000134824 | 9415 | FADS2 | Fatty acid desaturase 2 | 0.58 | 0.0002 |
| 28 | ENSG00000126767 | 2002 | ELK1 | ELK1, member of ETS oncogene family | 0.42 | 0.0002 |
| 29 | ENSG00000144712 | 23066 | CAND2 | Cullin-associated and neddylation-dissociated 2 | 0.31 | 0.0006 |
| 30 | ENSG00000011600 | 7305 | TYROBP | TYRO protein tyrosine kinase binding protein | 0.59 | 0.0007 |
Among other significantly changed differential genes, versican (#1, 4.5-fold), tensin-like protein (#4, 8.7-fold), SIGLEC1 (#7, 3.6-fold), glipican 6 (#15, 5.1-fold) and EPSTI1 (#18, 3.3-fold) expression all tie to cell adhesion, which itself is a phenotypic difference between stable CHRFAM7A- and GFP-transduced cells (Figure 5). Interestingly, there are also two genes encoding antisense (PAX-AS1) and microRNA with differential expression that are nearly as high (44.3- and 40.6-fold) as the 55-fold change elicited by the CHRFAM7A transgene. Finally, it is also interesting to note that six of the most significantly altered genes are tied to interferon activity including IFI6 (#5), IFI44 (#6), IFIT2 (#11) IF44L (#13), IFIT1 (#14) and IFI27 (#19).
We use GO enrichment analyses of the gene expression changes induced by stable CHRFAM7A expression to assess biological processes, cellular components and molecular functions (see Materials and Methods). As presented in Table 2, the most significantly enriched GO terms in each of the up- and down-regulated differentially expressed genes included the Type L interferon pathway, immune system processes and changes in cell adhesion. The latter is compatible with the phenotypic change detected microscopically. KEGG pathway assessments were used in a second test of differential gene expression and the most affected genes attributed to known pathways of cancer, leukocyte transendothelial migration and focal adhesion (Table 3 and Supplementary Figures S1, S2, S3).
Table 2.
Top five significantly enriched GO terms in differentially expressed genes.
| Enriched GO terms | P value | False discovery rate | |
|---|---|---|---|
| Upregulated by CHRFAM7A (DESeq) | Type I interferon signaling pathway | 6.93E-019 | 5.23E-015 |
| Cellular response to type I interferon | 6.93E-019 | 5.23E-015 | |
| Response to type I interferon | 8.79E-019 | 5.23E-015 | |
| Response to stimulus | 7.83E-017 | 2.28E-013 | |
| Defense response to virus | 8.82E-017 | 2.28E-013 | |
| Upregulated by CHRFAM7A (Cuffdiff) | Response to chemical | 1.44E-11 | 6.18E-008 |
| Response to organic substance | 1.72E-11 | 6.15E-008 | |
| Defense response | 3.74E-10 | 1.11E-006 | |
| Immune system process | 2.07E-007 | 4.62E-006 | |
| Biological regulation | 2.08E-009 | 4.63E-006 | |
| Downregulated by CHRFAM7A (DESeq) | Molecular function | 7.90E-011 | 1.41E-006 |
| Binding | 1.61E-010 | 1.44E-006 | |
| Cytosolic ribosome | 8.17E-010 | 2.82E-006 | |
| Cytosolic large ribosomal subunit | 9.47E-010 | 2.82E-006 | |
| Biological process | 9.49E-010 | 2.82E-006 | |
| Downregulated by CHRFAM7A (Cuffdiff) | Calcium-dependent cell-cell adhesion | 3.40E-005 | 1.59E-001 |
| Extracellular region | 3.41E-005 | 1.59E-001 | |
| Cell adhesion | 3.54E-005 | 1.59E-001 | |
| Biological adhesion | 3.57E-005 | 1.59E-001 | |
| Establishment of protein localization to membrane | 1.07E-004 | 3.83E-001 |
Table 3.
Top five most significantly enriched KEGG pathways.
| Sample comparison | Enriched pathways | P value | False discovery rate |
|---|---|---|---|
| CHRFAM7A (DESeq) | Ribosome | 3.35E-003 | 4.40E-001 |
| Pathways in cancer | 5.68E-003 | 4.40E-001 | |
| Hepatitis C | 7.37E-003 | 4.40E-001 | |
| Colorectal cancer | 1.16E-002 | 5.20E-001 | |
| Leukocyte transendothelial migration | 1.78E-002 | 6.06E-001 | |
| CHRFAM7A (Cuffdiff) | Cytokine-receptor interactions | 1.64E-002 | 4.05E-001 |
| Osteoclast differentiation | 1.74E-002 | 4.05E-001 | |
| RIG-I-like receptor signaling | 1.78E-002 | 4.05E-001 | |
| Insulin signaling pathway | 2.23E-002 | 4.05E-001 | |
| TGF-β signaling pathway | 2.76E-002 | 4.05E-001 |
DISCUSSION
Most genes are shared among all species but taxonomically restricted genes are only found in the genomes of specific taxa (for example, primates) and still others are species-specific, for example those uniquely localized to the human genome (29,44–52). While the emergence of human-specific genes is neither new nor unexpected, historically their study has been the focus of evolutionary biologists interested in the human-specific biology that relates to cognition, behavior, reproduction and mental health (30,53,54). Interestingly, the existence of human-specific genes is rarely invoked to explain unexpected human inflammatory responses to disease. Yet, human immunity is highly adaptive to the environment, the human inflammatory response is both highly plastic and highly significant to species survival, particularly after injury.
To date, there are more than 300 human-specific genes that have been identified, although their number, identity and significance remain contentious (30,31). Generally, these genes are thought to arise from segmental duplication of previously existing genes, human-specific alternative splicing that creates new open reading frames regulated by cognate promoters, mutational events during humanoid evolution or through the endogenization of integrated viral sequences (55–57). The evolutionary advantages that these changes confer drive their retention and presumably their activities outweigh “off-target” effects that might mediate disease. In hominid evolution, one such selection is likely tracked to burn injury and the control of fire, which is a uniquely human activity that dates back some 3 million years. To this end, it is interesting to note that it is controversial as to whether or not animal models mimic the programmed inflammatory response to burn injury (58,59).
In support of the existence of unexpected and off-target consequences to the emergence of new genes, newly evolved genes appear to be represented disproportionately in gene sets associated with complex diseases when they are compared with older and more evolutionarily conserved genes (31). Thus, it is thought that human-specific genes might confer subtle but differential responsiveness to human cells when compared with other species (60–62). Ironically, the absence of these human-specific genes in the genomes of animal models makes it impossible to study their contribution to human disease. CHRFAM7A is a case in point. It contributes human-specific activities that are tied to a widely distributed nicotinic ligand-gated ion channel called α7nAChR (CHRNA7) that plays a central role as a neurotransmitter receptor in the nervous system and in the regulation of inflammation.
The studies presented here are the first to establish a clear and unambiguous functional consequence to stable CHRFAM7A gene overexpression in a human leukocyte cell line. RNAseq of these stably transduced cells demonstrated increased CHRNA7 gene expression and increased bungarotoxin binding, which differs from the reported effects of transient overexpression (12–14). While the significance of this difference is not known, the studies establish a clear and unambiguous coregulatory link between CHRFAM7A and the α7nAChR neurotransmitter receptor encoded by CHRNA7. Furthermore, they may reflect the importance of the ratio of CHRFAM7A to CHRNA7 at the leukocyte cell surface in regulating the inflammation response (Figure 6). Pathway analyses of the changes in gene expression in these cells using both GO and KEGG algorithms show that CHRFAM7A has functional effects on leukocytes that tie the neurotransmitter receptor to leukocyte adhesion, trafficking and transformation. These sequelae may be direct effects of CHRFAM7A expression or indirect effects caused by downstream genes affected by CHRFAM7A.
Figure 6.
Specifying the human vagus-mediated inflammatory response. The identification of a gene encoding a human-specific subunit of α7nAChR raises the possibility that in humans, the canonical regulation of inflammation by vagus nerve may be mediated by cell surface α7nAChR homopentamers composed of CHRNA7 (A) or CHRFAM7A (B) on human leukocytes or alternatively, heteropentameric receptors composed of a combination CHRFAM7A and CHRFAM7A subunits (C) The latter would alter ligand tropism, specificity, binding kinetics and cell responsiveness.
In the experiments described here, we show that CHRFAM7A is normally expressed in human leukocytes (see Figures 1, 2) and at levels similar to those of CHRNA7. In view of its capacity to regulate α7nAChR activity after transient transfection in vitro (12–14), the data shown imply that CHRFAM7A has the potential to modulate human leukocyte function and by inference, α7nAChR-regulated inflammation (16). Interestingly, we also find that the stable transduction of CHRFAM7A increases basal expression of CHRNA7, thereby establishing the existence of a concomitant and compensatory response to high CHRFAM7A expression. These data point to the possibility that the ratio of CHRFAM7A and CHRNA7 expression may be important in defining the human α7nAChR response. To that end, we also established the identity of the THP1 CHRFAM7A variant, deduced the amino acid difference that distinguishes it from α7nAChR and mapped a unique promoter that differentially regulates CHRFAM7A from CHRNA7 in leukocytes (see Figure 4). Finally, we showed that stable CHRFAM7A expression affects several molecular pathways of cell function including cell adhesion, cell growth and cell trafficking. Inasmuch as there are no analogous or independently regulated CHRFAM7A-like genes in the genomes of other species (3,9), these findings implicate the existence of a human-specific mechanism in human leukocytes to gauge the human inflammatory response.
CONCLUSION
Together these data provide compelling evidence supporting the hypothesis that CHRFAM7A has a significant role in human leukocyte cell biology. CHRFAM7A has been shown to have the capacity to form cell surface heteropolymers with the wild-type α7nAChR (see Figure 6) and is reported in some models either to exert a dominant negative effect on α7nAChR, regulating the appearance of α7nAChR on the cell surface, or to alter ligand tropism (12–14). As a role for α7nAChRs in leukocyte homeostasis is unequivocal (16), CHRFAM7A might then confer leukocytes with a human-specific responsiveness to trophic stimuli that remains to be defined. In one example, efferent signaling of the vagus nerve has a central role in regulating inflammation and α7nAChR activation is an obligatory intermediate, at least in mice (2,16–19,29,63,64). If the vagus nerve were presumed to have a similarly critical role in maintaining immune homeostasis in humans, then CHRFAM7A would likely gauge the immunomodulatory properties of human α7nAChR in a human-specific manner.
The human chromosome 15q13–14 locus contains the 10-exon human CHRNA7 gene that encodes the α7 subunit of the prototypic homopentameric α7nAChR ligand-gated ion channel. This locus, however, also underwent significant rearrangement during human evolution, leading to the emergence of the CHRFAM7A gene (reviewed in [3,9]). While the CHRFAM7A gene is structurally similar to CHRNA7 (4), it has lost five CHRNA7 exons and acquired five exons of a partially duplicated FAM7-UL kinase gene that was once on human chromosome 3. The in-frame fusion of these three partially duplicated genes led to the emergence of CHRFAM7A (3,9). Since its discovery in 1998 (4), numerous studies have shown that, like CHRNA7, CHRFAM7A is expressed in the central nervous system (CNS) and several genomic analyses have associated it and a 2-bp mutation with neuropsychiatric disorders including schizophrenia (65), bipolar disorder (66) and autism (67).
The natural ligand(s) responsible for CHRFAM7A activation is presumed to include acetylcholine, but its ligand selectivity is not known compared with other nAChRs, each of which have been described extensively (68). Normally, acetylcholine is thought to be the intrinsic ligand that activates the cell surface α7nAChR homopentameric channel, but there are numerous alternative ligands including choline, nicotine and even peptide hormone-like ligands (69–73) that can activate α7nAChR. The discovery of an alternative α7nAChR in human leukocytes raises the possibility that heteropentamers composed of CHRNA7 and CHRFAM7A proteins could modulate the cellular immune response to proinflammatory challenges (see Figure 6). In this way, the responsiveness of the nuclear factor kappa-B (NF-κB) and Jak2/STAT3 inflammatory signaling pathways (74,75) that has been normally ascribed to the homopentameric product of CHRNA7 may be altered by ligand binding to cells that express homopentameric CHRFAM7A or CHRNA7–CHRFAM7A heteropentamers on the cell surface. Interestingly, this subunit heterogeneity is well recognized for growth factors (for example, PDGF, activin and inhibin), receptors (for example, transforming growth factor [TGF]β, platelet-derived growth factor [PDGF] and fibroblast growth factor [FGF] receptors), integrins (for example, α/β/γ subunits) and other α/βAChR subunits from distinct genes. In the case of CHRFAM7A-encoded heteropentamers, they appear unique because CHRFAM7A only exists in the human genome and therefore, could define a currently unknown, human-specific response.
On a final note, it is worth noting that there is a mutagenized form of CHRFAM7A that has been more extensively studied in mental health research on the presumption that CHRFAM7A might have evolved as an alternative ligand-gated ion-channel-regulating neuronal function (76–78). There are no studies describing the possible effect of this mutant gene on human leukocyte function, let alone the evolutionary pressures that originally led to the emergence, selection and retention of CHRFAM7A in humans. The findings presented here add to a growing body of evidence that collectively underscores the need to dissect the functional link between genes that arose for human speciation and the off-target emergence of complex human disease. In light of genetic analyses that associate newly emerged genes with complex disease, it is incumbent on investigators to use emerging models of human disease that can serve to study the contribution of human-specific disease. Just as mouse models were adapted specifically to study viruses with human-specific tropism (for example, HIV), analogous models will be needed to help determine the contribution of human-specific and taxonomically restricted genes to human biology, physiology and the pathobiology of human diseases tied to inflammation.
Supplemental Data
ACKNOWLEDGMENTS
The authors wish to thank Emelie Amburn, Annemarie Hageny and James Putnam for their expert technical assistance and support from the National Institutes of Health (CA170140) and a Department of Defense Grant administered by the American Burn Association (W81XWH-10-1-0527). The research presented here originated with seed funding from an NIH P20 Center for Exploratory Wound Healing Research (NIGM78421).
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Cite this article as: Costantini TW, et al. (2015) A human-specific α7-nicotinic acetylcholine receptor gene in human leukocytes: identification, regulation and the consequences of CHRFAM7A expression. Mol. Med. 21:323–36.
REFERENCES
- 1.Richman DP, Arnason BG. Nicotinic acetylcholine receptor: evidence for a functionally distinct receptor on human lymphocytes. Proc Natl Acad Sci U S A. 1979;76:4632–5. doi: 10.1073/pnas.76.9.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheadle GA, Costantini TW, Bansal V, Eliceiri BP, Coimbra R. Cholinergic signaling in the gut: a novel mechanism of barrier protection through activation of enteric glia cells. Surg Infect (Larchmt) 2014;15:387–93. doi: 10.1089/sur.2013.103. [DOI] [PubMed] [Google Scholar]
- 3.Sinkus ML, et al. The human CHRNA7 and CHRFAM7A genes: a review of the genetics, regulation, and function. Neuropharmacology. 2015;96(Pt B):274–88. doi: 10.1016/j.neuropharm.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gault J, et al. Genomic organization and partial duplication of the human alpha7 neuronal nicotinic acetylcholine receptor gene (CHRNA7) Genomics. 1998;52:173–85. doi: 10.1006/geno.1998.5363. [DOI] [PubMed] [Google Scholar]
- 5.Zody MC, et al. Analysis of the DNA sequence and duplication history of human chromosome 15. Nature. 2006;440:671–5. doi: 10.1038/nature04601. [DOI] [PubMed] [Google Scholar]
- 6.Le Novere N, Corringer PJ, Changeux JP. The diversity of subunit composition in nAChRs: evolutionary origins, physiologic and pharmacologic consequences. J Neurobiol. 2002;53:447–56. doi: 10.1002/neu.10153. [DOI] [PubMed] [Google Scholar]
- 7.Locke DP, et al. Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster. Genome Biol. 2003;4:R50. doi: 10.1186/gb-2003-4-8-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riley B, Williamson M, Collier D, Wilkie H, Makoff A. A 3-Mb map of a large Segmental duplication overlapping the alpha7-nicotinic acetylcholine receptor gene (CHRNA7) at human 15q13–q14. Genomics. 2002;79:197–209. doi: 10.1006/geno.2002.6694. [DOI] [PubMed] [Google Scholar]
- 9.Costantini TW, Dang X, Coimbra R, Eliceiri BP, Baird A. CHRFAM7A, a human-specific and partially duplicated alpha7-nicotinic acetylcholine receptor gene with the potential to specify a human-specific inflammatory response to injury. J Leukoc Biol. 2015;97:247–57. doi: 10.1189/jlb.4RU0814-381R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villiger Y, et al. Expression of an alpha7 duplicate nicotinic acetylcholine receptor-related protein in human leukocytes. J Neuroimmunol. 2002;126:86–98. doi: 10.1016/s0165-5728(02)00057-7. [DOI] [PubMed] [Google Scholar]
- 11.Benfante R, et al. Expression of the alpha7 nAChR subunit duplicate form (CHRFAM7A) is down-regulated in the monocytic cell line THP-1 on treatment with LPS. J Neuroimmunol. 2011;230:74–84. doi: 10.1016/j.jneuroim.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Araud T, et al. The chimeric gene CHRFAM7A, a partial duplication of the CHRNA7 gene, is a dominant negative regulator of alpha7*nAChR function. Biochem Pharmacol. 2011;82:904–14. doi: 10.1016/j.bcp.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lucas-Cerrillo AM, et al. Function of partially duplicated human alpha77 nicotinic receptor subunit CHRFAM7A gene: potential implications for the cholinergic anti-inflammatory response. J Biol Chem. 2011;286:594–606. doi: 10.1074/jbc.M110.180067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, et al. The duplicated alpha7 subunits assemble and form functional nicotinic receptors with the full-length alpha7. J Biol Chem. 2014;289:26451–63. doi: 10.1074/jbc.M114.582858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tracey KJ. The inflammatory reflex. Nature. 2002;420:853–9. doi: 10.1038/nature01321. [DOI] [PubMed] [Google Scholar]
- 16.Tracey KJ. Physiology and immunology of the cholinergic antiinflammatory pathway. J Clin Invest. 2007;117:289–96. doi: 10.1172/JCI30555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tracey KJ. Reflex control of immunity. Nat Rev Immunol. 2009;9:418–28. doi: 10.1038/nri2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson U, Tracey KJ. Reflex principles of immunological homeostasis. Annu Rev Immunol. 2012;30:313–35. doi: 10.1146/annurev-immunol-020711-075015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olofsson PS, et al. alpha7 nicotinic acetylcholine receptor (alpha7nAChR) expression in bone marrow-derived non-T cells is required for the inflammatory reflex. Mol Med. 2012;18:539–43. doi: 10.2119/molmed.2011.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olofsson PS, Rosas-Ballina M, Levine YA, Tracey KJ. Rethinking inflammation: neural circuits in the regulation of immunity. Immunol Rev. 2012;248:188–204. doi: 10.1111/j.1600-065X.2012.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costantini TW, et al. Vagal nerve stimulation protects against burn-induced intestinal injury through activation of enteric glia cells. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1308–18. doi: 10.1152/ajpgi.00156.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costantini TW, et al. Efferent vagal nerve stimulation attenuates gut barrier injury after burn: modulation of intestinal occludin expression. J Trauma. 2010;68:1349–54. doi: 10.1097/TA.0b013e3181dccea0. discussion 1354–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costantini TW, et al. Targeting alpha-7 nicotinic acetylcholine receptor in the enteric nervous system: a cholinergic agonist prevents gut barrier failure after severe burn injury. Am J Pathol. 2012;181:478–86. doi: 10.1016/j.ajpath.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Lopez NE, et al. Vagal nerve stimulation decreases blood-brain barrier disruption after traumatic brain injury. J Trauma Acute Care Surg. 2012;72:1562–6. doi: 10.1097/TA.0b013e3182569875. [DOI] [PubMed] [Google Scholar]
- 25.Costantini TW, et al. Burn-induced gut barrier injury is attenuated by phosphodiesterase inhibition: effects on tight junction structural proteins. Shock. 2009;31:416–22. doi: 10.1097/SHK.0b013e3181863080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costantini TW, et al. Role of p38 MAPK in burn-induced intestinal barrier breakdown. J Surg Res. 2009;156:64–9. doi: 10.1016/j.jss.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Costantini TW, et al. Targeting the gut barrier: identification of a homing peptide sequence for delivery into the injured intestinal epithelial cell. Surgery. 2009;146:206–12. doi: 10.1016/j.surg.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matteoli G, et al. A distinct vagal anti-inflammatory pathway modulates intestinal muscularis resident macrophages independent of the spleen. Gut. 2014;63:938–48. doi: 10.1136/gutjnl-2013-304676. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, et al. Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature. 2003;421:384–8. doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]
- 30.Zhang YE, Long M. New genes contribute to genetic and phenotypic novelties in human evolution. Curr Opin Genet Dev. 2014;29:90–6. doi: 10.1016/j.gde.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper DN, Kehrer-Sawatzki H. Exploring the potential relevance of human-specific genes to complex disease. Hum Genomics. 2011;5:99–107. doi: 10.1186/1479-7364-5-2-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rapaport F, et al. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol. 2013;14:R95. doi: 10.1186/gb-2013-14-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Lin N, Zhang B. An iteration normalization and test method for differential expression analysis of RNA-seq data. BioData Min. 2014;7:15. doi: 10.1186/1756-0381-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang ZH, et al. A comparative study of techniques for differential expression analysis on RNA-Seq data. PloS One. 2014;9:e103207. doi: 10.1371/journal.pone.0103207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochem Biophys Res Commun. 2004;316:1050–8. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 39.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cartharius K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–42. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 41.Dang X, Eliceiri BP, Baird A, Costantini TW. CHRFAM7A: a human-specific alpha7-nicotinic acetylcholine receptor gene shows differential responsiveness of human intestinal epithelial cells to LPS. FASEB J. 2015;29:2292–302. doi: 10.1096/fj.14-268037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marks MJ, Collins AC. Characterization of nicotine binding in mouse brain and comparison with the binding of alpha-bungarotoxin and quinuclidinyl benzilate. Mol Pharmacol. 1982;22:554–64. [PubMed] [Google Scholar]
- 43.Orr-Urtreger A, et al. Mice deficient in the alpha7 neuronal nicotinic acetylcholine receptor lack alpha-bungarotoxin binding sites and hippocampal fast nicotinic currents. J Neurosci. 1997;17:9165–71. doi: 10.1523/JNEUROSCI.17-23-09165.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chimpanzee S, Analysis C. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 45.Hedlund M, et al. N-glycolylneuraminic acid deficiency in mice: implications for human biology and evolution. Mol Cell Biol. 2007;27:4340–6. doi: 10.1128/MCB.00379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang H, et al. Evolutionary conservation and selection of human disease gene orthologs in the rat and mouse genomes. Genome Biol. 2004;5:R47. doi: 10.1186/gb-2004-5-7-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knowles DG, McLysaght A. Recent de novo origin of human protein-coding genes. Genome Res. 2009;19:1752–9. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4:865–75. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 49.Long M, et al. Origin of new genes: evidence from experimental and computational analyses. Genetica. 2003;118:171–82. [PubMed] [Google Scholar]
- 50.Nahon JL. Birth of ‘human-specific’ genes during primate evolution. Genetica. 2003;118:193–208. doi: 10.1023/a:1024157714736. [DOI] [PubMed] [Google Scholar]
- 51.Stahl PD, Wainszelbaum MJ. Human-specific genes may offer a unique window into human cell signaling. Sci Signal. 2009;2:pe59. doi: 10.1126/scisignal.289pe59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tu Z, et al. Further understanding human disease genes by comparing with housekeeping genes and other genes. BMC Genomics. 2006;7:31. doi: 10.1186/1471-2164-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang YE, Landback P, Vibranovski M, Long M. New genes expressed in human brains: implications for annotating evolving genomes. Bioessays. 2012;34:982–991. doi: 10.1002/bies.201200008. [DOI] [PubMed] [Google Scholar]
- 54.Zhang YE, Landback P, Vibranovski MD, Long M. Accelerated recruitment of new brain development genes into the human genome. PLoS Biol. 2011;9:e1001179. doi: 10.1371/journal.pbio.1001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterston RH, Lander ES, Sulston JE. On the sequencing of the human genome. Proc Natl Acad Sci U S A. 2002;99:3712–6. doi: 10.1073/pnas.042692499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–64. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 58.Seok J, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–12. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2015;112:1167–72. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davis MM. Immunology taught by humans. Sci Transl Med. 2012;4:117fs112. doi: 10.1126/scitranslmed.3003385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A. 2008;105:6987–92. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–8. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 63.Lowry DM, et al. The vagus nerve alters the pulmonary dendritic cell response to injury. J Surg Res. 2014;192:12–8. doi: 10.1016/j.jss.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 64.Morishita K, Costantini TW, Eliceiri B, Bansal V, Coimbra R. Vagal nerve stimulation modulates the dendritic cell profile in posthemorrhagic shock mesenteric lymph. J Trauma Acute Care Surg. 2014;76:610–7. doi: 10.1097/TA.0000000000000137. discussion 617–8. [DOI] [PubMed] [Google Scholar]
- 65.Freedman R, et al. Linkage disequilibrium for schizophrenia at the chromosome 15q13–14 locus of the alpha7-nicotinic acetylcholine receptor subunit gene (CHRNA7) Am J Med Genet. 2001;105:20–2. [PubMed] [Google Scholar]
- 66.Hong CJ, Lai IC, Liou LL, Tsai SJ. Association study of the human partially duplicated alpha7 nicotinic acetylcholine receptor genetic variant with bipolar disorder. Neurosci Lett. 2004;355:69–72. doi: 10.1016/j.neulet.2003.10.043. [DOI] [PubMed] [Google Scholar]
- 67.Casey JP, et al. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Gen. 2012;131:565–79. doi: 10.1007/s00439-011-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wessler I, Kirkpatrick CJ. Acetylcholine beyond neurons: the non-neuronal cholinergic system in humans. Br J Pharmacol. 2008;154:1558–71. doi: 10.1038/bjp.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arredondo J, Chernyavsky AI, Grando SA. Nicotinic receptors mediate tumorigenic action of tobacco-derived nitrosamines on immortalized oral epithelial cells. Cancer Biol Ther. 2006;5:511–7. doi: 10.4161/cbt.5.5.2601. [DOI] [PubMed] [Google Scholar]
- 70.Arredondo J, Chernyavsky AI, Jolkovsky DL, Pinkerton KE, Grando SA. Receptor-mediated tobacco toxicity: cooperation of the Ras/Raf-1/MEK1/ERK and JAK-2/STAT-3 pathways downstream of alpha7 nicotinic receptor in oral keratinocytes. FASEB J. 2006;20:2093–101. doi: 10.1096/fj.06-6191com. [DOI] [PubMed] [Google Scholar]
- 71.Chernyavsky AI, Arredondo J, Galitovskiy V, Qian J, Grando SA. Upregulation of nuclear factor-kappaB expression by SLURP-1 is mediated by alpha7-nicotinic acetylcholine receptor and involves both ionic events and activation of protein kinases. Am J Physiol Cell Physiol. 2010;299:C903–11. doi: 10.1152/ajpcell.00216.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Papke RL. Merging old and new perspectives on nicotinic acetylcholine receptors. Biochem Pharmacol. 2014;89:1–11. doi: 10.1016/j.bcp.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Papke RL, Chojnacka K, Horenstein NA. The minimal pharmacophore for silent agonism of the alpha7 nicotinic acetylcholine receptor. J Pharmacol Exp Ther. 2014;350:665–80. doi: 10.1124/jpet.114.215236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Jonge WJ, et al. Stimulation of the vagus nerve attenuates macrophage activation by activating the Jak2-STAT3 signaling pathway. Nat Immunol. 2005;6:844–51. doi: 10.1038/ni1229. [DOI] [PubMed] [Google Scholar]
- 75.Yoshikawa H, et al. Nicotine inhibits the production of proinflammatory mediators in human monocytes by suppression of I-kappaB phosphorylation and nuclear factor-kappaB transcriptional activity through nicotinic acetylcholine receptor alpha7. Clin Exp Immunol. 2006;146:116–23. doi: 10.1111/j.1365-2249.2006.03169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sinkus ML, et al. A 2-base pair deletion polymorphism in the partial duplication of the alpha7 nicotinic acetylcholine gene (CHRFAM7A) on chromosome 15q14 is associated with schizophrenia. Brain Res. 2009;1291:1–11. doi: 10.1016/j.brainres.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neri M, Bonassi S, Russo P. Genetic variations in CHRNA7 or CHRFAM7 and susceptibility to dementia. Curr Drug Targets. 2012;13:636–43. doi: 10.2174/138945012800398928. [DOI] [PubMed] [Google Scholar]
- 78.Rozycka A, et al. Association study of the 2-bp deletion polymorphism in exon 6 of the CHRFAM7A gene with idiopathic generalized epilepsy. DNA Cell Biol. 2013;32:640–7. doi: 10.1089/dna.2012.1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.