Abstract
Borrelia burgdorferi, the etiologic agent of Lyme disease, produces a variety of proteins that promote survival and colonization in both the Ixodes species vector and various mammalian hosts. We initially identified BB0744 (also known as p83/100) by screening for B. burgdorferi strain B31 proteins that bind to α1β1 integrin and hypothesized that, given the presence of a signal peptide, BB0744 may be a surface-exposed protein. In contrast to this expectation, localization studies suggested that BB0744 resides in the periplasm. Despite its subsurface location, we were interested in testing whether BB0744 is required for borrelial pathogenesis. To this end, a bb0744 deletion was isolated in a B. burgdorferi strain B31 infectious background, complemented, and queried for the role of BB0744 following experimental infection. A combination of bioluminescent imaging, cultivation of infected tissues, and quantitative PCR (qPCR) demonstrated that Δbb0744 mutant B. burgdorferi bacteria were attenuated in the ability to colonize heart tissue, as well as skin locations distal to the site of infection. Furthermore, qPCR indicated a significantly reduced spirochetal load in distal skin and joint tissue infected with Δbb0744 mutant B. burgdorferi. Complementation with bb0744 restored infectivity, indicating that the defect seen in Δbb0744 mutant B. burgdorferi was due to the loss of BB0744. Taken together, these results suggest that BB0744 is necessary for tissue tropism, particularly in heart tissue, alters the ability of B. burgdorferi to disseminate efficiently, or both. Additional studies are warranted to address the mechanism employed by BB0744 that alters the pathogenic potential of B. burgdorferi.
INTRODUCTION
In 2013, the CDC estimated that the number of Americans diagnosed with Lyme disease was ∼300,000 per year, indicating that infection with Borrelia burgdorferi represents a significant public health concern in the United States, particularly in areas where the disease is endemic (1, 2). The etiologic agent of Lyme disease, B. burgdorferi, is an extracellular pathogenic spirochetal bacterium that has adapted to invade and colonize various organs within mammalian hosts following transmission by an infected Ixodes sp. tick (3–6). Although a skin rash and influenza-like symptoms are typical symptoms of early Lyme disease, untreated patients can present with lymphocytomas, myocarditis, meningitis, arthritis, and a large variety of other debilitating inflammatory symptoms (7–12). The mechanisms underlying the dissemination of B. burgdorferi to the various organs it colonizes during infection have yet to be discerned.
Some factors that may influence the severity and type of symptoms seen in Lyme disease patients can be inferred from the results of mouse model studies and include the location of the initial infection, the immune response of the host, and the amount of spirochetes able to disseminate and colonize affected organs (13–19). The ability of B. burgdorferi to evade the immune response and colonize tissues lies within the numerous lipoproteins that adorn its outer surface (20). Many of these lipoproteins have been characterized as ECM (extracellular matrix) adhesins, including those that bind to decorin (DbpA), fibronectin (BBK32), glycosaminoglycans (Bgp; BBK32; DbpA), and integrins (BBB07, P66), as well as many others with unknown host ligands (21–27). B. burgdorferi also expresses on its surface a variable surface antigen, VlsE, and five different factor H binding proteins designated complement regulator-acquiring surface proteins, which are involved in the evasion of complement-dependent killing (28, 29).
A study by Motameni et al. showed a correlation between myocarditis and arthritis severity and the location of the B. burgdorferi injection (16). The results imply that the available route of dissemination of the spirochetes has an impact on the extent of infection. Previously, studies have focused primarily on the route of dissemination through either connective tissues or the bloodstream (13, 30–35). BBK32 has been implicated as one of the proteins involved in blood vessel dissemination, and spirochetes have been visualized adhering to and transmigrating across blood vessel epithelial cell layers during infection (36). However, there is increasing evidence that B. burgdorferi may also migrate through the lymphatic system (16, 23, 37).
Lymph nodes are rapidly and consistently colonized by B. burgdorferi in both early and late stages of infection (37, 38). The time in which lymph nodes, joints, and other connective tissues become infected is relative to their proximity to the inoculation site (31, 37). B. burgdorferi may travel through the lymphatic system not only for dissemination but also to interfere with the immune response and gain a survival advantage for spirochetes infecting all tissues (39). Recent studies have also found that, in contrast to BBK32, DbpA is involved in transmission via the lymphatic system, indicating that distinct modalities may be operative in borrelial dissemination (23, 32, 33, 36). It is probable that B. burgdorferi has adapted to multiple routes of dissemination, as quick systemic infection is an important function for a pathogen that travels from host to host by tick bite at random locations.
This study focused on the protein p83/100, which is encoded by the chromosomal gene bb0744. The impact and role of BB0744 during infection have not been previously studied, but it is a known serodiagnostic antigen for Lyme infection (40–42). BB0744 has also been observed forming complexes with other proteins isolated in outer membrane vesicles, suggesting an association with the outer membrane envelope (43). In this study, we show that BB0744 is required for optimal tissue tropism and/or normal dissemination in mice infected by needle inoculation given that B. burgdorferi strains lacking this gene exhibit a defect in colonization to both distal skin sites and the heart and exhibit reduced bacterial loads in lymph node and joint tissues.
MATERIALS AND METHODS
Bacterial strains.
All of the strains used in this study are listed in Table 1. Escherichia coli Mach1-T1R cells were utilized for cloning, and C41(DE3) cells (Lucigen, Middleton, WI) were used for expression of recombinant BB0744. E. coli strains were grown at 37°C with aeration in lysogeny broth (LB). Strains were maintained under antibiotic selection with gentamicin at 5 μg/ml, spectinomycin at 100 μg/ml, kanamycin at 50 μg/ml, chloramphenicol at 25 μg/ml, ampicillin at 100 μg/ml, and carbenicillin at 100 μg/ml.
TABLE 1.
Plasmids and bacterial strains used in this study
| Strain or plasmid | Description or genotype | Source or reference |
|---|---|---|
| Plasmids | ||
| pCR2.1-TOPO | Kanr Carbr | Invitrogen |
| pKFSS1 | Specr in E. coli, Strr in B. burgdorferi; borrelial shuttle vector | 46 |
| p744KO | Kanr Carbr Specr/Strr; 1.5-kb regions up- and downstream of bb0744 flanking a Specr/Strr cassette ligated into pCR2.1-TOPO | This study |
| pCR-Blunt II-TOPO | Kanr | Invitrogen |
| p744Comp | Kanr Gentr; 1.5-kb regions up- and downstream of bb0744 flanking bb0744 and a Gentr cassette ligated into pCR-Blunt II-TOPO | This study |
| pBSV2G | Gentr; borrelial shuttle vector | 47 |
| pBBE22luc | Kanr, plasmid containing bbe22 and B. burgdorferi codon-optimized firefly luc gene under control of strong borrelial promoter (PflaB-luc) | 31 |
| pCR2.1-recA | Kanr; recA gene in pCR2.1-TOPO | 67 |
| pCR2.1-β-actin | Kanr; Actb gene in pCR2.1-TOPO | 68 |
| pGEM-T Easy | Ampr; TA cloning vector | Promega |
| pProEX-HTb | Ampr; recombinant expression construct with N-terminal, TEVa protease-cleavable His6 tag | Life Technologies |
| pRARE | Camr; plasmid expressing six rare tRNAs to improve expression of genes containing rare E. coli codons | EMD Millipore |
| pProEX::BB0744 | Ampr; pProEX-HTb containing bb0744; construct lacks first 22 amino acids of BB0744 | This study |
| B. burgdorferi strains | ||
| ML23 | B. burgdorferi strain B31 clonal isolate missing lp25 | 55 |
| ML23/pBBE22luc | Kanr | 31 |
| SH100 | ML23 Δbb0744::Strr | This study |
| SH200 | ML23 bb0744-Gentr; Strs | This study |
| SH100 pBBE22luc | ML23 Δbb0744::Strr containing pBBE22luc; Strr Kanr | This study |
| SH200 pBBE22luc | ML23 bb0744-Gentr; Strs containing pBBE22luc; Gentr Kanr | This study |
| 5A4 | Clonal isolate of strain B31 containing all plasmids | 69 |
| 5A4NP1 | 5A4 bbe02::Kanr | 70 |
| 297 | Clinical isolate | 71 |
| E. coli strains | ||
| Mach1-T1R | F− ϕ80 lacZΔM15 ΔlacX74 hsdR (rK− mK+) ΔrecA1398 endA1 tonA | Invitrogen |
| C41(DE3) | F− ompT hsdSB (rB− mB−) gal dcm(DE3) | Lucigen |
TEV, tobacco etch virus.
B. burgdorferi strains were grown in BSK-II medium supplemented with 6% normal rabbit serum (Pel-Freez, Rogers, AR) lacking gelatin at 32°C with 1% CO2 (44). Borrelial strains were maintained with antibiotic selection with kanamycin at 300 μg/ml, streptomycin at 50 μg/ml, or gentamicin at 50 μg/ml. The use of infectious B. burgdorferi in this study was reviewed and approved by the Institutional Biosafety Committee at Texas A&M University.
Plasmid constructs.
All of the plasmids utilized in this study are listed in Table 1. All DNA fragments were amplified by PCR with TaKaRa PrimeSTAR GXL DNA polymerase. Plasmid p744KO was constructed via the Gibson assembly method (45). A streptomycin resistance cassette (PflgB-aadA; Strr) was amplified from plasmid pKFSS1 (46). A fragment of pCR2.1-TOPO (Invitrogen) was amplified for use as the vector backbone. The 1.5-kb DNA fragments immediately upstream and downstream of bb0744 were both amplified from a ML23 genomic DNA sample. The primers were designed such that the 5′ end of the bb0744 upstream fragment contained a 20-bp overlap at the 3′ end of the pCR2.1 fragment and its 3′ end contained a 20-bp overlap at the 5′ end of the Strr cassette. The primers for the bb0744 downstream fragment were similarly designed to overlap the 5′ end of the pCR2.1 fragment and the 3′ end of the Strr cassette. The bb0744 upstream fragment, bb0744 downstream fragment, and Strr cassette were combined with Gibson assembly master mix (New England BioLabs) into one product and amplified by PCR. This fragment was ligated into the pCR2.1 backbone to create complete plasmid p744KO, which was chemically transformed into Mach1-T1R.
For cis complementation of bb0744, plasmid p744Comp was constructed. Specifically, DNA fragments containing the 1.5-kb sequence upstream of bb0744 and bb0744 itself and a 1.5-kb sequence downstream of bb0744 were amplified from B. burgdorferi genomic DNA. A gentamicin resistance cassette (PflgB-aacC) was amplified from plasmid pBSV2G (47). These three fragments were amplified with 20-bp overlapping pieces and annealed to each other by the method of overlap extension PCR (48). This product was ligated into pCR Blunt II-TOPO and chemically transformed into Mach1-T1R.
The bb0744 open reading frame (ORF) of B. burgdorferi strain 297 was amplified with primers 5′ BB0744 ORF and 3′ BB0744 ORF (Integrated DNA Technologies, Coralville, IA) with genomic DNA as the template. The primers incorporated BamHI and NotI restriction sites at the 5′ and 3′ ends, respectively. The 5′ primer was also designed to omit the N-terminal 22 amino acids of the ORF, which was predicted to contain a signal peptide by SignalP 4.1. The amplicon was TA cloned into pGEM-T Easy (Promega Corp., Madison, WI) and confirmed by sequencing. The construct was excised with BamHI and NotI and ligated into the pProEX-HTb expression vector (Life Technologies) digested with the same enzymes to generate pProEX::BB0744.
Expression and purification of recombinant BB0744.
To produce recombinant BB0744, C41(DE3) was transformed with pProEX::BB0744 and pRARE (EMD Millipore, Billerica, MA). Cultures were induced for 3 h with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Recombinant BB0744, which includes an N-terminal His6 tag, was affinity purified with HisPur Ni-nitrilotriacetic acid (NTA) resin (Thermo Scientific, Rockford, IL) under native conditions in accordance with the manufacturer's protocol. Fractions containing recombinant BB0744 were then buffer exchanged into 20 mM Tris-HCl (pH 8)—350 mM NaCl and concentrated with an Amicon Ultra-15 centrifugal filtration unit (EMD Millipore) with a 50-kDa molecular mass cutoff. Recombinant BB0744 was resolved on a HiPrep 16/60 Sephacryl S-300 HR gel filtration column with an Äkta UPC-10 fast-performance liquid chromatography (FPLC) system (GE Healthcare Biosciences, Piscataway, NJ). The protein was eluted with 20 mM Tris-HCl (pH 8)–350 mM NaCl, and samples of the fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to assess the yield. Fractions containing recombinant BB0744 were pooled, buffer exchanged into 20 mM Tris-HCl (pH 8), and concentrated as described above. Concentrated proteins were purified further by FPLC and MonoQ 5/50 GL anion-exchange column chromatography (GE Healthcare Biosciences). Recombinant BB0744 that bound to the MonoQ column was eluted with a linear gradient up to 1 M NaCl buffered in 20 mM Tris-HCl (pH 8). On the basis of SDS-PAGE, recombinant BB0744 was purified to apparent homogeneity (i.e., >95%).
Generation of polyclonal antisera.
BB0744-specific polyclonal antiserum was generated in 3- to 4-week-old female Sprague-Dawley rats (Harlan, Indianapolis, IN) as described by Groshong et al. (49). Briefly, 25 μg of recombinant BB0744 protein was combined with complete Freund's adjuvant (Sigma, St. Louis, MO). Rats were injected intraperitoneally with the emulsified antigen-adjuvant mixture and then boosted twice with 25 μg of recombinant BB0744 protein emulsified with incomplete Freund's adjuvant (Sigma). Sera were collected 2 weeks after the final boost. The University of Arkansas for Medical Sciences (UAMS) is accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC), and all immunization protocols were approved by the UAMS Institutional Animal Care and Use Committee (IACUC).
Membrane protein phase partitioning.
Membrane protein phase partitioning was performed as previously described (49). B. burgdorferi cells were collected at the mid-log growth phase and washed with phosphate-buffered saline (PBS). Cells were then sonicated and incubated overnight in 2% Triton X-114 at 4°C. Samples were centrifuged to remove insoluble material and then phase partitioned by warming to 37°C. The detergent and aqueous phases were separated and washed, and protein was precipitated in cold acetone. Precipitated protein was prepared in SDS-PAGE buffer and analyzed by Western blotting. OspA was included as a positive control for membrane localization (colorimetric detection), and BB0796 was used as a positive control for aqueous localization (chemiluminescent detection) (20). BB0744 was detected by colorimetric assay.
IFAs for surface accessibility.
Immunofluorescence assays (IFAs) were performed as described by Groshong et al. (49). Briefly, B. burgdorferi cells were grown to mid-log growth phase and harvested by centrifugation. For permeabilized samples, spirochetes were washed with PBS-MgCl2 and immediately applied to silylated slides. The cells were allowed to air dry and then fixed by heating to 65°C for 1 h and incubation in acetone. The samples were then blocked for 1 h and incubation with primary antibodies (anti-OspA monoclonal, chicken anti-FlaB, and rat anti-BB0744 antibodies) for 1 h. For nonpermeabilized samples, spirochetes were washed and incubated in solution with primary antibodies for 1 h. Samples were then applied to silylated slides and fixed with 4% formaldehyde. The slides were then incubated with fluorescent-dye-conjugated secondary antibodies (anti-mouse antibody conjugated with Alexa 594, anti-chicken antibody conjugated with Alexa 647, and anti-rat antibody conjugated with Alexa 448; Life Technologies) and fixed again with formaldehyde. Cells were visualized with a Nikon Ti-U microscope and a 60× oil immersion objective, and imaging was performed with a D5-QilMc digital camera (Nikon, Melville, NY).
Proteinase K accessibility assay.
Proteinase K assays were performed as previously described (49). B. burgdorferi cells were pelleted following growth to mid-log phase and washed with PBS. The samples were then divided. Samples were either mock treated with PBS or treated with proteinase K (50, 51). Proteinase K accessibility assays were also performed in the presence or absence of 0.05% Triton X-100 (Promega). Phenylmethanesulfonyl fluoride was used to stop digestion after 1 h, and the samples were placed in SDS-PAGE buffer, separated by SDS-PAGE, and analyzed by Western blotting. FlaB was used as a negative (periplasmic) control, and OspA was used as a positive (outer surface) control.
SDS-PAGE and immunoblot analysis.
B. burgdorferi protein lysates were resolved by SDS-PAGE, and gels were either stained with Coomassie brilliant blue R-250 (Sigma-Aldrich, St. Louis, MO, USA) or transferred to polyvinylidene difluoride membranes and immunoblotted. Primary antibodies were used at the following dilutions: rat anti-BB0744 at 1:5,000, chicken anti-FlaB IgY at 1:25,000, rat anti-OspA at 1:25, mouse anti-OspC at 1:1,000, and rat anti-BB0796 at 1:1,500 (49). Appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (anti-mouse IgG-HRP, anti-rat IgG-HRP [Invitrogen, Carlsbad, CA], and donkey anti-chicken IgY antibodies at 1:2,500) were used to detect immune complexes. The membranes were washed extensively in PBS with 0.2% Tween 20 and developed with the Western Lightning Chemiluminescent Reagent plus system (PerkinElmer, Waltham, MA, USA) for chemiluminescent detection or with 4-chloro-1-naphthol as the substrate for colorimetric detection.
Transformation of B. burgdorferi.
B. burgdorferi strain ML23 was made competent and transformed by electroporation (52, 53). For the bb0744 deletion and subsequent cis complementation, linearized versions of p744KO and p744Comp (Table 1), respectively, were used for electroporation. All transformants were selected for with appropriate antibiotics. The resulting bb0744 deletion mutant was designated SH100, and its corresponding cis complement was termed SH200. These strains were additionally screened for the presence or absence of bb0744 by PCR product size and DNA sequencing as previously described (54). Once validated, strains SH100 and SH200 were transformed with pBBE22luc under kanamycin selection and transformants were screened by PCR for the presence of the shuttle vector (31). Strains were then evaluated for plasmid content, and only those that maintained the complete plasmid set of the parent strain were used. SH200 was tested for sensitivity to streptomycin to rule out a mixed population.
In vitro bioluminescence assays.
ML23/pBBE22luc, SH100/pBBE22luc, and SH200/pBBE22luc were grown to mid-log phase and concentrated to 108 cells/ml in duplicate in a 96-well plate. Each sample was treated with a final concentration of 667 μM d-luciferin (Research Products International Corp., Mt. Prospect, IL) in PBS and immediately measured for luminescence in a FluorChem E Digital Darkroom (ProteinSimple).
Infectivity studies.
All animal experiments were performed in accordance with AAALAC guidelines. The Texas A&M University IACUC granted approval for all mouse infections conducted. Four- to 6-week-old female BALB/c mice were used to enhance the signal for bioluminescence imaging. The mice were infected intradermally within the abdominal skin with an inoculum of either 103 or 105 cells of B. burgdorferi strain ML23/pBBE22luc, SH100/pBBE22luc, or SH200/pBBE22luc. The mice were then imaged as described below. Twenty-one days postinfection, the mice were sacrificed and pinna, abdominal skin, inguinal lymph node, heart, spleen, bladder, and tibiotarsal joint tissues were aseptically removed and placed directly into BSK-II medium with appropriate antibiotics. Qualitative analysis of infection was performed by incubating these tissue-inoculated cultures at 32°C and 1% CO2 for 3 weeks. The presence of B. burgdorferi was scored by dark-field microscopy (55).
In vivo bioluminescent imaging.
To detect luciferase activity, mice were intraperitoneally injected with d-luciferin prior to imaging with an IVIS Spectrum live-animal imaging system (Caliper Life Sciences, Hopkinton, MA) (31), with the exception of one mouse that did not receive d-luciferin and served as a negative control for background luminescence. Mice were imaged at 1 h and 1, 4, 7, 10, 14, and 21 days after B. burgdorferi infection by methods described previously (31). Luminescence was measured with 1- and 10-min exposures to obtain images for quantification and visual representation, respectively. Regions of interest (ROI) were selected to measure luminescence in photons per second from the 1-min exposure images by using an equal area of the whole body for all of the mice in all of the experiments. The luminescence for mice that received d-luciferin was averaged and normalized by subtracting the background luminescence on the negative-control mouse. All images from the 10-min exposures were corrected for the background and set on a color scale with 7.5 × 103 and 1 × 105 photons per second as the minimum and maximum, respectively.
Quantitative PCR (qPCR).
Quantitative analysis of pinna, inoculation site skin (abdominal skin), inguinal lymph node, and tibiotarsal joint tissues from infected mice was performed by extracting DNA with the Roche High Pure PCR Template preparation kit as described previously (38). B. burgdorferi genome copies were enumerated along with mammalian genome copies with iTaq Universal SYBR Green Supermix (Bio-Rad) and the Applied Biosystems 7900HT Fast real-time PCR system. Approximately 100 ng of total DNA was used in each reaction mixture with either a primer set for the amplification of the B. burgdorferi recA gene (38, 56) or a primer set for amplification of the Mus musculus β-actin gene (57). Borrelial and mammalian genomic copy numbers were determined for each sample by normalizing the threshold cycle (CT) values of individual tissues to a standard curve established with 10 to 105 copies of pCR2.1-recA and pCR2.1-β-actin (Table 1). All samples were assayed in triplicate, and the standard curve for each plate had a linear regression coefficient of determination (R2) of ≥0.98.
Statistical analysis.
To determine significant differences between samples in the real-time qPCR analysis, a one-way analysis of variance (ANOVA) of the strains in each tissue type was performed with a Tukey multiple-comparison test. For the bioluminescence signal data, two-way ANOVAs were performed with Bonferroni posttests. Statistical significance was accepted when the P values were <0.05 for all of the statistical analyses employed.
RESULTS
BB0744 is located subsurface in the cell envelope.
Screening for α1β1 integrin binding proteins by a Western overlay approach coupled with mass spectroscopy identified BB0744 as a potential adhesin to these mammalian host structures. This binding is likely due to an RGD integrin binding motif at residues 146 to 148 (not shown; 21). BB0744 is a 700-residue protein with a predicted molecular mass of 81 kDa and has historically been referred to as p83/100 (58). In addition, BB0744 is a well-established antigen that is used to diagnose human Lyme borreliosis (40–42). The deduced amino acid sequence of BB0744 predicts an amino-terminal signal peptide that would be subject to cleavage by leader peptidase I. However, the sequence lacks evidence of predicted transmembrane segments that could be used to integrate the protein into a membrane bilayer. Nevertheless, the presence of an amino-terminal signal sequence, coupled with the atypical cell envelope properties of B. burgdorferi, suggested that BB0744 might be an outer membrane protein. Additional support of surface localization of BB0744 comes from its presence in outer membrane vesicle preparations from B. burgdorferi (not shown) and a prior study showing that BB0744 (referred to as p83/100) was associated with membrane-associated proteins (59, 60), as well as those proteins that were deemed soluble (60). Given these linkages to the borrelial cell envelope and the putative integrin-binding motif, we addressed the location of BB0744 in B. burgdorferi by three independent methods.
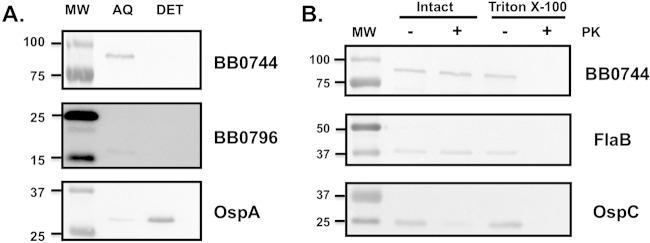
First, whole-cell extracts of B. burgdorferi strain B31 derivative 5A4 were partitioned into aqueous and detergent phase proteins with the detergent Triton X-114. BB0744 was present in the aqueous phase along with control soluble protein BB0796 (20) and absent from the detergent phase, whereas the cell surface lipoprotein OspA associated with the detergent phase as expected (Fig. 1A).
FIG 1.
Determination of the cell envelope localization of BB0744. (A) BB0744 is a soluble protein. B. burgdorferi 5A4 cells were partitioned into detergent (DET) and aqueous (AQ) phases with Triton X-114. Precipitated protein samples were resolved by SDS-PAGE and analyzed by Western blotting. OspA was included as a positive control for a membrane-localized protein (DET lane), and BB0796 was used as a positive control as an aqueous-phase protein (AQ lane). BB0744 was detected only in the aqueous phase. (B) BB0744 is not protease accessible in intact borrelial cells. B. burgdorferi 5A4 cells were suspended in PBS alone (Intact lanes) or PBS containing Triton X-100 (Triton X-100 lanes). Minus and plus signs signify the presence of added proteinase K (PK). The resulting B. burgdorferi samples were resolved by SDS-PAGE and analyzed by Western blotting. FlaB and OspC were used as controls for subsurface and surface-exposed proteins, respectively. No decrease in BB0744 was observed after proteinase K treatment. Lanes MW contained prestained protein molecular size markers (sizes are in kilodaltons).
Second, the whole-cell extracts of B. burgdorferi were collected from cells treated with PBS or proteinase K. Proteins on the outer surface of the bacteria were cleaved by proteinase K, while those in the periplasm or cytoplasm would be protected. The presence of the surface-exposed OspC lipoprotein, the subsurface endoflagellar structural protein FlaB, and BB0744 was detected by Western blotting of both samples. The amount of OspC was reduced because of cleavage by proteinase K, while both periplasmic FlaB and BB0744 remained unaltered (Fig. 1B). As a control, Triton X-100 was added to samples and treated with proteinase K to ensure that these proteins were not insensitive to proteolysis (Fig. 1B). The protection of BB0744 from proteinase K was not affected by altering the temperature, carbon dioxide, or pH of the culture conditions (data not shown). In addition, no other culture condition, including those that induce high levels of mammalian specific antigens (notably, OspC and DbpA) resulted in a change in BB0744 expression (not shown).
Lastly, borrelial cells were either permeabilized by heat fixation or left untreated prior to incubation with antibodies against surface-exposed OspA, subsurface FlaB, or BB0744. Appropriate fluorescent-dye-conjugated secondary antibodies showed that OspA was fluorescently labeled on both permeabilized and nonpermeabilized cells, whereas BB0744 was detected only in permeabilized cells, similar to FlaB, a borrelial protein that is known to be periplasmically localized (Fig. 2). Taken together, these results suggest that BB0744 is not present on the cell surface of B. burgdorferi under any of the in vitro conditions tested.
FIG 2.
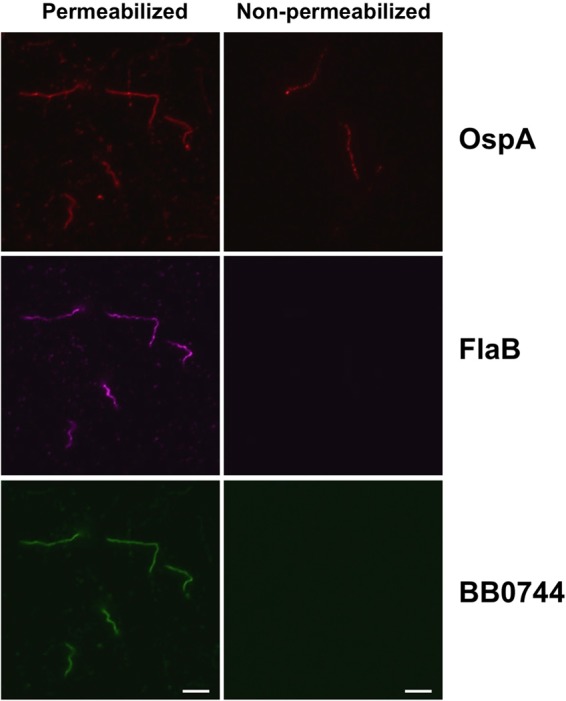
Surface exposure of BB0744. B. burgdorferi 5A4 cells were either washed or permeabilized by heat fixation prior to incubation with antibodies to the proteins listed on the right. Nonpermeabilized cells were then fixed to silylated slides with formaldehyde, and all cells were incubated with appropriate fluorescent secondary antibodies (anti-mouse Alexa 594 for mouse antibody to OspA, anti-chicken Alexa 647 for chicken antibody to FlaB, and anti-rat Alexa 488 for rat antibody to BB0744). FlaB served as a periplasmic protein control, and OspA was used as an outer surface protein control. BB0744 was labeled only in permeabilized cells. Scale bars, 10 μm.
Construction of Δbb0744 mutant and complemented strains.
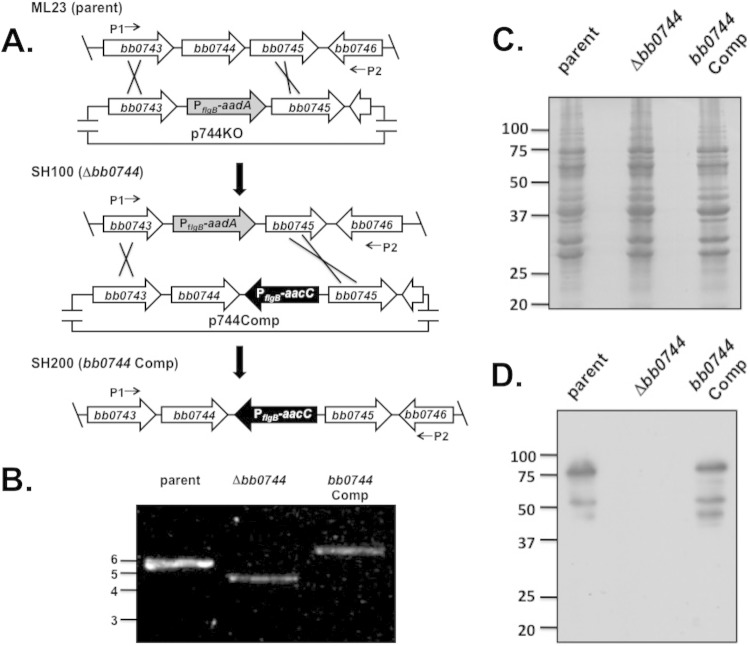
Despite the subsurface localization of BB0744, its presence within the cell envelope prompted us to determine whether BB0744 played a role in borrelial pathogenesis. To this end, bb0744 was deleted from the chromosome by homologous recombination and replaced with a streptomycin resistance (Strr) cassette in B. burgdorferi strain ML23 (55, 61) (Fig. 3A). Resulting transformants were screened by PCR for evidence of the bb0744 deletion (Δbb0744::Strr) with oligonucleotide primers (listed in Table 2) that amplify a 5.1-kb fragment in the parent strain and a 4.3-kb fragment in the resulting Δbb0744::Strr strain (Fig. 3B). Each Δbb0744::Strr strain obtained was then screened for its total borrelial plasmid profile. Only those strains that retained the complete plasmid profile were utilized. Western immunoblot analysis with a monospecific polyclonal antibody to BB0744 demonstrated that BB0744 was not produced in putative deletion strain SH100 (Fig. 3C and D).
FIG 3.
Construction of bb0744 knockout and complemented strains. (A) Construction of the Δbb0744::Strr mutant strain and cis complementation strategies. Representation of the bb0744 locus in the B. burgdorferi B31 strain ML23 (parent) chromosome and subsequent replacement of intact bb0744 with a streptomycin resistance (Strr) cassette (PflgB-aadA) by homologous recombination with linearized p744KO, resulting in strain SH100 (Δbb0744). For complementation, the Strr cassette of the bb0744 knockout mutant is replaced with an intact copy of bb0744 linked to a gentamicin resistance cassette (PflgB-aacC) to yield strain SH200 (bb0744 Comp). Lines with arrows depict the locations of primers P1 and P2 (Table 2), which were used to confirm the deletion mutation and subsequent complemented (Comp) strain. Note that the gene depictions shown are not to scale. (B) Confirmation of the Δbb0744::Strr strain and cis-complemented strains. The configuration of the bb0744 locus was determined by PCR for the ML23/pBBE22luc, SH100/pBBE22luc, and SH200 strains with the oligonucleotide primers depicted in panel A. The values to the left are DNA size markers (sizes are in kilobases). (C, D). The Δbb0744::Strr strain does not produce BB0744 protein. B. burgdorferi whole-cell lysates were separated by SDS-PAGE, immunoblotted, and probed with antisera to BB0744. The Coomassie-stained gel used as a loading control is shown in panel C. The presence of BB0744 was detected with antiserum specific for BB0744 in panel D.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ to 3′)a | Description |
|---|---|---|
| P1 | GGTACCGAGCTCGGATCCACAAACTCGTTTGGAAGTATAAGCT | Primer pair used to amplify 1.5-kb region upstream of bb0744 with 20-bp 5′ flanking region homologous to insertion site of pCR2.1-TOPO and 3′ flanking region homologous to 5′ region of PflgB-aadA |
| bb0744US-R | TTCGCCATTCGGTACCGCGTAAAAGAAATTCTCCTATAAATTTAATTATAATCTACC | |
| pflgB-SpecF-KpnI | ACGCGGTACCGAATGGCGAATGGCGCGGCC | Primer pair used to amplify PflgB-Strr cassette from pKFSS1 |
| pflgB-SpecR-KpnI | ACGCGGTACCCCACGAAGTCCCGGGAGAACCC | |
| bb0744DS-F | GACTTCGTGGGGTACCGCGTCTTTACAGTTAGATTTAATTTGTATAAATCGT | Primer pair used to amplify 1.5-kb region upstream of bb0744 with 20-bp 5′ flanking region homologous to insertion site of PflgB-aadA and 3′ flanking region homologous to 5′ region of pCR2.1-TOPO |
| P2 | CTAGATGCATGCTCGAGCGGATTTCAAAACCAATTGAAACATTGC | |
| 744KO seq1 | TTTACTTCTTGAAGAGATCCTTAATTC | Primers used for sequencing and verification of fragments and final product of p744KO and p744Comp prior to transformation into B. burgdorferi |
| 744KO seq2 | CAAAATATCTATAAAGCTTGCCTCTTG | |
| 744KOseq3 | GTAGGTCGAAAAACGGCAAA | |
| 744KOseq4 | AAAATTAACAAACTCTTGGAGCAAA | |
| 744KOseq5 | GACAATATTCATGAAAGTGATTCCA | |
| 744KOseq6 | TTCTAAAGCTTCTAGCAAAGAAAAA | |
| 744KOseq7 | CTGCAAATCTTCCAGGCTCT | |
| aadA-F seq | CCCGTCATACTTGAAGCTAGACAG | |
| aadA-R seq | ATTTGCCGACTACCTCCTTGGTG | |
| bb0744US-744-R | CGGCTCTAGCCGAATGCGCATTACTTAACTTTCTTTAAAGTATTTACATCTAAA | Primers used in conjunction with P1 and P2, respectively, to amplify fragments containing 1.5-kb region upstream of bb0744 and bb0744 and 1.5-kb region downstream of bb0744 with 20-bp 5′ and 3′ flanking regions homologous to PflgB-aacC |
| aacC-bb0744DS-F | TTCGAGCCCATAATGGATCCCTTTACAGTTAGATTTAATAATTTGTATAAATCGT | |
| flgB-aacC-F | CCTAGGTAATACCCGAGCTTCA | Primer pair used to amplify PflgB-Gentr cassette from pBSV2G |
| flgB-aacC-R | ACGCGTAAGCCGATCTCG | |
| b-Actin-F | ACGCAGAGGGAAATCGTGCGTGAC | Primer pair used for enumeration of copies of mouse β-actin via qPCR (72) |
| b-Actin-R | ACGCGGGAGGAAGAGGATGCGGCAGTG | |
| nTM17FrecA | GTGGATCTATTGTATTAGATGAGGCT | Primer pair used for enumeration of copies of B. burgdorferi recA via qPCR (38) |
| nTM17RrecA | GCCAAAGTTCTGCAACATTAACACCT | |
| 5′ BB0744 ORF | GGATCCAGAGAAGTTGATAGGGAAAAATTAAAGGAC | Primer pair used to amplify bb0744 ORF for recombinant protein expression |
| 3′ BB0744 ORF | GCGGCCGCTATTACTTAACTTTCTTTAAAGTATTTACATC |
Restriction sites are underlined.
All attempts to complement bb0744 in trans were unsuccessful. Therefore, we complemented bb0744 in cis by integrating bb0744 and a linked gentamicin resistance cassette into ML23 Δbb0744::Strr (Fig. 3A). Transformants were screened for gentamicin resistance (Genr) and streptomycin sensitivity. Isolates that replace the Δbb0744::Strr allele with intact bb0744 linked to the Genr marker exhibit a 6.1-kb amplified product following PCR analysis with primers P1 and P2 (Fig. 3A and B). The resulting strain was designated SH200. Western immunoblot analysis showed that the complemented strain expressed BB0744 protein (Fig. 3D). The ML23 bb0744 knockout (SH100) and complemented strain (SH200) were then transformed with the shuttle vector pBBE22luc. The presence of the bbe22 and bbe23 genes on pBBE22luc complements lp25-deficient ML23 and restores infectivity (62). Plasmid pBBE22luc also contains the borrelial codon-optimized firefly luciferase gene, luc, (63) to enable bioluminescent tracking of the spirochetes (31, 54).
There was no statistically significant difference among the growth rates of ML23/pBBE22luc, SH100/pBBE22luc, and SH200/pBBE22luc, indicating that the expression of bb0744 is not important for in vitro growth or replication (not shown). No overt morphological alterations were observed. Lastly, a qualitative test for the expression of luciferase by each strain was performed that verified comparable levels of luminescence in all of the strains tested (data not shown).
Assessment of the bb0744 deletion in the ML23 background.
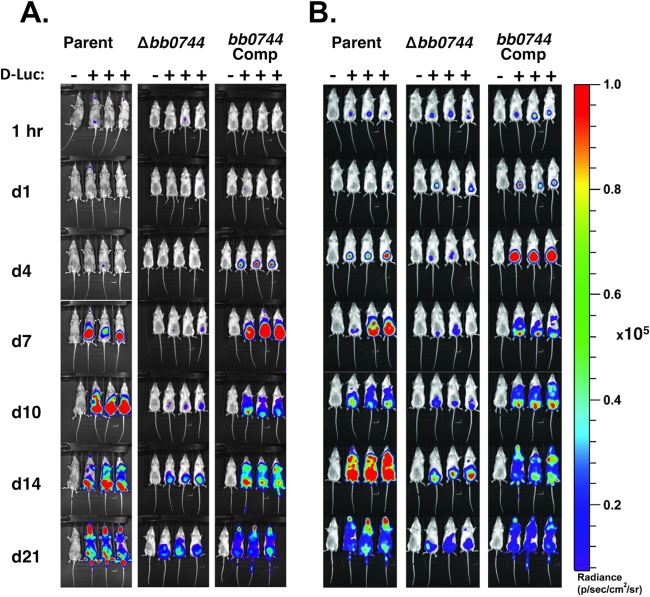
In previous studies, bioluminescent imaging served as a powerful tool to determine the role of specific gene products in the colonization and dissemination of B. burgdorferi (31, 54). To assess the role of BB0744 in borrelial pathogenesis in a temporal and spatial manner, BALB/c mice were infected intradermally with a low (103) or high (105) dose of ML23/pBBE22luc, SH100/pBBE22luc, or SH200/pBBE22luc. Low-level luminescence was detected during the exposures of mice infected with the high dose at the 1-h postinfection time point for all of the strains tested, indicating that all of the mice were infected. When looking at total body luminescence, mice infected with either dose of ML23/pBBE22luc showed a luminescence peak corresponding to the highest concentration of live B. burgdorferi at day 7, with a secondary peak at day 14, as previously observed (31). Surprisingly, mice infected with the low dose of SH200/pBBE22luc showed a much higher peak at day 7 and mice infected with the high dose had a luminescence peak as early as day 4. However, both bb0744-expressing strains have prominent luminescence peaks. Comparatively, SH100/pBBE22luc-infected mice had low levels of luminescence with a minor peak appearing only on day 14 (Fig. 4; see Fig. S1 in the supplemental material).
FIG 4.
Bioluminescent temporal and spatial tracking of a B. burgdorferi Δbb0744 mutant strain in living mice. BALB/c mice were infected with a dose of 103 (A) or 105 (B) cells of ML23/pBBE22luc (parent), SH100/pBBE22luc (Δbb0744), or SH200/pBBE22luc (bb0744 complemented [Comp]). Mice indicated by plus signs were treated with d-luciferin (d-Luc) at the time points indicated (d, day). Mice indicated by minus signs were not given a substrate and were used to assess the background. The images obtained are 10-min exposures, and all of the images shown are adjusted to the same scale, as indicated by the color spectrum scale on the right.
When the imaging data are analyzed, a clear difference between the patterns of infection with ML23/pBBE22luc and SH100/pBBE22luc emerges. In the mice infected with the high dose, on day 4 for SH200/pBBE22luc and day 7 for ML23/pBBE22luc, a high level of fluorescence is observed on the abdomens of the mice radiating from the site of B. burgdorferi injection. On day 10 for both strains, the luminescence is decreased because of partial clearing of spirochetes by virtue of the burgeoning humoral response to B. burgdorferi (13, 61, 64). On day 14, the signal returns but is now distributed more widely across the entire torso and extends to the limbs and head. On day 21, while the signal is reduced again, it is also more concentrated in the head, limbs, and lower abdomen (Fig. 4B). In contrast, throughout the SH100/pBBE22luc infection cycle, the luminescence signal only leaves the confines of the lower abdomen to spread to the lower limbs. A high signal level is never observed on the upper torso or head of mice infected with either dose of SH100/pBBE22luc. This pattern of infection suggests that the bb0744 knockout was not attenuated in its ability to infect and subsist in mice but rather exhibits a dissemination defect in moving away from the initial site of infection.
At day 21, all infected mice were sacrificed and their tissues were processed for cultivation and qPCR. Cultivation of tissues from mice infected with 103 cells of parental infectious strain ML23/pBBE22luc or SH200/pBBE22luc demonstrated infection of all tissues (Table 3). While SH100/pBBE22luc was capable of colonizing the joint, bladder, lymph node, and skin near the site of infection independently of the dose, no spirochetes were observed in the cultures of heart, spleen, or ear tissue of any mice infected with either the low or the high dose (Table 3). One possible explanation for this inability to spread may be impaired motility. To address this possibility, the Δbb0744::Strr mutant was compared to its parent strain by using motility agarose. No significant difference in motility or swarming was evident (not shown), suggesting that the defect in colonization was not due to compromised locomotion of the bb0744 mutant.
TABLE 3.
Infectivity of Δbb0744 derivative in the ML23/pBBE22luc background
| Strain and inoculum dose (no. of cells) | No. of cultures positive/total |
|||||||
|---|---|---|---|---|---|---|---|---|
| Pinnaa | Skinb | Lymph node | Heart | Spleen | Bladder | Joint | All sites | |
| ML23/pBBE22luc (parent) | ||||||||
| 103 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 28/28 |
| 105 | 4/4 | 3/4 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 27/28 |
| SH100/pBBE22luc (Δbb0744) | ||||||||
| 103 | 0/4 | 4/4 | 4/4 | 0/4 | 0/4 | 4/4 | 4/4 | 16/28 |
| 105 | 0/4 | 4/4 | 4/4 | 0/4 | 0/4 | 4/4 | 4/4 | 16/28 |
| SH200/pBBE22luc (bb0744 complemented) | ||||||||
| 103 | 4/4 | 3/4 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 27/28 |
| 105 | 4/4 | 4/4 | 4/4 | 4/4 | 3/4 | 4/4 | 4/4 | 27/28 |
Represents cultivation from a distal skin site, specifically, the ear pinna.
Represents the site of inoculation on the abdominal skin.
qPCR was also performed with total DNA extracted from mouse ear pinna, abdominal skin, lymph node, and tibiotarsal joint and was normalized to mouse tissue as previously described (31, 38) (Fig. 5). No significant difference in borrelial genome copy numbers between any strains within the abdominal skin was observed. However, there were significant differences in the ear pinna, lymph node, and joint tissues between the Δbb0744::Strr mutant and its parent or complemented derivatives. In particular, and consistent with the cultivation data, the qPCR detected no genomic copies of SH100/pBBE22luc in the ear pinna tissue (Fig. 5). Taken together, these data suggest that BB0744 is needed for optimal tissue tropism and/or dissemination during experimental infection by B. burgdorferi, given that significantly lower numbers of spirochetes are observed in joint tissue and distal skin sites, respectively.
FIG 5.
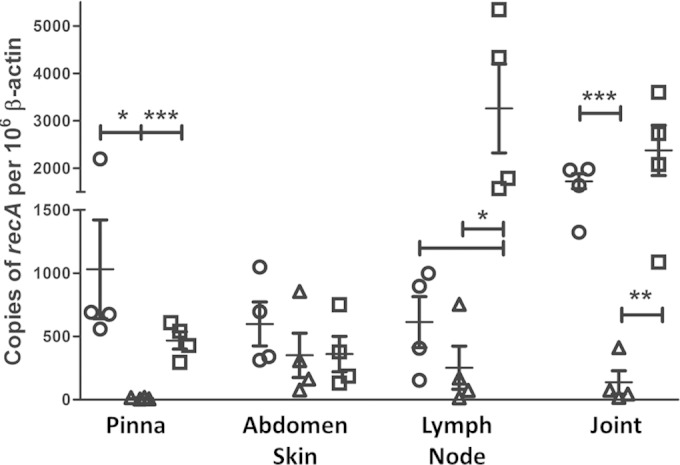
Loss of BB0744 affects the B. burgdorferi load in distal tissues. Quantitative PCR analysis of joint, lymph node, skin, and ear tissues was performed. Circles, ML23/pBBE22luc; triangles, SH100/pBBE22luc; squares, SH200/pBBE22luc. Copies of recA were quantified and represent the number of total B. burgdorferi genomes. To normalize the data to host tissue, β-actin copy numbers were also enumerated and data points are expressed as numbers of copies of B. burgdorferi genomes per 106 copies of β-actin. Bars indicate the average values of the samples tested. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
The ability to adhere to the surface of host cells is an essential step in the colonization process for all pathogenic bacteria. As a part of its life cycle, B. burgdorferi is transmitted between the tick vector and mammalian host and disseminates to various tissues during the infection process. Tissues contain diversity within their ECM, with various subunits and ratios of collagens, fibronectin, integrins, fibrinogen, and laminin (65). ECM-binding adhesins have been characterized in many pathogens, and some have been discovered in B. burgdorferi, including DbpA, BBK32, P66, Bgp, BBB07, and BBA33 (21–27, 54). B. burgdorferi has an outer surface rich in lipoproteins, many of which are hypothesized to interact with host structures and provide for optimal adhesion and/or immune evasion.
This study was started as part of screening for B. burgdorferi proteins that could engage host integrins. Despite the linkage of BB0744, also known as p83/100 (58), as a serodiagnostic marker for B. burgdorferi infection (40–42), little is known about a role for this protein in borrelial pathogenesis. Because of its RGD integrin-binding motif and the ability of BB0744 to bind to β1 integrins (21), specifically α1β1 (not shown), we hypothesized that BB0744 may use a leader peptidase I signal sequence to be exported to the cell envelope and be surface exposed within the outer membrane of B. burgdorferi. The localization of this protein in outer membrane vesicles provided further evidence that BB0744 might be surface exposed (43). However, multiple approaches suggested that, while exported, BB0744 was not surface exposed, despite its association with outer membrane vesicle preparations during integrin binding screening (not shown). The localization studies instead suggested that BB0744 might be anchored to the inner leaflet of the outer membrane or interact with outer membrane proteins within the periplasm. Despite the observation that BB0744 appears to be a periplasmic protein, thus reducing the possibility that BB0744 interacts directly with host proteins, the deletion of bb0744 still resulted in a tissue-specific infectivity defect (Table 3; Fig. 4 and 5). As a corollary to this, the inability of the bb0744 mutant strain to infect heart and distal skin sites implies that the functional activity associated with BB0744 may be the consequence of BB0744 envelope proteins that are required for normal surface protein conformation or lipid structures, particularly for heart, lymph node, and joint colonization. Further experiments are required to identify the proteins that BB0744 specifically recognizes.
Infection of BALB/c mice with the ML23 background bb0744 knockout strain (SH100/pBBE22luc) demonstrated a loss of infectivity in the heart, spleen, and ear when cultivation of infected tissues was the readout independent of the dose (Table 3). Interestingly, the qPCR results demonstrate that even tissues that are culture positive for SH100/pBBE22luc exhibit a lighter bacterial load than the parental and/or complemented strain, with the exception of the site of inoculation where the numbers are not significantly different (Fig. 5). One reasonable explanation for the phenotype observed is a defect in the bb0744 mutant's ability to bind to and colonize the host structures found in these tissues. However, this would not account for the mutant's being unable to bind cartilage in the ear yet still being able to infect joint cartilage, unless the ligands are not structures common to both of these locations. It is important to note that two additional genetic backgrounds lacking bb0744, specifically, strains B31 5A4NP1 and 297, exhibited a similar infectivity pattern in which heart tissue and distal skin sites were not infected (data not shown). These results indicated that the infectivity defect associated with the loss of BB0744 was not restricted to the B31 ML23 background alone and provide independent validation regarding the phenotype observed.
The combination of the cultivation data and bioluminescent imaging shows that SH100/pBBE22luc colonizes the joints posterior to the site of B. burgdorferi injection, as well as the regional (inguinal) lymph nodes (Table 3 and Fig. 4). These data also demonstrate that over the course of 3 weeks, in contrast to the parental and complemented strains, the SH100/pBBE22luc spirochetes are unable to move anterior to a point near the midline of the transverse plane of the mouse torso and never gain access to the upper torso, anterior limbs, or head (Fig. 4). We speculate that this location may reflect a limited ability to disseminate for reasons that are not yet known.
Another alternative for this observed phenotype posits that replication is delayed (or the survival rate is lower) during infection and, with slower infection kinetics, the spirochetes are unable to spread to the mouse anterior. However, this theory loses merit when infectivity is compared to that of the parental strain. SH100/pBBE22luc is able to invade the joints in the same time frame as its parent, ML23/pBBE22luc, yet is still unable to infect the spleen (Table 3). Wild-type virulent B. burgdorferi infects the spleen before the joints (13, 61). These data support the idea that either the kinetics of dissemination are impaired or B. burgdorferi may disseminate to target sites at differential rates, which reduces the spread of the spirochete to skin sites peripheral to the site of inoculation. Although this may be the case, particularly for spread throughout the skin, we cannot exclude the possibility that the mutant is also impaired for colonization at specific tissues. Assessment of alterations in tissue tropism will require additional validation but is feasible considering the variability in the tissues that B. burgdorferi can target during infection and the unique structures associated with these different tissues.
There are three primary mechanisms that may explain how a periplasmically localized protein like BB0744 may be required for the pathogenic potential of B. burgdorferi. (i) BB0744 is part of a metabolic or stress response pathway that regulates the expression of various surface proteins. (ii) BB0744 has a role in targeting surface proteins to the outer membrane or even “flipping” them to the outer leaflet, e.g., functioning or augmenting the activity of a membrane “flippase.” (iii) The size of BB0744 suggests that it may act as a linker between outer and inner membrane proteins. All of these theories need to be tested in future experiments.
Carditis is a common clinical feature of Lyme disease, with physical symptoms and medical complications linked to untreated infections (7, 12, 66). As SH100/pBBE22luc is unable to infect the heart during infection, it is possible that BB0744 is needed indirectly for the stabilization of an important cell envelope structure(s) that provides adhesion to host cells and thus promotes colonization of the heart and the subsequent development of carditis. We are currently exploring these possibilities to decipher how BB0744 affects these unique pathogenic properties.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steve Norris for generously providing borrelial strain 5A4. We acknowledge Johannes Eble and Hui Zhi for helpful discussions and Katherine Biancardi for technical assistance. We also acknowledge Daniel Voth for his assistance with microscopy and Allen Gies in the UAMS Sequencing Core Facility for technical assistance. Finally, we thank Darrin Akins for providing the BB0796-specific antiserum used in these studies.
This work was supported by Public Health Service grants AI087678 (to J.S.B.) and AI095935 (to J.T.S.) from the National Institute of Allergy and Infectious Diseases, as well as the UAMS Center for Microbial Pathogenesis and Host Inflammatory Responses (P20-GM103625).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00828-15.
REFERENCES
- 1.Kuehn BM. 2013. CDC estimates 300,000 US cases of Lyme disease annually. JAMA 310:1110. doi: 10.1001/jama.2013.278331. [DOI] [PubMed] [Google Scholar]
- 2.Hinckley AF, Connally NP, Meek JI, Johnson BJ, Kemperman MM, Feldman KA, White JL, Mead PS. 2014. Lyme disease testing by large commercial laboratories in the United States. Clin Infect Dis 59:676–681. doi: 10.1093/cid/ciu397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapiro ED. 2014. Lyme disease. N Engl J Med 370:1724–1731. doi: 10.1056/NEJMcp1314325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10:87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steere AC, Coburn J, Glickstein L. 2004. The emergence of Lyme disease. J Clin Invest 113:1093–1101. doi: 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanek G, Wormser GP, Gray J, Strle F. 2012. Lyme borreliosis. Lancet 379:461–473. doi: 10.1016/S0140-6736(11)60103-7. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention (CDC). 2013. Three sudden cardiac deaths associated with Lyme carditis—United States, November 2012–July 2013. MMWR Morb Mortal Wkly Rep 62:993–996. [PMC free article] [PubMed] [Google Scholar]
- 8.Eiffert H, Karsten A, Schlott T, Ohlenbusch A, Laskawi R, Hoppert M, Christen H-J. 2004. Acute peripheral facial palsy in Lyme disease—a distal neuritis at the infection site. Neuropediatrics 35:267–273. doi: 10.1055/s-2004-821174. [DOI] [PubMed] [Google Scholar]
- 9.Hansen K, Crone C, Kristoferitsch W. 2013. Lyme neuroborreliosis. Handb Clin Neurol 115:559–575. doi: 10.1016/B978-0-444-52902-2.00032-1. [DOI] [PubMed] [Google Scholar]
- 10.Li X, McHugh GA, Damle N, Sikand VK, Glickstein L, Steere AC. 2011. Burden and viability of Borrelia burgdorferi in skin and joints of patients with erythema migrans or Lyme arthritis. Arthritis Rheum 63:2238–2247. doi: 10.1002/art.30384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Strle K, Wang P, Acosta DI, McHugh GA, Sikand N, Strle F, Steere AC. 2013. Tick-specific borrelial antigens appear to be upregulated in American but not European patients with Lyme arthritis, a late manifestation of Lyme borreliosis. J Infect Dis 208:934–941. doi: 10.1093/infdis/jit269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wenger N, Pellaton C, Bruchez P, Schläpfer J. 2012. Atrial fibrillation, complete atrioventricular block and escape rhythm with bundle-branch block morphologies: an exceptional presentation of Lyme carditis. Int J Cardiol 160:e12–14. doi: 10.1016/j.ijcard.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Barthold SW, Persing DH, Armstrong AL, Peeples RA. 1991. Kinetics of Borrelia burgdorferi dissemination and evolution of disease after intradermal inoculation of mice. Am J Pathol 139:263–273. [PMC free article] [PubMed] [Google Scholar]
- 14.Campfield BT, Nolder CL, Marinov A, Bushnell D, Davis A, Spychala C, Hirsch R, Nowalk AJ. 2014. Follistatin-like protein 1 is a critical mediator of experimental Lyme arthritis and the humoral response to Borrelia burgdorferi infection. Microb Pathog 73:70–79. doi: 10.1016/j.micpath.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lochhead RB, Ma Y, Zachary JF, Baltimore D, Zhao JL, Weis JH, O'Connell RM, Weis JJ. 2014. MicroRNA-146a provides feedback regulation of Lyme arthritis but not carditis during infection with Borrelia burgdorferi. PLoS Pathog 10:e1004212. doi: 10.1371/journal.ppat.1004212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motameni A-R, Bates TC, Juncadella IJ, Petty C, Hedrick MN, Anguita J. 2005. Distinct bacterial dissemination and disease outcome in mice subcutaneously infected with Borrelia burgdorferi in the midline of the back and the footpad. FEMS Immunol Med Microbiol 45:279–284. doi: 10.1016/j.femsim.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Oosting M, Buffen K, van der Meer JWM, Netea MG, Joosten LAB. 25 June 2014. Innate immunity networks during infection with Borrelia burgdorferi. Crit Rev Microbiol doi: 10.3109/1040841X.2014.929563. [DOI] [PubMed] [Google Scholar]
- 18.Ritzman AM, Hughes-Hanks JM, Blaho VA, Wax LE, Mitchell WJ, Brown CR. 2010. The chemokine receptor CXCR2 ligand KC (CXCL1) mediates neutrophil recruitment and is critical for development of experimental Lyme arthritis and carditis. Infect Immun 78:4593–4600. doi: 10.1128/IAI.00798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaible UE, Wallich R, Kramer MD, Museteanu C, Simon MM. 1991. A mouse model for Borrelia burgdorferi infection: pathogenesis, immune response and protection. Behring Inst Mitt 88:59–67. [PubMed] [Google Scholar]
- 20.Lenhart TR, Kenedy MR, Yang X, Pal U, Akins DR. 2012. BB0324 and BB0028 are constituents of the Borrelia burgdorferi β-barrel assembly machine (BAM) complex. BMC Microbiol 12:60. doi: 10.1186/1471-2180-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behera AK, Durand E, Cugini C, Antonara S, Bourassa L, Hildebrand E, Hu LT, Coburn J. 2008. Borrelia burgdorferi BBB07 interaction with integrin alpha3beta1 stimulates production of pro-inflammatory mediators in primary human chondrocytes. Cell Microbiol 10:320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris G, Ma W, Maurer LM, Potts JR, Mosher DF. 2014. Borrelia burgdorferi protein BBK32 binds to soluble fibronectin via the N-terminal 70 kDa region, causing fibronectin to undergo conformational extension. J Biol Chem 289:22490–22499. doi: 10.1074/jbc.M114.578419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai DM, Samuels DS, Feng S, Hodzic E, Olsen K, Barthold SW. 2013. The early dissemination defect attributed to disruption of decorin-binding proteins is abolished in chronic murine Lyme borreliosis. Infect Immun 81:1663–1673. doi: 10.1128/IAI.01359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ristow LC, Miller HE, Padmore LJ, Chettri R, Salzman N, Caimano MJ, Rosa PA, Coburn J. 2012. The β3-integrin ligand of Borrelia burgdorferi is critical for infection of mice but not ticks. Mol Microbiol 85:1105–1118. doi: 10.1111/j.1365-2958.2012.08160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell TM, Johnson BJB. 2013. Lyme disease spirochaetes possess an aggrecan-binding protease with aggrecanase activity. Mol Microbiol 90:228–240. [DOI] [PubMed] [Google Scholar]
- 26.Salo J, Loimaranta V, Lahdenne P, Viljanen MK, Hytönen J. 2011. Decorin binding by DbpA and B of Borrelia garinii, Borrelia afzelii, and Borrelia burgdorferi sensu stricto. J Infect Dis 204:65–73. doi: 10.1093/infdis/jir207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Defoe G, Coburn J. 2001. Delineation of Borrelia burgdorferi p66 sequences required for integrin alpha(IIb)beta(3) recognition. Infect Immun 69:3455–3459. doi: 10.1128/IAI.69.5.3455-3459.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coutte L, Botkin DJ, Gao L, Norris SJ. 2009. Detailed analysis of sequence changes occurring during vlsE antigenic variation in the mouse model of Borrelia burgdorferi infection. PLoS Pathog 5:e1000293. doi: 10.1371/journal.ppat.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraiczy P, Stevenson B. 2013. Complement regulator-acquiring surface proteins of Borrelia burgdorferi: structure, function and regulation of gene expression. Ticks Tick-Borne Dis 4:26–34. doi: 10.1016/j.ttbdis.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cassatt DR, Patel NK, Ulbrandt ND, Hanson MS. 1998. DbpA, but not OspA, is expressed by Borrelia burgdorferi during spirochetemia and is a target for protective antibodies. Infect Immun 66:5379–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyde JA, Weening EH, Chang M, Trzeciakowski JP, Höök M, Cirillo JD, Skare JT. 2011. Bioluminescent imaging of Borrelia burgdorferi in vivo demonstrates that the fibronectin-binding protein BBK32 is required for optimal infectivity. Mol Microbiol 82:99–113. doi: 10.1111/j.1365-2958.2011.07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imai DM, Feng S, Hodzic E, Barthold SW. 2013. Dynamics of connective-tissue localization during chronic Borrelia burgdorferi infection. Lab Invest J Tech Methods Pathol 93:900–910. doi: 10.1038/labinvest.2013.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moriarty TJ, Shi M, Lin Y-P, Ebady R, Zhou H, Odisho T, Hardy P-O, Salman-Dilgimen A, Wu J, Weening EH, Skare JT, Kubes P, Leong J, Chaconas G. 2012. Vascular binding of a pathogen under shear force through mechanistically distinct sequential interactions with host macromolecules. Mol Microbiol 86:1116–1131. doi: 10.1111/mmi.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Troy EB, Lin T, Gao L, Lazinski DW, Camilli A, Norris SJ, Hu LT. 2013. Understanding barriers to Borrelia burgdorferi dissemination during infection using massively parallel sequencing. Infect Immun 81:2347–2357. doi: 10.1128/IAI.00266-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wormser GP, Brisson D, Liveris D, Hanincová K, Sandigursky S, Nowakowski J, Nadelman RB, Ludin S, Schwartz I. 2008. Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J Infect Dis 198:1358–1364. doi: 10.1086/592279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norman MU, Moriarty TJ, Dresser AR, Millen B, Kubes P, Chaconas G. 2008. Molecular mechanisms involved in vascular interactions of the Lyme disease pathogen in a living host. PLoS Pathog 4:e1000169. doi: 10.1371/journal.ppat.1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tunev SS, Hastey CJ, Hodzic E, Feng S, Barthold SW, Baumgarth N. 2011. Lymphoadenopathy during Lyme borreliosis is caused by spirochete migration-induced specific B cell activation. PLoS Pathog 7:e1002066. doi: 10.1371/journal.ppat.1002066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weening EH, Parveen N, Trzeciakowski JP, Leong JM, Höök M, Skare JT. 2008. Borrelia burgdorferi lacking DbpBA exhibits an early survival defect during experimental infection. Infect Immun 76:5694–5705. doi: 10.1128/IAI.00690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hastey CJ, Elsner RA, Barthold SW, Baumgarth N. 2012. Delays and diversions mark the development of B cell responses to Borrelia burgdorferi infection. J Immunol 188:5612–5622. doi: 10.4049/jimmunol.1103735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasiah C, Rauer S, Gassmann GS, Vogt A. 1994. Use of a hybrid protein consisting of the variable region of the Borrelia burgdorferi flagellin and part of the 83-kDa protein as antigen for serodiagnosis of Lyme disease. J Clin Microbiol 32:1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rauer S, Kayser M, Neubert U, Rasiah C, Vogt A. 1995. Establishment of enzyme-linked immunosorbent assay using purified recombinant 83-kilodalton antigen of Borrelia burgdorferi sensu stricto and Borrelia afzelii for serodiagnosis of Lyme disease. J Clin Microbiol 33:2596–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilske B, Habermann C, Fingerle V, Hillenbrand B, Jauris-Heipke S, Lehnert G, Pradel I, Rössler D, Schulte-Spechtel U. 1999. An improved recombinant IgG immunoblot for serodiagnosis of Lyme borreliosis. Med Microbiol Immunol 188:139–144. doi: 10.1007/s004300050116. [DOI] [PubMed] [Google Scholar]
- 43.Yang X, Promnares K, Qin J, He M, Shroder DY, Kariu T, Wang Y, Pal U. 2011. Characterization of multiprotein complexes of the Borrelia burgdorferi outer membrane vesicles. J Proteome Res 10:4556–4566. doi: 10.1021/pr200395b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zückert WR. 2007. Laboratory maintenance of Borrelia burgdorferi. Curr Protoc Microbiol Chapter 12:Unit 12C.1. [DOI] [PubMed] [Google Scholar]
- 45.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 46.Frank KL, Bundle SF, Kresge ME, Eggers CH, Samuels DS. 2003. aadA confers streptomycin resistance in Borrelia burgdorferi. J Bacteriol 185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elias AF, Bono JL, Kupko JJ 3rd, Stewart PE, Krum JG, Rosa PA. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol 6:29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- 48.Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 49.Groshong AM, Fortune DE, Moore BP, Spencer HJ, Skinner RA, Bellamy WT, Blevins JS. 2014. BB0238, a presumed tetratricopeptide repeat-containing protein, is required during Borrelia burgdorferi mammalian infection. Infect Immun 82:4292–4306. doi: 10.1128/IAI.01977-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Norris SJ, Carter CJ, Howell JK, Barbour AG. 1992. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun 60:4662–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Probert WS, Allsup KM, LeFebvre RB. 1995. Identification and characterization of a surface-exposed, 66-kilodalton protein from Borrelia burgdorferi. Infect Immun 63:1933–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samuels DS. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol Biol 47:253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyde JA, Weening EH, Skare JT. 2011. Genetic transformation of Borrelia burgdorferi. Curr Protoc Microbiol Chapter 12:Unit 12C.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhi H, Weening EH, Barbu EM, Hyde JA, Höök M, Skare JT. 2015. The BBA33 lipoprotein binds collagen and impacts Borrelia burgdorferi pathogenesis. Mol Microbiol 96:68–83. doi: 10.1111/mmi.12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Labandeira-Rey M, Skare JT. 2001. Decreased Infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect Immun 69:446–455. doi: 10.1128/IAI.69.1.446-455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liveris D, Wang G, Girao G, Byrne DW, Nowakowski J, McKenna D, Nadelman R, Wormser GP, Schwartz I. 2002. Quantitative detection of Borrelia burgdorferi in 2-millimeter skin samples of erythema migrans lesions: correlation of results with clinical and laboratory findings. J Clin Microbiol 40:1249–1253. doi: 10.1128/JCM.40.4.1249-1253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pal U, Li X, Wang T, Montgomery RR, Ramamoorthi N, deSilva AM, Bao FK, Yang XF, Pypaert M, Pradhan D, Kantor FS, Telford S, Anderson JF, Fikrig E. 2004. TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell 119:457–468. doi: 10.1016/j.cell.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 58.Rössler D, Eiffert H, Jauris-Heipke S, Lehnert G, Preac-Mursic V, Teepe J, Schlott T, Soutschek E, Wilske B. 1995. Molecular and immunological characterization of the p83/100 protein of various Borrelia burgdorferi sensu lato strains. Med Microbiol Immunol 184:23–32. doi: 10.1007/BF00216786. [DOI] [PubMed] [Google Scholar]
- 59.Nowalk AJ, Gilmore RD, Carroll JA. 2006. Serologic proteome analysis of Borrelia burgdorferi membrane-associated proteins. Infect Immun 74:3864–3873. doi: 10.1128/IAI.00189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nowalk AJ, Nolder C, Clifton DR, Carroll JA. 2006. Comparative proteome analysis of subcellular fractions from Borrelia burgdorferi by NEPHGE and IPG. Proteomics 6:2121–2134. doi: 10.1002/pmic.200500187. [DOI] [PubMed] [Google Scholar]
- 61.Labandeira-Rey M, Seshu J, Skare JT. 2003. The absence of linear plasmid 25 or 28-1 of Borrelia burgdorferi dramatically alters the kinetics of experimental infection via distinct mechanisms. Infect Immun 71:4608–4613. doi: 10.1128/IAI.71.8.4608-4613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purser JE, Lawrenz MB, Caimano MJ, Howell JK, Radolf JD, Norris SJ. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol 48:753–764. doi: 10.1046/j.1365-2958.2003.03452.x. [DOI] [PubMed] [Google Scholar]
- 63.Blevins JS, Revel AT, Smith AH, Bachlani GN, Norgard MV. 2007. Adaptation of a luciferase gene reporter and lac expression system to Borrelia burgdorferi. Appl Environ Microbiol 73:1501–1513. doi: 10.1128/AEM.02454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lazarus JJ, Meadows MJ, Lintner RE, Wooten RM. 2006. IL-10 deficiency promotes increased Borrelia burgdorferi clearance predominantly through enhanced innate immune responses. J Immunol 177:7076–7085. doi: 10.4049/jimmunol.177.10.7076. [DOI] [PubMed] [Google Scholar]
- 65.Coburn J, Fischer JR, Leong JM. 2005. Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol Microbiol 57:1182–1195. doi: 10.1111/j.1365-2958.2005.04759.x. [DOI] [PubMed] [Google Scholar]
- 66.Semmler D, Blank R, Rupprecht H-J. 2010. Complete AV block in Lyme carditis: an important differential diagnosis. Clin Res Cardiol 99:519–526. doi: 10.1007/s00392-010-0152-8. [DOI] [PubMed] [Google Scholar]
- 67.Shaw DK, Hyde JA, Skare JT. 2012. The BB0646 protein demonstrates lipase and haemolytic activity associated with Borrelia burgdorferi, the aetiological agent of Lyme disease. Mol Microbiol 83:319–334. doi: 10.1111/j.1365-2958.2011.07932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maruskova M, Esteve-Gassent MD, Sexton VL, Seshu J. 2008. Role of the BBA64 locus of Borrelia burgdorferi in early stages of infectivity in a murine model of Lyme disease. Infect Immun 76:391–402. doi: 10.1128/IAI.01118-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Purser JE, Norris SJ. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A 97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawabata H, Norris SJ, Watanabe H. 2004. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect Immun 72:7147–7154. doi: 10.1128/IAI.72.12.7147-7154.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steere AC, Grodzicki RL, Craft JE, Shrestha M, Kornblatt AN, Malawista SE. 1984. Recovery of Lyme disease spirochetes from patients. Yale J Biol Med 57:557–560. [PMC free article] [PubMed] [Google Scholar]
- 72.Pal U, Wang P, Bao F, Yang X, Samanta S, Schoen R, Wormser GP, Schwartz I, Fikrig E. 2008. Borrelia burgdorferi basic membrane proteins A and B participate in the genesis of Lyme arthritis. J Exp Med 205:133–141. doi: 10.1084/jem.20070962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.