Abstract
Intracellular bacteria use a variety of strategies to evade degradation and create a replicative niche. Legionella pneumophila is an intravacuolar pathogen that establishes a replicative niche through the secretion of more than 300 effector proteins. The function of most effectors remains to be determined. Toxicity in yeast has been used to identify functional domains and elucidate the biochemical function of effectors. A library of L. pneumophila effectors was screened using an expression plasmid that produces low levels of each protein. This screen identified the effector SdeA as a protein that confers a strong toxic phenotype that inhibits yeast replication. The toxicity of SdeA was suppressed in cells producing the effector SidJ. The effector SdeA is a member of the SidE family of L. pneumophila effector proteins. All SidE orthologs encoded by the Philadelphia isolate of Legionella pneumophila were toxic to yeast, and SidJ suppressed the toxicity of each. We identified a conserved central region in the SidE proteins that was sufficient to mediate yeast toxicity. Surprisingly, SidJ did not suppress toxicity when this central region was produced in yeast. We determined that the amino-terminal region of SidE was essential for SidJ-mediated suppression of toxicity. Thus, there is a genetic interaction that links the activity of SidJ and the amino-terminal region of SidE, which is required to modulate the toxic activity displayed by the central region of the SidE protein. This suggests a complex mechanism by which the L. pneumophila effector SidJ modulates the function of the SidE proteins after translocation into host cells.
INTRODUCTION
Legionella pneumophila is a Gram-negative, facultative intravacuolar pathogen that is the causative agent of Legionnaires' disease in humans (1–3). L. pneumophila is phagocytosed by host cells and creates a vacuole that supports replication by inhibiting the fusion of host endocytic vesicles and subverting the transport of secretory vesicles (4–7). The bacterially encoded Dot/Icm type IV secretion system (T4SS) is essential for the creation of this Legionella-containing vacuole (LCV) (8, 9). Proteins secreted by the T4SS, known as effectors, have been shown to both decorate the LCV and modify the location and function of host proteins, especially Rab GTPases (8, 10, 11). Determining the function of these effector proteins has been complicated by the observation that deletion of a single effector or groups of effectors does not typically result in a strong replication defect within host cells (12–14).
The use of the yeast Saccharomyces cerevisiae as a system to study L. pneumophila effector proteins has been successful in previous studies (15–19). Examples include studies on the effector protein RalF, which was shown to inhibit yeast replication when overproduced, and the effector proteins YlfA and YlfB were both identified in an early yeast toxicity screen (16). Effector toxicity in yeast has also been used to study effector activities. The L. pneumophila effector AnkX is capable of transferring phosphocholine onto Rab1 family members (20), and this activity results in yeast toxicity due to phosphocholination of the Rab1 homolog Ypt1 (21). The effector Lem3 was identified because it has the ability to inhibit AnkX toxicity in yeast, which is because Lem3 has an activity that promotes dephosphocholination of Rab1 family members (21). Thus, yeast is an effective model organism that can be used for both the identification and the elucidation of effector function.
Several recent studies have increased the number of identified T4SS effectors to more than 300 (22–28). We created a plasmid library containing the majority of effector proteins expressed from a constitutive low-expression vector. Using this library, we have identified SidE family members as being toxic to yeast and have found that the SidJ protein is capable of counteracting this toxicity. Thus, the SidE and SidJ proteins are predicted to work in concert to regulate host processes during infection by L. pneumophila. These data are consistent with a recent study that showed a functional interaction between SdeA and SidJ in yeast and on the LCV (29). In addition, we show here that SdeA contains distinct toxicity and SidJ suppression regions. The ability of SidJ to counteract SidE family member toxicity and the presence of specific regions for each of these activities suggest that these proteins functionally interact during infection.
MATERIALS AND METHODS
Strains and plasmid construction.
Effector genes were amplified from Legionella pneumophila serogroup 1 strain Lp01 (30) genomic DNA by PCR and recombined into the Gateway recombination system (Life Technologies) donor vector pDONR223 (31). Correct effector gene recombination was confirmed by sequencing (Keck Sequencing, Yale University). Effector genes were then recombined into Saccharomyces cerevisiae expression vector pDEST22 or pDEST32 (Life Technologies). Truncated effectors were constructed by PCR using primers designed to amplify the desired region of the gene from the sequenced pDONR223 plasmid. Products were then recombined into pDONR223, sequenced, and inserted into pDEST32 as described above. During plasmid construction, products of recombination reactions were transformed into Escherichia coli strain Top10 by calcium chloride transformation. Saccharomyces strains MATa Y8800 and MATα Y8930 (leu2-3,112 trp1-901 his3Δ200 ura3-52 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ cyh2R) (32) were transformed individually with pDEST22 and pDEST32 plasmids as described below. Effector overexpression experiments utilized the glucose-galactose-inducible plasmid pGML10 (33). Effectors were amplified from sequenced pDONR223 plasmids by PCR using primers that contained 5′ BamHI and 3′ SalI restriction sites. PCR products and pGML10 were then digested with BamHI and SalI and digestion products were ligated with T4 ligase (NEB). Products of ligation reactions were subsequently transformed into Top10 cells, confirmed by sequencing, and then transformed into yeast strain BY4742.
Media.
L. pneumophila strains were grown on charcoal yeast extract plates [1% yeast extract, 1% N-(2-acetamido)-2-aminoethanesulfonic acid (ACES; pH 6.9), 3.3 mM l-cysteine, 0.33 mM Fe(NO3)3, 1.5% Bacto agar, 0.2% activated charcoal] at 37°C. Wild-type Saccharomyces was grown on YPAD medium (1% yeast extract, 2% Bacto peptone, 100 mg/liter adenine hemisulfate, 2% Bacto agar, and 2% glucose) at 30°C. Saccharomyces transformed with pDEST22 or pDEST32 was grown at 30°C on synthetic complete (SC) medium (0.06% −Ade, −His, −Leu, −Trp dropout mix [Clontech], 0.67% yeast nitrogen base, 2% Bacto agar, and 2% glucose) supplemented with 0.08 mg/ml adenine, 0.8 mM histidine, 0.8 mM leucine, or 0.32 mM tryptophan as appropriate for growth. Experiments testing effector overexpression were plated on SC medium with 2% galactose and 1% raffinose as a carbon source in place of glucose. Escherichia coli strain Top10 was cultivated in Luria-Bertani medium with antibiotics when necessary at the following concentrations: ampicillin, 100 μg/ml; spectinomycin, 100 μg/ml; and gentamicin, 10 μg/ml. Liquid cultures were grown in broth medium of the same composition as described above but lacking agar.
Saccharomyces transformation.
Saccharomyces samples were transformed with plasmids by the lithium acetate/single-stranded carrier DNA/polyethylene glycol method (34). Overnight cultures of a strain grown in YPAD or SC liquid medium were pelleted and then resuspended in a T-mix solution (300 mM lithium acetate and 0.8 mg of boiled sonicated salmon sperm DNA/ml). Next, 5 to 10 μl of plasmid was added to resuspended cells for a total transformation volume of 50 μl. Transformations were incubated at 42°C for 1 h and 30 min. Cells were pelleted, and the transformation mix was aspirated. Cells were resuspended in 50 μl of distilled H2O. Then, 5 to 10 μl of resuspended cells was plated on the appropriate selective medium. Plates were incubated at 30°C for 2 to 4 days.
RESULTS
L. pneumophila effector toxicity in yeast.
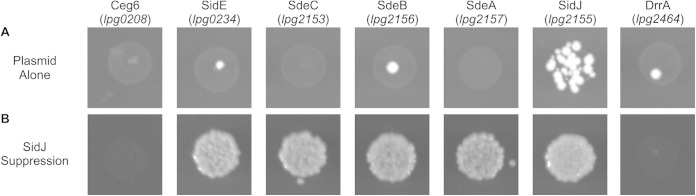
During the construction of a yeast two-hybrid library of effector proteins, we transformed 300 individually cloned effector proteins encoded by gateway vectors pDEST22 and pDEST32 (Life Technologies) (Fig. 1 and Table 1) (35). Effectors were amino-terminally tagged with the DNA binding or activation domain of Gal4 and were constitutively expressed from the PADH1 promoter. Effector expression, although not measured, should be considerably lower than when overexpressed from a glucose-galactose expression system. Effector transformation yielded a variety of phenotypes, from no change in viability to lethality (Fig. 1). The observation of effector toxicity at low levels of expression indicated that some L. pneumophila effectors possessed a much stronger toxic phenotype in yeast than previously appreciated. L. pneumophila effectors YlfA (lpg2298) and YlfB (lpg1884) (Fig. 1, labeled “$” and “#”, respectively) were previously identified by Campodonico et al. (16) as toxic when overexpressed in yeast; however, when the Ylf proteins were expressed at low levels they did not display a pronounced toxic phenotype compared to the effector proteins SidE (lpg0234) and SdeA (lpg2157) (Fig. 1, labeled “*” and “@”, respectively). Importantly, effectors previously identified as being toxic when produced at high levels in yeast, such as AnkX (lpg0695) (Fig. 1, labeled “&”), also conferred a growth defect when produced in yeast by these pDEST vectors, indicating that this transformation approach had the capacity to identify toxic effectors (21). In an attempt to verify that apparent differences in yeast viability were the result of effector transformation, as opposed to plasmid transformation efficiency, several rounds of transformations of putatively toxic effectors were attempted using different concentrations of plasmid DNA encoding the effectors (data not shown). From this screen we identified two effectors that were highly toxic to yeast, which were the effectors Ceg6 (lpg0208) and SdeA (lpg2157).
FIG 1.
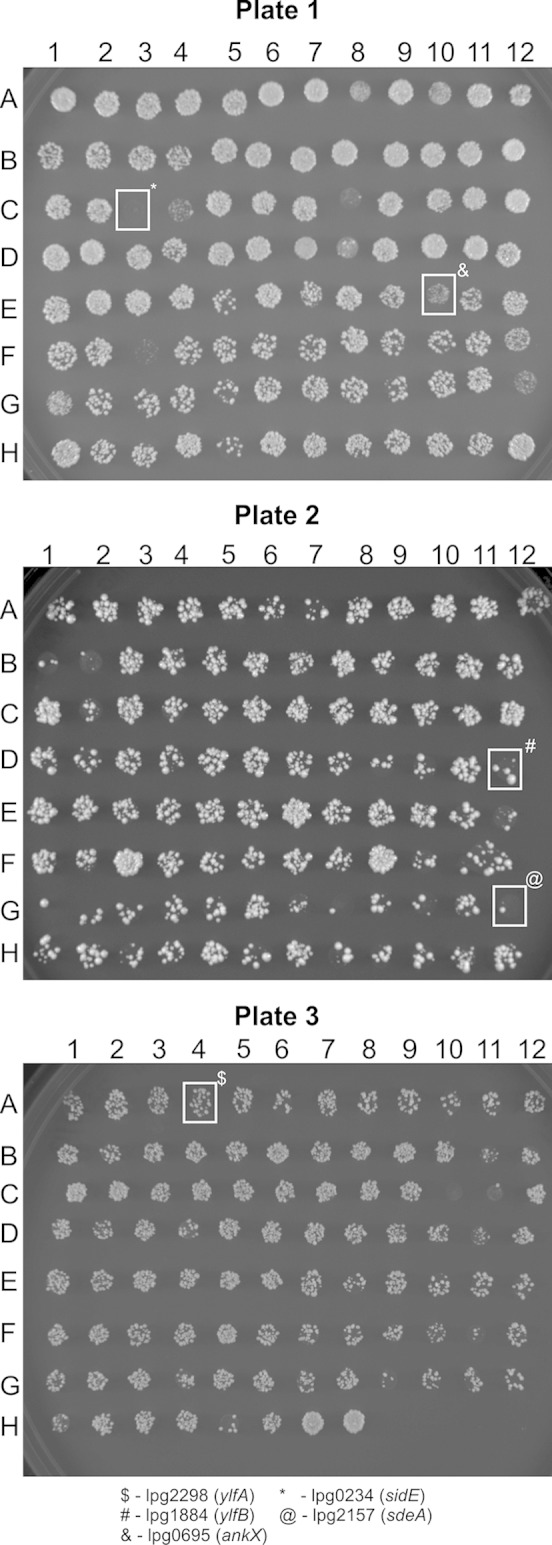
L. pneumophila effector toxicity in yeast. Effectors encoded from the constitutive low-expression plasmid pDEST22 were transformed into yeast and then plated on SC −Trp selective medium. Plates were incubated at 30°C. Transformed effectors are indicated in Table 1. Of specific note, spotted transformation marked with a “$” represents cells transformed with ylfA (lpg2298), transformation marked with a “#” represents cells transformed with ylfB (lpg1884), transformation marked with a “&” represents cells transformed with ankX (lpg0695), transformation marked with a “*” represents cells transformed with sidE (lpg0234), and transformation marked with a “@” represents cells transformed with sdeA (lpg2157).
TABLE 1.
Legionella gene numbers of effectors transformed into Saccharomycesa
| Plate no. and row |
Legionella gene (lpg locus no. or gene name) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| Plate 1 | ||||||||||||
| A | 0008 | 0012 | 0021 | 0030 | 0038 | 0045 | 0059 | 0080 | 0081 | 0086 | 0096 | 0103 |
| B | 0107 | 0126 | 0130 | 0135 | 0160 | 0171 | 0172 | 0181 | 0191 | 0195 | 0196 | 0209 |
| C | 0210 | 0227 | 0234 | 0240 | 0246 | 0254 | 0260 | 0275 | 0276 | 0284 | 0285 | 0294 |
| D | 0364 | 0365 | 0375 | 0376 | 0390 | 0393 | 0401 | 0402 | 0403 | 0405 | 0422 | 0436 |
| E | 0439 | 0515 | 0518 | 0519 | 0563 | 0621 | 0634 | 0642 | 0645 | 0695 | 0716 | 0717 |
| F | 0733 | 0796 | 0898 | 0921 | 0926 | 0940 | 0944 | 0945 | 0963 | 0967 | 0968 | 0969 |
| G | 1083 | 1101 | 1106 | 1108 | 1109 | 1110 | 1111 | 1120 | 1121 | 1124 | 1129 | 1137 |
| H | 1144 | 1147 | 1148 | 1152 | 1154 | 1158 | 1171 | 1183 | 1227 | 1273 | 1290 | rab1A |
| Plate 2 | ||||||||||||
| A | 1312 | 1316 | 1317 | 1328 | 1354 | 1356 | 1368 | 1408 | 1449 | 1453 | 1483 | 1484 |
| B | 1488 | 1489 | 1496 | 1551 | 1578 | 1588 | 1598 | 1602 | 1621 | 1625 | 1639 | 1642 |
| C | 1654 | 1660 | 1663 | 1666 | 1667 | 1683 | 1684 | 1685 | 1687 | 1692 | 1701 | 1702 |
| D | 1716 | 1717 | 1718 | 1751 | 1752 | 1797 | 1798 | 1803 | 1822 | 1851 | 1871 | 1884 |
| E | 1888 | 1890 | 1907 | 1924 | 1933 | 1947 | 1948 | 1949 | 1950 | 1953 | 1958 | 1959 |
| F | 1960 | 1961 | 1962 | 1963 | 1964 | 1965 | 1966 | 1969 | 1972 | 1975 | 1976 | 1978 |
| G | 1986 | 2050 | 2073 | 2131 | 2137 | 2144 | 2147 | 2148 | 2149 | 2154 | 2155 | 2157 |
| H | 2160 | 2164 | 2176 | 2199 | 2200 | 2206 | 2215 | 2216 | 2222 | 2223 | 2224 | 2239 |
| Plate 3 | ||||||||||||
| A | 2248 | 2271 | 2283 | 2298 | 2300 | 2311 | 2327 | 2328 | 2344 | 2351 | 2359 | 2370 |
| B | 2372 | 2375 | 2382 | 2391 | 2392 | 2395 | 2400 | 2406 | 2407 | 2409 | 2410 | 2416 |
| C | 2420 | 2422 | 2424 | 2425 | 2434 | 2443 | 2444 | 2455 | 2456 | 2461 | 2464 | 2465 |
| D | 2482 | 2490 | 2498 | 2504 | 2505 | 2508 | 2509 | 2511 | 2523 | 2526 | 2527 | 2529 |
| E | 2538 | 2539 | 2541 | 2546 | 2555 | 2556 | 2577 | 2588 | 2591 | 2603 | 2628 | 2637 |
| F | 2638 | 2678 | 2692 | 2694 | 2718 | 2720 | 2744 | 2745 | 2793 | 2804 | 2806 | 2813 |
| G | 2815 | 2819 | 2826 | 2828 | 2830 | 2831 | 2832 | 2844 | 2862 | 2874 | 2884 | 2885 |
| H | 2888 | 2907 | 2912 | 2936 | 2975 | 2999 | icmS | icmW | ||||
Suppression of effector toxicity.
We next screened for effectors that were capable of suppressing the toxic activity displayed by Ceg6 and SdeA. As a control, we found that yeast toxicity mediated by AnkX was suppressed by Lem3 (data not shown), which was predicted based on previous studies (21, 36–38). Yeast strains producing individual effectors were pooled, and a plasmid encoding Ceg6 or SdeA was transformed into the yeast pool. The prediction was that colonies would only be produced by yeasts producing an effector capable of counteracting the toxic activity of Ceg6 or SdeA. Candidate suppressors were identified using this approach (Fig. 2A). Sequencing of the effectors encoded by the surviving yeast identified the following candidates capable of suppressing SdeA or Ceg6 function: for toxic effector Ceg6 (lpg0208) we identified the suppressing effectors RavO (lpg1129) and MavM (lpg2577), and for toxic effector SdeA (lpg2157) we identified the suppressing effector SidJ (lpg2155). Candidates were screened by directly transforming Ceg6 or SdeA into yeast containing a plasmid encoding the effector predicted to function as a suppressor; yeast encoding Rab1A was used as a negative control (Fig. 2B). This analysis identified SidJ (lpg2155) as an effector capable of suppressing SdeA toxicity. All other candidates failed to show suppression when directly challenged with the toxic effector. The genes sidJ and sdeA are carried together on a 20-kb stretch of DNA on the L. pneumophila chromosome that also carried two other SidE family members: sdeB and sdeC (29, 39). The previously identified examples of effectors capable of suppressing the toxicity of other effectors have revealed genetic linkage, which provides further support for the hypothesis that SidJ may regulate the toxic activity displayed by SdeA.
FIG 2.
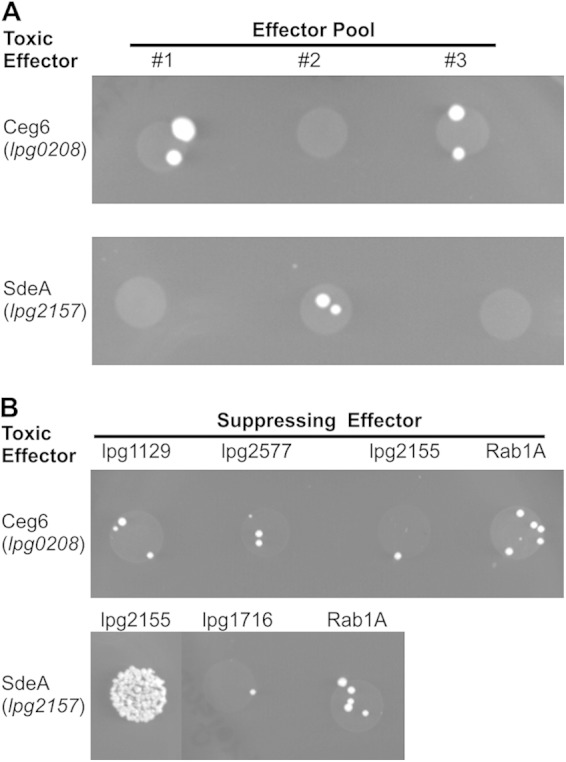
Effector-based suppression of SdeA and Ceg6 toxicity. (A) Yeast cells were individually transformed with each member of the L. pneumophila effector repertoire expressed from the pDEST22 plasmid and grown on selective media (data not shown). Each individually transformed cell was grown in liquid culture selecting for pDEST22 plasmid and then pooled. Pools were then transformed with either pDEST32-lpg0208(ceg6) or pDEST32-lpg2157(sdeA). Transformations were plated on SC −Leu, −Trp medium, which selects for both plasmids, and incubated at 30°C. (B) Effectors identified as suppressing Ceg6 or SdeA toxicity through the pooled assay by sequencing were individually transformed into yeast; Rab1A was used as a negative control. Transformed bacteria then underwent a second transformation with Ceg6 or SdeA and were plated on SC −Leu, −Trp medium, which selects for both plasmids. Transformations were incubated at 30°C.
As previously mentioned, SdeA is a member of a protein family that shares homology to the L. pneumophila effector SidE (lpg0234) (39). Given the high degree of sequence homology between all of the orthologs in the SidE family of proteins, we investigated whether the other members of the SidE family were toxic to yeast. The effectors SidE, SdeB, and SdeC each showed a similar level of toxicity when expressed in yeast (Fig. 3A). The effector SidJ was able to suppress the toxicity of each SidE family member (Fig. 3B), which indicates a conserved mechanism of both SidE-mediated toxicity and SidJ-mediated suppression. The effector SidJ was unable to inhibit toxicity conferred by Ceg6 or DrrA (Fig. 2B and 3B), which suggests that the ability of SidJ to interfere with the toxicity of L. pneumophila effectors is specific for the SidE family of proteins.
FIG 3.
SidJ suppression of SidE family toxicity. (A) The indicated effectors were transformed into yeast and expressed from plasmid pDEST32. Transformations were grown on SC −Leu plates at 30°C. (B) SidJ, encoded from pDEST22, was transformed into wild-type yeast, and transformants were selected on SC −Trp (data not shown). These transformants were then transformed with the indicated effectors encoded from the pDEST32 plasmid. Transformants were plated on SC −Leu, −Trp medium selecting for both plasmids, and incubated at 30°C.
To determine whether the levels of SdeA and SidJ were important for the observed toxicity and suppression, we used a glucose-galactose-inducible yeast expression system encoded by the plasmid pGML10 (33). Plasmids containing either effector could be successfully transformed if the yeast were plated on medium that repressed effector expression; however, when the resulting colonies of yeast were incubated in medium that induced effector expression, there was no growth detected for yeast producing SdeA alone or for yeast producing SidJ alone (Fig. 4). This suggests that SdeA and SidJ are both toxic to yeast when produced at high levels. This observed toxicity and suppression are consistent with recently published data showing a similar relationship between SdeA and SidJ (29).
FIG 4.
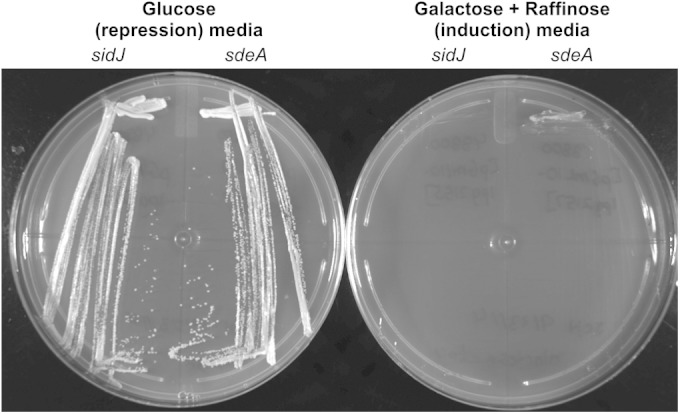
Overexpression of SdeA and SidJ. Saccharomyces strains were transformed with sdeA and sidJ expressed from a glucose-galactose-inducible expression system on pGML10. Transformed cells were plated on medium that repressed protein expression, i.e., SC −Leu with 2% glucose. Single colonies were then streaked onto repressing (glucose) and induction (galactose + raffinose) media. Plates were incubated at 30°C.
A conserved central region in SidE proteins confers toxicity to yeast.
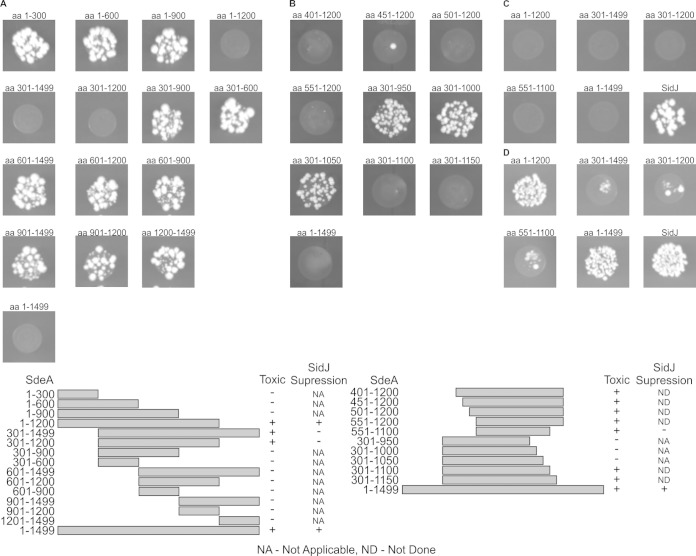
All of the members of the SidE family of proteins are large, greater than 1,400 amino acids, making it difficult to determine potential function. Deletion analysis was used to identify a minimal region in SdeA that was sufficient for yeast toxicity (Fig. 5A). This analysis revealed that deletion of the 300 amino-terminal residues and/or deletion of 300 carboxyl-terminal residues of SdeA did not affect yeast toxicity. Smaller deletions of 50 amino acids were then used to further delineate a minimal region in SdeA that conferred yeast toxicity (Fig. 5B and C). This analysis revealed that a central region between amino acids 551 and 1100 in SdeA was both necessary and sufficient for yeast toxicity. Pairwise protein sequence alignment of the minimal toxicity region of SdeA with other SidE homologs revealed a >60% homology within this region (data not shown), which is consistent with data suggesting that there is a conserved mechanism by which SidE proteins cause yeast toxicity.
FIG 5.
SdeA has distinct toxicity and SidJ suppression regions. (A to C) Truncations of SdeA were made by PCR amplification of genes from the pDONR223 vector and then recombined into pDEST32 vector by Gateway recombination, as described in Materials and Methods. Plasmids were transformed into wild-type yeast and plated on SC −Leu medium at 30°C. Specific truncations are indicated above the transformation. SidJ was transformed as a control to observe the full transformation efficiency (see panel C). (D) Yeast cells were transformed with SidJ (data not shown), and then the indicated SdeA truncation was transformed. Transformations were plated on SC −Leu, −Trp medium, which selects for both plasmids, followed by incubation at 30°C. aa, amino acids.
The amino-terminal region of SidE is required for SidJ-mediated suppression of yeast toxicity.
We next examined the ability of SidJ to suppress yeast toxicity mediated by the conserved central region of SdeA. Unexpectedly, SidJ was unable to suppress yeast toxicity mediated by the SdeA551−1100 protein (Fig. 5D). The panel of SdeA deletion mutants was used to identify the region of SdeA that was necessary for SidJ-mediated suppression of toxicity. These data show that SdeA deletions lacking the amino-terminal 300 amino acids were toxic to yeast and that this toxicity was not suppressed by SidJ (Fig. 5D). In contrast, deletion of the carboxyl-terminal 300 amino acids in SdeA had no effect on suppression of yeast toxicity by SidJ. Further deletions of SdeA from the amino terminus that were toxic in yeast could also not be suppressed by SidJ (Fig. 5D). This result mitigates the possibility that the observed inability of SidJ to counteract SdeA toxicity is an artifact of the deletion construct and indicates that there is a genetic interaction between SidJ and the amino-terminal region of SdeA that regulates the toxic activity displayed by SdeA. Because the sdeA gene encodes a very large protein that is produced at low levels in yeast, it was difficult to detect steady-state levels of the SdeA protein by immunoblot analysis. However, the ability of SidJ to suppress the toxicity of all SdeA truncation derivatives except those that have deletions in the amino-terminal region suggests that a genetic interaction between SidJ and the amino-terminal region of SdeA is required for the suppression of toxicity. Multiple sequence alignment of the amino terminus of SdeA with SidE family members in L. pneumophila confirms that this region is highly conserved in all of the SidE proteins (Fig. 6), which would be consistent with this region being important for the genetic interaction that leads to SidJ-mediated suppression of toxicity mediated by SidE proteins. Deletion proteins that consisted of 200-amino-acid truncations from either the amino or the carboxyl terminus of SidJ resulted in proteins that failed to suppress SdeA toxicity (data not shown), which suggests that there is not a minimal domain in SidJ that is sufficient for suppressing toxicity mediated by SidE proteins. From these data we conclude that SidJ is an effector that modulates a toxic activity encoded by a central region in all SidE family members and that SidJ-mediated control of SidE function requires an amino-terminal region in the SidE proteins.
FIG 6.
SidE proteins have a conserved amino-terminal domain. CLUSTAL W2 multiple-sequence alignment of the amino-terminal 300 amino acids from SdeA with the three other SidE family members.
DISCUSSION
Legionella pneumophila has served as an excellent model system for studying the mechanism by which T4SS effectors manipulate mammalian host cells. L. pneumophila secretes ∼300 effector proteins into the host cytosol via a T4SS that it uses to manipulate the process of vacuole maturation (8, 25). The identification of effector proteins that have important intracellular functions has been challenging because elimination of a single effector, or even large clusters of effectors, does not typically interfere with the ability of L. pneumophila to replicate intracellularly (13, 14). In the present study, we used screened effectors for toxic activities in yeast, an approach which has previously been shown to be an effective method for identifying effector functions that target eukaryotic pathways (15–19). Effectors were found to have a variety of effects on yeast viability, from no effect to lethality (Fig. 1). We selected effectors SdeA and Ceg6, which showed the most reproducible defects in yeast replication, for further study. Ceg6 was previously identified as an L. pneumophila gene containing an upstream PmrA binding site and was designated as being coregulated with effector-encoding genes (ceg) (40). Ceg6 was also predicted to have a serine/threonine kinase domain or motif, although no work has been done to further characterize the function of this effector. SdeA was originally identified as a SidE paralog and was subsequently shown to be translocated into the host cell by a Dot/Icm-dependent mechanism that requires the protein IcmS (27, 39). Deletion of SidE and its three paralogs—SdeA, SdeB, and SdeC—has been shown to have a modest effect on L. pneumophila intracellular replication, which could be complemented by expression of SdeA (39). Thus, modulation of cellular functions by SidE proteins is likely important for transport and replication of L. pneumophila inside host cells.
The effector repertoire of L. pneumophila was screened for effectors that could suppress toxicity mediated by SdeA and Ceg6. The effector SidJ was capable of specifically counteracting the toxicity of SdeA and the other members of the SidE family of effector proteins (Fig. 2 and 3). The ability of infectious bacteria to regulate the function of their effectors with other effectors has been previously established in L. pneumophila, as well as in the type III secretion system in Salmonella (41–44). In L. pneumophila, effector functions displayed by DrrA (SidM) have been shown to be modulated by multiple other effectors (10). The Rab1 guanine nucleotide exchange factor (GEF) activity displayed by DrrA can be counteracted by the Rab1 GTPase activating protein activity displayed by the effector LepB (42–44). DrrA also has a domain that adenosine monophosphorylates (AMPylates) Rab1, which is reversed by the de-AMPylation activity of the effector SidD (20, 21, 37, 45). Another example of effectors regulating the activity of other effectors in L. pneumophila involves the phosphocholinase/decholinase activity of AnkX and Lem3, respectively (20, 21, 38). What is important about these examples is that the effectors with opposing activities can be linked genetically, but the effectors themselves do not interact physically with each other. This is not the case for the first classic example of an L. pneumophila effector modulating the activity of another effector, which is the targeting of the effector SidH by the ubiquitin-ligase activity of the effector LubX to promote the proteasomal degradation of SidH inside the host cell (46). Importantly, the genes encoding most effectors that interact with each other functionally are closely linked on the L. pneumophila chromosome. Indeed, the observation that SidJ is encoded at a genetic locus that includes the genes encoding SdeA, SdeB, and SdeC (47) is consistent with data indicating that SidJ is an effector that modulates the activity the SidE proteins after translocation into host cells.
Consistent with our data, a recent study identified SidJ as an effector capable of regulating the SidE family of proteins (29). Similar to our data, it has been shown that SidJ suppresses the toxicity of SdeA in yeast and that overexpression of either SidJ or SdeA singularly was toxic to yeast. It was also shown that SidJ will suppress SidE-mediated toxicity in mammalian HEK293 cells and that sidJ mutants display an intracellular replication defect in mammalian cells (29, 39). A previous study showed that a mutant of L. pneumophila having the genes encoding all of the SidE proteins and the gene encoding the SidJ protein deleted displayed an intracellular replication defect, which was complemented by production of SdeA alone on a plasmid (39). Intriguingly, recent data showed that overproduction of SdeA in a mutant with a single deletion in sidJ resulted in a more significant growth defect compared to the parental sidJ mutant (29). Thus, it appears that delivery of SdeA alone into the host cell is sufficient to promote intracellular replication when the SidJ protein is absent. In contrast, delivering large amounts of SdeA protein in combination with the other SidE family members into host cells in the absence of SidJ exacerbates the intracellular replication defect of a sidJ mutant. This would be consistent with a role for SidJ in modulating the function of SidE family members inside the host cytosol because the requirement for SidJ for efficient intracellular replication becomes more pronounced as the amount of SidE proteins delivered into cells increases.
Although the mechanism by which SidJ suppresses SdeA function remains unclear, recent data indicate that SidE proteins are localized on or near the membrane of the LCV, and that SidJ either directly or indirectly promotes release of SidE proteins from the LCV environment (29). Here, we identified regions in the SdeA protein required for toxicity in yeast and for SidJ-mediated suppression of toxicity (Fig. 5). Deletion analysis showed that the central region of SdeA is sufficient for robust toxicity when transformed into yeast. Importantly, toxicity mediated by the central region of SdeA was not suppressed by SidJ. However, SidJ did suppress toxicity to SdeA proteins that contained the amino-terminal region, indicating that the amino-terminal region of SdeA receives SidJ-mediated signals that control toxicity (Fig. 7).
FIG 7.
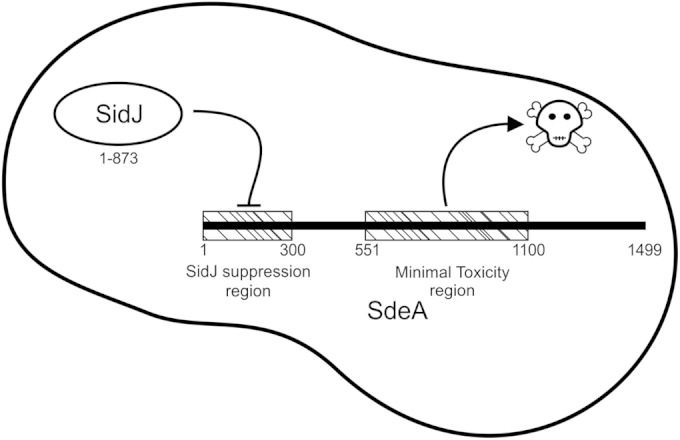
Functional regions of SdeA. The amino terminus of SdeA contains the residues necessary for SidJ suppression of toxicity, which we call the SidJ suppression region. The central region of SdeA contains all of the residues sufficient for toxicity, which we call the minimal toxicity region. The remaining regions of the proteins are of unknown function.
The mechanism by which SidJ can interfere with SdeA toxicity through the amino-terminal region is not clear. We did not detect a direct interaction between SidJ and SdeA using yeast two-hybrid analysis and coimmunoprecipitation approaches, which was consistent with previous data (29). Two conserved aspartic acid residues in SidJ were shown to be necessary for function, and it was suggested that SidJ may have enzymatic activity. Based on these studies, one possibility is that SidJ regulates a conformational or posttranslational modification to the amino-terminal region of SdeA and that this results in autoinhibition of the toxicity region. Further support for this hypothesis comes from the observation that SidJ stimulates release of SidE proteins from the LCV (29), which could result from a modification to the amino-terminal region of SidE. Several L. pneumophila effectors have been found to have enzymatic activities that are regulated through autoinhibition. The GEF activity of RalF is autoinhibited by a C-terminal domain that functions as a cap that senses host membranes (48–50), and VipD is a phospholipase that is autoinhibited by an amino-terminal domain that binds to the host protein Rab5 (51–53). Thus, future studies on the mechanism by which SidJ regulates SidE function could reveal additional strategies by which the activities of L. pneumophila effectors are controlled spatially and temporally during infection.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grant R37 AI041699.
We thank Lara Kohler for providing the pGML10 plasmid used for the overexpression of effectors.
REFERENCES
- 1.Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, Sharrar RG, Harris J, Mallison GF, Martin SM, McDade JE, Shepard CC, Brachman PS. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N Engl J Med 297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 2.McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, Dowdle WR. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med 297:1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- 3.Isberg RR, O'Connor TJ, Heidtman M. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horwitz MA. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med 158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kagan JC, Roy CR. 2002. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol 4:945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- 6.Swanson MS, Isberg RR. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun 63:3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR. 2001. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J Cell Sci 114:4637–4650. [DOI] [PubMed] [Google Scholar]
- 8.Hubber A, Roy CR. 2010. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 9.Segal G, Feldman M, Zusman T. 2005. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol Rev 29:65–81. doi: 10.1016/j.femsre.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Hardiman CA, McDonough JA, Newton HJ, Roy CR. 2012. The role of Rab GTPases in the transport of vacuoles containing Legionella pneumophila and Coxiella burnetii. Biochem Soc Trans 40:1353–1359. doi: 10.1042/BST20120167. [DOI] [PubMed] [Google Scholar]
- 11.Sherwood RK, Roy CR. 2013. A Rab-centric perspective of bacterial pathogen-occupied vacuoles. Cell Host Microbe 14:256–268. doi: 10.1016/j.chom.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorer MS, Kirton D, Bader JS, Isberg RR. 2006. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog 2:e34. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connor TJ, Adepoju Y, Boyd D, Isberg RR. 2011. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A 108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Connor TJ, Boyd D, Dorer MS, Isberg RR. 2012. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science 338:1440–1444. doi: 10.1126/science.1229556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belyi Y, Tabakova I, Stahl M, Aktories K. 2008. Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J Bacteriol 190:3026–3035. doi: 10.1128/JB.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campodonico EM, Chesnel L, Roy CR. 2005. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol Microbiol 56:918–933. doi: 10.1111/j.1365-2958.2005.04595.x. [DOI] [PubMed] [Google Scholar]
- 17.Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen X, Banga S, Liu Y, Xu L, Gao P, Shamovsky I, Nudler E, Luo ZQ. 2009. Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol 11:911–926. doi: 10.1111/j.1462-5822.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shohdy N, Efe JA, Emr SD, Shuman HA. 2005. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci U S A 102:4866–4871. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, Roy CR. 2011. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477:103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Y, Arnold RJ, Luo ZQ. 2011. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci U S A 108:21212–21217. doi: 10.1073/pnas.1114023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. 2009. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog 5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, Kalachikov S, Shuman HA. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol 187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O'Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. 2011. The E block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol 13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isaac DT, Isberg R. 2014. Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol 9:343–359. doi: 10.2217/fmb.13.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, Pupko T, Segal G. 2013. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci U S A 110:E707-715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo ZQ, Isberg RR. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A 101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu W, Banga S, Tan Y, Zheng C, Stephenson R, Gately J, Luo ZQ. 2011. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One 6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong KC, Sexton JA, Vogel JP. 2015. Spatiotemporal regulation of a Legionella pneumophila T4SS substrate by the Metaeffector SidJ. PLoS Pathog 11:e1004695. doi: 10.1371/journal.ppat.1004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger KH, Isberg RR. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 31.Rual JF, Hirozane-Kishikawa T, Hao T, Bertin N, Li S, Dricot A, Li N, Rosenberg J, Lamesch P, Vidalain PO, Clingingsmith TR, Hartley JL, Esposito D, Cheo D, Moore T, Simmons B, Sequerra R, Bosak S, Doucette-Stamm L, Le Peuch C, Vandenhaute J, Cusick ME, Albala JS, Hill DE, Vidal M. 2004. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res 14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das J, Vo TV, Wei X, Mellor JC, Tong V, Degatano AG, Wang X, Wang L, Cordero NA, Kruer-Zerhusen N, Matsuyama A, Pleiss JA, Lipkin SM, Yoshida M, Roth FP, Yu H. 2013. Cross-species protein interactome mapping reveals species-specific wiring of stress-response pathways. Sci Signal 6:ra38. doi: 10.1126/scisignal.2003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iha H, Tsurugi K. 1998. Shuttle-vector system for Saccharomyces cerevisiae designed to produce C-terminal-Myc-tagged fusion proteins. Biotechniques 25:936–938. [DOI] [PubMed] [Google Scholar]
- 34.Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96. doi: 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 35.Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, Rineer J, Greenberg JJ, Steshenko V, Park SH, Zhao B, Teplitskaya E, Edwards JR, Pampou S, Georghiou A, Chou I-C, Iannuccilli W, Ulz ME, Kim DH, Geringer-Sameth A, Goldsberry C, Morozov P, Fischer SG, Segal G, Qu X, Rzhetsky A, Zhang P, Cayanis E, De Jong PJ, Ju J, Kalachikov S, Shuman HA, Russo JJ. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 36.Goody PR, Heller K, Oesterlin LK, Muller MP, Itzen A, Goody RS. 2012. Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J 31:1774–1784. doi: 10.1038/emboj.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neunuebel MR, Chen Y, Gaspar AH, Backlund PS Jr, Yergey A, Machner MP. 2011. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333:453–456. doi: 10.1126/science.1207193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan Y, Luo ZQ. 2011. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475:506–509. doi: 10.1038/nature10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bardill JP, Miller JL, Vogel JP. 2005. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol 56:90–103. doi: 10.1111/j.1365-2958.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- 40.Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. 2007. The response regulator PmrA is a major regulator of the Icm/Dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol 63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
- 41.Kubori T, Galan JE. 2003. Temporal regulation of salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115:333–342. doi: 10.1016/S0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- 42.Ingmundson A, Delprato A, Lambright DG, Roy CR. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 43.Machner MP, Isberg RR. 2007. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318:974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- 44.Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol 8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 45.Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. 2010. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 46.Kubori T, Shinzawa N, Kanuka H, Nagai H. 2010. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog 6:e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Luo ZQ. 2007. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun 75:592–603. doi: 10.1128/IAI.01278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alix E, Chesnel L, Bowzard BJ, Tucker AM, Delprato A, Cherfils J, Wood DO, Kahn RA, Roy CR. 2012. The capping domain in RalF regulates effector functions. PLoS Pathog 8:e1003012. doi: 10.1371/journal.ppat.1003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Folly-Klan M, Alix E, Stalder D, Ray P, Duarte LV, Delprato A, Zeghouf M, Antonny B, Campanacci V, Roy CR, Cherfils J. 2013. A novel membrane sensor controls the localization and ArfGEF activity of bacterial RalF. PLoS Pathog 9:e1003747. doi: 10.1371/journal.ppat.1003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Folly-Klan M, Sancerne B, Alix E, Roy CR, Cherfils J, Campanacci V. 2015. On the use of Legionella/Rickettsia chimeras to investigate the structure and regulation of Rickettsia effector RalF. J Struct Biol 189:98–104. doi: 10.1016/j.jsb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 51.Gaspar AH, Machner MP. 2014. VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc Natl Acad Sci U S A 111:4560–4565. doi: 10.1073/pnas.1316376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ku B, Lee KH, Park WS, Yang CS, Ge J, Lee SG, Cha SS, Shao F, Heo WD, Jung JU, Oh BH. 2012. VipD of Legionella pneumophila targets activated Rab5 and Rab22 to interfere with endosomal trafficking in macrophages. PLoS Pathog 8:e1003082. doi: 10.1371/journal.ppat.1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lucas M, Gaspar AH, Pallara C, Rojas AL, Fernandez-Recio J, Machner MP, Hierro A. 2014. Structural basis for the recruitment and activation of the Legionella phospholipase VipD by the host GTPase Rab5. Proc Natl Acad Sci U S A 111:E3514–E3523. doi: 10.1073/pnas.1405391111. [DOI] [PMC free article] [PubMed] [Google Scholar]