Abstract
The food-borne pathogen Salmonella enterica serovar Typhimurium benefits from acute inflammation in part by using host-derived nitrate to respire anaerobically and compete successfully with the commensal microbes during growth in the intestinal lumen. The S. Typhimurium genome contains three nitrate reductases, encoded by the narGHI, narZYV, and napABC genes. Work on homologous genes present in Escherichia coli suggests that nitrate reductase A, encoded by the narGHI genes, is the main enzyme promoting growth on nitrate as an electron acceptor in anaerobic environments. Using a mouse colitis model, we found, surprisingly, that S. Typhimurium strains with defects in either nitrate reductase A (narG mutant) or the regulator inducing its transcription in the presence of high concentrations of nitrate (narL mutant) exhibited growth comparable to that of wild-type S. Typhimurium. In contrast, a strain lacking a functional periplasmic nitrate reductase (napA mutant) exhibited a marked growth defect in the lumen of the colon. In E. coli, the napABC genes are transcribed maximally under anaerobic growth conditions in the presence of low nitrate concentrations. Inactivation of narP, encoding a response regulator that activates napABC transcription in response to low nitrate concentrations, significantly reduced the growth of S. Typhimurium in the gut lumen. Cecal nitrate measurements suggested that the murine cecum is a nitrate-limited environment. Collectively, our results suggest that S. Typhimurium uses the periplasmic nitrate reductase to support its growth on the low nitrate concentrations encountered in the gut, a strategy that may be shared with other enteric pathogens.
INTRODUCTION
Pathogenic bacteria are supremely adept at infiltrating barriers to colonization in order to thrive in specific niches. For intestinal pathogens, the challenges include surviving passage through the gastrointestinal tract, neutralizing host defenses, and competing with the commensal microflora for space and nutrients. While it is clear that many bacterial virulence proteins help pathogens by thwarting host immunity, how bacteria have adapted metabolically to the host environment is more nebulous.
The food-borne pathogen Salmonella enterica serovar Typhimurium is well equipped for survival and proliferation in the harsh environment of the gut. Once ingested, S. Typhimurium traverses the intestinal tract and localizes to the epithelium in the distal regions of the gut (1, 2). Here, an invading population initiates translocation through the epithelium using a type 3 secretion system (T3SS) encoded on Salmonella pathogenicity island 1 (SPI-1), termed T3SS-1 (1, 3). T3SS-1 subsequently injects a cocktail of effector proteins that promote epithelial invasion, allowing S. Typhimurium to traverse into the underlying lamina propria (4). Patrolling macrophages and dendritic cells detect the invading bacteria and, upon activation of microbial pattern recognition receptors, release proinflammatory cytokines to induce a robust immune response (summarized in references 5, 6, and 7). This response includes chemokine production to recruit neutrophils (8) that, upon migration to infected intestinal sites, discharge antimicrobial reactive oxygen species (9). Additionally, increased inducible nitric oxide synthase (iNOS) production leads to the release of nitric oxide (NO) into the intestinal lumen (10–12). This host response to S. Typhimurium invasion is purported to eliminate the pathogen from tissue, but several studies have indicated beneficial effects for the bacterial population residing in the lumen.
S. Typhimurium encodes multiple metabolic pathways that enable it to adapt quickly to the altered luminal landscape caused by the host inflammatory response. A flexible metabolic network is particularly evident in S. Typhimurium nitrate respiratory pathways (Fig. 1A). S. Typhimurium encounters nitrate as a by-product of inflammation of the gut, where iNOS-derived NO and reactive oxygen species react to form nitrate (11). The pathogen is capable of chemotaxis toward, and respiration of, nitrate, thereby enhancing its growth in the intestinal lumen (11, 14). S. Typhimurium encodes three nitrate reductases, which are also present in the closely related species Escherichia coli and support growth in the lumen of the inflamed gut (11, 15, 16). Most studies on the regulation and functional characterization of these nitrate reductases have been performed with E. coli. This body of work suggests that nitrate reductase A, encoded by narGHI, is the major nitrate reductase. Nitrate reductase Z, encoded by narZYV, is a homologue synthesized constitutively at low levels and may function during the transition to anaerobic environments, before narGHI expression is induced by anaerobiosis (17). Nitrate reductases A and Z are oriented to the cytosol and are capable of proton translocation across the cytoplasmic membrane, thereby generating a net proton gradient during respiration (17–20). The periplasmic nitrate reductase, encoded by napABC, on the other hand, is oriented to the periplasm, and there is no evidence that this enzyme mediates proton translocation (21, 22). The physiological role of NapABC remains unclear, since it does not appear to be a major player during growth on nitrate in vitro. For instance, batch culture experiments show that E. coli expressing narGHI exhibits a strong growth advantage over a strain with a narGHI mutation, while a strain expressing napABC has no growth advantage over the respective single mutant (23). These results are corroborated by in vivo work showing that E. coli strains lacking NarGHI, but not E. coli strains lacking NapABC, have growth defects relative to the wild type (wt) in the mouse intestine (24).
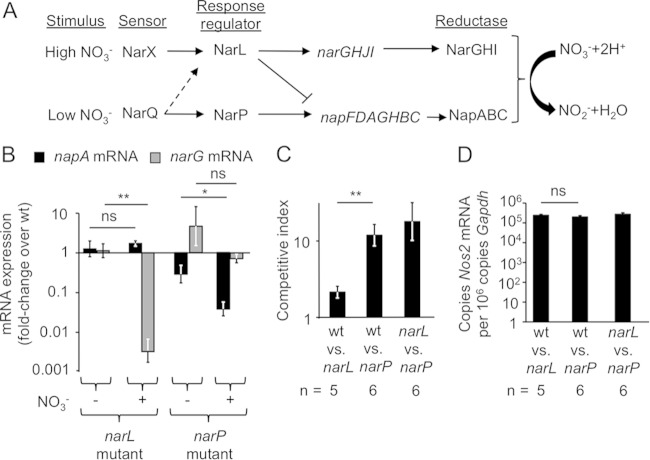
FIG 1.
Functional NarP contributes to the growth of S. Typhimurium in the intestine during infection. (A) Schematic of nitrate sensing and regulation. Environmental nitrate concentrations are sensed by the 2-component histidine kinases NarX and NarQ, which subsequently phosphorylate the response regulators NarL and NarP, respectively, although some cross talk exists between response regulator pairs. NarL and NarP alter the transcription of the nitrate reductases NarGHI and NapABC, which function to catalyze the reduction of nitrate to nitrite for respiration. (B) Levels of napA and narG mRNA transcripts in a narL (left) or narP (right) mutant were determined relative to the transcript levels detected in wild-type S. Typhimurium grown anaerobically in the presence (+) or absence (−) of nitrate (0.4 mM). Experiments were performed in triplicate. **, P < 0.01; *, P < 0.05; ns, not statistically significantly different. (C) Mice were infected with a mixture of the indicated S. Typhimurium strains in equal amounts. Four days after infection, mice were euthanized, and the competitive index was determined by dividing the output ratio by the input ratio. (D) Nos2 mRNA expression in the cecal mucosa was determined by quantitative real-time PCR.
The synthesis of nitrate reductase A (NarGHI) and the periplasmic nitrate reductase (NapABC) is controlled by two 2-component systems, NarXL and NarQP, respectively, and is repressed under aerobic conditions (25). NarX senses high nitrate concentrations, leading to phosphorylation of the response regulator NarL, which induces narGHI transcription while inhibiting napABC transcription. NarQ responds to low nitrate concentrations, phosphorylating NarP, which, in turn, leads to the upregulation of napABC transcription (25, 26). As a result, the periplasmic nitrate reductase is synthesized optimally under anaerobic conditions in the presence of low levels of nitrate, while its synthesis is repressed when concentrations of nitrate are high. In contrast, nitrate reductase A is synthesized optimally in anaerobic environments containing high concentrations of nitrate (Fig. 1A). Neither two-component system affects narZYV transcription, since the encoded nitrate reductase Z is synthesized constitutively at low levels, thereby ensuring that enzymatic activity is available when bacteria transit from an aerobic to an anaerobic environment.
While the molecular mechanisms governing the regulation and enzymatic function of nitrate reductases are well studied in Escherichia coli, understanding of the physiological relevance of these reductases during S. Typhimurium infection is lacking (17, 23, 24, 27, 28). Characterization of nitrate reductases in E. coli would predict that NarGHI is the major player conferring a nitrate respiration-dependent benefit during S. Typhimurium infection. In this study, we tested this prediction by investigating the contributions of individual S. Typhimurium nitrate reductases to the intestinal lifestyle of this pathogen.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
For the E. coli and S. Typhimurium strains used in this study, see Table 1. Strains were routinely grown in Luria-Bertani (LB) broth (catalog no. 244620; BD Biosciences, San Jose, CA) or on LB plates. Antibiotics in bacterial cultures were used at the following concentrations: for carbenicillin (Carb), 0.1 mg/ml; for chloramphenicol (Cm), 0.015 mg/ml; for kanamycin (Kan), 0.1 mg/ml; and for streptomycin (Strep), 0.1 mg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. Typhimurium | ||
| SL1344 | ST4/74 his | 48, 49 |
| SW284 | Strain IR715; ΔphoN::Cmr | 50 |
| CAL51 | ΔnarZ | 11 |
| CAL55 | ΔnapA ΔnarZ narG::pCAL5 | 11 |
| CAL63 | ΔphoN::Kanr | 11 |
| CAL64 | ΔnapA ΔnarZ narG::pCAL5 ΔphoN::Kanr | 11 |
| CAL65 | narG::pCAL5 | 11 |
| CAL128 | ΔphoN::Cmr | This study |
| CAL139 | ΔphoN::Kanr narP::pCAL35 | This study |
| CAL156 | ΔnarL::Kanr | This study |
| CAL160 | ΔnarL | This study |
| CAL173 | ΔphoN::Kanr ΔnarL | This study |
| CAL277 | ΔphoN::Kanr ΔnapA | This study |
| CAL315 | ΔnarZ ΔphoN::Cmr | This study |
| E. coli | ||
| DH5α λpir | F− endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 ϕ80lacZΔM15 λpir | 51 |
| TOP10 | ϕ80lacZΔM15 lacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| S17-1 λpir | C600:RP4 2-(Tet::Mu) (Kan::Tn7) λpir recA1 thi pro hsdR (rK− mK+) | 51a |
| Plasmids | ||
| pCR2.1 | Cloning vector | Invitrogen |
| pGP704 | ori(R6K) mobRP4 Carbr | 52 |
| pRDH10 | sacRB ori(R6K) mobRP4 Tetr Cmr | 53 |
| pEP185.2 | ori(R6K) mobRP4 Cmr | 54 |
| pUC4 KSAC | ori(pMB1) Carbr Kanr | Pharmacia (GE Healthcare) |
| pCAL5 | pGP704 with internal region of narG | 11 |
| pCAL32 | pCR2.1 with narP internal region inserted | This study |
| pCAL35 | pEP185.2 with narP internal region inserted | This study |
| pCAL36 | pCR2.1 with regions upstream and downstream of narL | This study |
| pCAL39 | pRDH10 with regions upstream and downstream of narL | This study |
| pCAL42 | pRDH10 with regions upstream and downstream of narL; Kanr cassette inserted between flanking regions | This study |
Mutant construction.
To construct an S. Typhimurium SL1344 wild-type strain carrying a selectable Cm resistance marker, we generated a phage lysate of SW284 (11), an S. Typhimurium strain carrying a Cmr cassette inserted into the phoN gene, using the methods described previously (11). Transfer of this cassette using P22 phage transduction into S. Typhimurium SL1344 generated strain CAL128, which is referred to below as the wild-type strain.
To construct an S. Typhimurium narP mutant, an internal narP region was amplified via PCR, inserted into the cloning vector pCR2.1 to generate pCAL32, and propagated in E. coli TOP10 (Invitrogen, Carlsbad, CA). After the insert was sequenced to confirm that there were no mutations in the amplified region, pCAL32 and plasmid pEP185.2 were digested with XbaI and SacI (New England BioLabs, Ipswich, MA). The XbaI/SacI-digested insert from pCAL32 was ligated into pEP185.2 to generate pCAL35. This plasmid was propagated first in E. coli DH5α λpir and then in E. coli S17-1 λpir for conjugation with S. Typhimurium CAL63 (ΔphoN::Kanr). Single homologous recombination events were selected for by growth on Cm and Kan plates. Insertion into the narP locus was confirmed via PCR, and this strain was designated CAL139.
To construct an S. Typhimurium narL mutant, approximately 1 kb regions upstream and downstream of narL were PCR amplified and digested with XbaI. The two flanking regions were ligated and PCR amplified to obtain an approximately 2 kb region that was inserted into pCR2.1 to generate pCAL36. After the insert was sequenced for accuracy, pCAL36 and the suicide plasmid pRDH10 were digested with SalI (New England BioLabs, Ipswich, MA). The narL flanking region fragment from pCAL36 was ligated into pRDH10 to generate pCAL39, which was propagated in E. coli DH5α λpir. A Kanr cassette from pUCKSAC was inserted into the XbaI restriction site between the narL flanking regions to generate pCAL42. E. coli S17-1 λpir was transformed by pCAL42 and conjugated with S. Typhimurium SL1344. Single homologous recombination events were selected for by growth on Strep and Kan plates. To select for strains that had lost the integrated plasmid, leaving the Kanr cassette in place of narL, we used sucrose selection as described in reference 11. The strain that successfully replaced narL with the Kanr cassette was designated CAL156. To remove the Kanr cassette and generate a nonpolar mutant, CAL156 was conjugated with E. coli S17-1 λpir containing pCAL39. After selecting for plasmid integration on Cm and Kan plates, we performed sucrose selection to identify a strain that had lost both the integrated plasmid and the Kanr cassette. After confirming the nonpolar mutation via PCR, we designated the strain CAL160. To provide a selection marker for the narL mutant strain, CAL160 was transduced with a phage lysate from CAL63 to transfer the ΔphoN::Kanr mutation. This narL mutant strain with Kanr at the phoN locus was designated CAL173.
We had previously constructed single nonpolar napA and napZ mutants (11). To provide these strains with a selective marker, CAL67 (11) was transduced with a phage lysate from CAL63 to transfer the ΔphoN::Kanr mutation, while CAL51 was transduced with a phage lysate from CAL128 to transfer the ΔphoN::Cmr mutation. These strains were designated CAL277 and CAL315, respectively.
Mouse experiments.
The Institutional Animal Care and Use Committee at the University of California, Davis, approved all animal experiments. We used the streptomycin model of gastroenteritis to reproducibly colonize mice with S. Typhimurium. In this model, mice first received a single dose of streptomycin sulfate (20 mg/mouse) (Calbiochem, San Diego, CA) intragastrically (i.g.). Mice were then inoculated i.g. with 100 μl of 1 × 108 S. Typhimurium CFU per mouse or, for mock-treated animals, with LB broth. In competitive infections, each strain was provided at a concentration of 5 × 107 CFU per mouse. At either 3 or 4 days postinfection, mice were euthanized. Colon contents were removed and stored in phosphate-buffered saline (PBS) on ice before being homogenized and plated on selective media for the enumeration of S. Typhimurium CFU. For RNA analysis, we collected the middle cecal section, removed fecal matter, and flash froze samples in liquid nitrogen for storage at −80°C.
Isolation of RNA from the mouse.
For the isolation of murine RNA, colon tissue sections were homogenized in a Mini-Beadbeater (BioSpec Products, Bartlesville, OK), and RNA was isolated by the TRI-Reagent method (Molecular Research Center, Inc., Cincinnati, OH) according to the manufacturer's protocol. Contaminating DNA was removed using the DNA-free kit (Applied Biosystems, Waltham, MA), and RNA was stored at −80°C.
For the isolation of bacterial RNA from the mouse, intestinal colon contents and the mucus layer (gently scraped from the colonic epithelial cells) were collected and were stored in RNAlater (Ambion, Carlsbad, CA) overnight at 4°C. Excess solution was subsequently removed from the pellet, and TRIzol (Invitrogen, Carlsbad, CA) was added. Next, chloroform was added, and the solution was transferred to 2-ml Phase Lock tubes (5 Prime, Hilden, Germany) and was centrifuged at 14,000 × g. The upper phase was removed, and RNA was precipitated with 100% ethanol at −20°C overnight. RNA was collected after centrifugation and was washed with 70% ethanol. Contaminating DNA was removed as described above.
In vitro RNA expression.
To quantify napA and narG induction in vitro in response to nitrate, S. Typhimurium was grown in M9 minimal medium (2 mM MgSO4, 0.2 mM CaCl2, M9 minimal salts [29], 0.4% glucose) supplemented with 40 μg/ml histidine. Overnight cultures of the strains to be tested were diluted 1:100 into fresh medium, and bacteria were grown in an anaerobic chamber (Shel Lab, Cornelius, OR) (atmosphere of 5% CO2, 5% H2, and 90% N2) at 37°C until the optical density reached 0.1 to 0.2. At this time, a 1 M sodium nitrate solution was added to the medium to reach a final concentration of 0.1 mM or 0.4 mM. The bacterial cultures were incubated for 5 min and were then immediately placed on ice to halt growth.
For the isolation of RNA, 1 ml of bacterial culture was processed by using the Aurum Total RNA minikit (Bio-Rad, Hercules, CA) according to the manufacturer's protocol. Contaminating DNA was removed as described above.
Quantitative PCR.
Isolated RNA was reverse transcribed using random hexamers and Moloney murine leukemia virus reverse transcriptase (Applied Biosystems, Waltham, MA). Quantitative real-time PCR was performed using SYBR green (Applied Biosystems, Waltham, MA) PCR mix and the appropriate primer sets (Table 2) at a final concentration of 0.25 mM. Absolute values were calculated using a series of standards and a standard curve. To generate standards, the primer sets mentioned above were used in standard PCR to amplify the target genes, which were subsequently inserted into pCR2.1 using the TOPO cloning kit (Life Technologies, Carlsbad, CA). The resulting plasmids were sequenced for accuracy and were quantified to create a set of standards ranging from 108 to 101 copies/ml diluted in a 0.02-mg/ml yeast RNA (Sigma, St. Louis, MO) solution.
TABLE 2.
Primers used in this study
| Gene | Organism | Primer sequencea |
|---|---|---|
| narP | S. Typhimurium | 5′-CCTGGAGCTCGTATTCGTCAATTACTG-3′ |
| 5′-ACGGTCTAGAGTAGATTACGAATGTGC-3′ | ||
| narL (upstream) | S. Typhimurium | 5′-ATTTGTCGACCGCAGAAGATGTCACG-3′ |
| 5′-CCACTCTAGACACCGTTGCTGGCTTCTCC-3′ | ||
| narL (downstream) | S. Typhimurium | 5′-CTGCTCTAGACGAAAGTACGGTCAAAGTG-3′ |
| 5′-TACGTCGACTGGTCTTACGCAACAG-3′ | ||
| napA (qPCR)b | S. Typhimurium | 5′-TGAAAGAGAAAGGACCAGAAGCG-3′ |
| 5′-TTGTTAGAGCGGAAACCAGCC-3′ | ||
| narG (qPCR) | S. Typhimurium | 5′-GAAGTGGAGTGGCGTGACAATG-3′ |
| 5′-AGCGGATGAATAAACGGATGC-3′ | ||
| 16S rRNA | S. Typhimurium | 5′-TGTTGTGGTTAATAACCGCA-3′ |
| 5′-GACTACCAGGGTATCTAATCC-3′ | ||
| Nos2 | Mouse | 5′-CCTCTTTCAGGTCACTTTGGTAGG-3′ |
| 5′-TTGGGTCTTGTTCAGCCACGG-3′ | ||
| Gapdh | Mouse | 5′-TGTAGACCATGTAGTTGAGGTCA-3′ |
| 5′-AGGTCGGTGTGAACGGATTTG-3′ |
Underlined sequences indicate engineered restriction sites.
qPCR, quantitative PCR.
Cecal nitrate measurements.
Mouse ceca were removed and were divided along their sagittal planes. Solid fecal matter was carefully removed from the lumen, while the mucus layer was gently scraped off the tissue. This layer was homogenized in 200 μl PBS and was placed immediately on ice. Samples were next centrifuged at 5,000 × g for 10 min at 4°C to remove remaining solid particles. The supernatant was removed and was filtered using a 0.2-μm Acrodisc syringe filter (Pall Life Sciences, Port Washington, NY). Measurement of cecal nitrate followed an adaptation of the Griess assay. In this assay, nitrate was first reduced to nitrite via vanadium(III) chloride by adding 50 μl of the cecal sample with 50 μl of Griess reagent 1 (0.5 M HCl, 0.2 mM VCl3, 1% sulfanilamide) and incubating the mixture at room temperature for 10 min. Next, 50 μl of Griess reagent 2 [0.1% (1-naphthyl)ethylenediamine dichloride] was added. Absorbance at 540 nm was measured immediately after the addition of Griess reagent 2. Nitrite immediately reacts with the added reagents, and the initial color change indicates available nitrite in the samples. The reduction of nitrate to nitrite via VCl3 occurs slowly, requiring incubation for 8 h at room temperature. Measurements of absorbance at 540 nm, along with a standard curve of known nitrate concentrations, were taken after the incubation. The initial absorbance (immediately after the addition of Griess reagent 2) was subtracted from the final absorbance to determine nitrate concentrations in the cecal mucus layer. Samples were tested in duplicate, and all measurements were standardized to the initial sample weight.
Histopathology.
The distal segment of the cecum was fixed in 10% buffered formalin phosphate and was stained with hematoxylin and eosin (H/E). A veterinary pathologist scored histopathological changes using blinded scoring of sections by a scheme described previously (30).
Statistical analysis.
Geometric mean values were determined throughout the study. Data on ratios were converted logarithmically to obtain a normal distribution prior to analysis. We used Student's t test to assess significance and considered a P value of 0.05 or less to be significant.
RESULTS
NarP, but not NarL, contributes to the growth of S. Typhimurium in the intestinal lumen.
Transcription of E. coli nitrate reductases is largely controlled through nitrate-dependent activation of the NarXL and NarQP two-component systems (Fig. 1A). To determine the relative contributions of the homologous two-component systems to S. Typhimurium fitness, we constructed S. Typhimurium mutant strains with defects in the response regulator narL or narP. In vitro analysis of these strains cultured anaerobically showed that neither mutant exhibited any significant difference in narG or napA mRNA expression from the wild-type (wt) strain when no nitrate was added to the medium (Fig. 1B). In the presence of nitrate (0.4 mM), narG expression was reduced only in the narL mutant, whereas napA expression was reduced only in the narP mutant. These data suggested that inactivation of narL and narP specifically affected the transcription of narG and napA, respectively, a finding consistent with previous reports on the function of these regulatory proteins in E. coli (Fig. 1A) (25, 26).
To test the significance of NarL and NarP for the growth of S. Typhimurium in the intestinal lumen during infection, we used a mouse colitis model (31), where mice were provided a single dose of streptomycin 1 day prior to infection with S. Typhimurium. Streptomycin pretreatment disrupts the resident microbiota, enabling S. Typhimurium to elicit acute cecal inflammation (reviewed in reference 32). Using this model for human gastroenteritis, we infected mice with 1:1 ratios of wild-type S. Typhimurium (CAL128) and either the narL mutant (CAL173) or the narP mutant (CAL139). Four days postinfection, mice were euthanized, and we quantified the ratios of the bacterial strains recovered. Interestingly, similar numbers of wild-type S. Typhimurium and a narL mutant were recovered from colon contents (Fig. 1C), suggesting that NarL was not necessary for in vivo growth in this model. In contrast, wild-type S. Typhimurium was recovered from the colon contents of mice in >10-fold-higher numbers than the narP mutant (Fig. 1C). Transcript levels of Nos2, the gene encoding iNOS, in the cecal mucosa were similar for the two groups (Fig. 1D), suggesting that differences in host-derived NO production were not responsible for the phenotypes observed. Collectively, these data indicated that, surprisingly, NarP contributed to S. Typhimurium growth in the intestinal lumen, while NarL was dispensable. To test this conclusion further, mice were infected with a mixture of equal parts of the narL and narP mutant strains. Higher recovery of the narL mutant (which encodes a functional narP gene) than the narP mutant (encoding a functional narL gene) further supported the conclusion that NarP played a more substantial role than NarL in the growth of S. Typhimurium in the intestinal lumen.
The periplasmic nitrate reductase NapABC boosts the growth of S. Typhimurium in the colon.
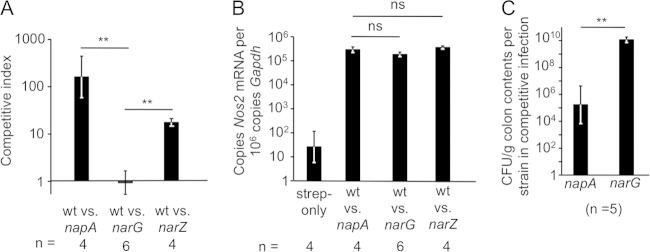
Next, we wanted to test whether the fitness advantage conferred by NarP arose from its role in regulating the transcription of genes important for anaerobic respiration, specifically the genes encoding the periplasmic nitrate reductase NapABC. To explore this notion, mice were infected with a mixture of equal parts of wild-type S. Typhimurium (CAL128) and an isogenic mutant carrying a mutation in either narG (CAL65) or napA (CAL277). In competitive infections, wild-type S. Typhimurium was recovered from colon contents in higher numbers than the napA mutant, while the wild type and the narG mutant were recovered in similar numbers (Fig. 2A). Infections with these different strain mixtures caused similar increases in Nos2 expression levels in the cecal mucosae (Fig. 2B). When mice were inoculated with a mixture of equal parts of a napA mutant and a narG mutant, the strain with a functional periplasmic nitrate reductase (narG mutant) grew to significantly higher levels in colon contents (Fig. 2C). We also sought to determine the contribution of NarZYV to the growth of S. Typhimurium in the intestinal lumen. In competitive infections with wild-type S. Typhimurium (CAL63) and a narZ mutant (CAL315), we recovered higher numbers of the wild-type strain than of the mutant (Fig. 2A).
FIG 2.
The periplasmic nitrate reductase NapABC provides a fitness advantage during competitive infection. (A) Mice were infected with mixtures of the indicated S. Typhimurium strains in equal amounts. Four days after infection, mice were euthanized, and the competitive index (output ratio over input ratio) was determined in colon contents. *, P < 0.05; **, P < 0.01; ns, not statistically significantly different; wt, the wild-type strain CAL128; napA, the napA mutant CAL277; narG, the narG mutant CAL65; narZ, the narZ mutant CAL315. (B) Nos2 mRNA levels in colonic tissues from the animals used in the experiment for which results are shown in panel A and in control animals treated with streptomycin and inoculated 1 day later with a sterile medium. (C) Mice were infected with a mixture of a napA mutant and a narG mutant in equal amounts. Four days after infection, mice were euthanized, and the CFU counts of both strains in colon contents were determined.
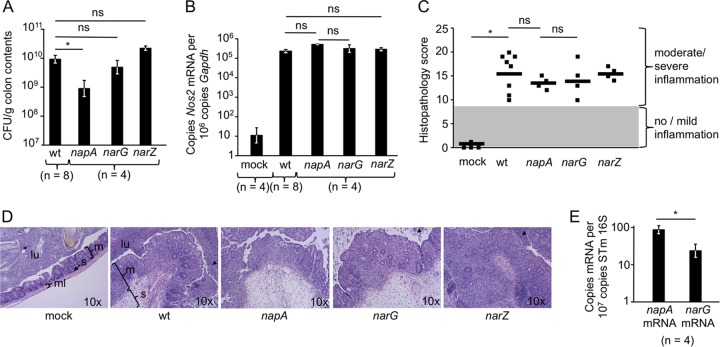
A growth defect caused by the napA mutation was also observed when mice were infected with individual bacterial strains. Mice were infected with either wild-type S. Typhimurium, a narG mutant, a narZ mutant, or a napA mutant. Interestingly, 4 days after infection, the wild type, the narG mutant, and the narZ mutant were recovered from colon contents in approximately 10-fold higher numbers than the napA mutant (Fig. 3A), although cecal Nos2 transcript levels were elevated by similar magnitudes during infections with the different S. Typhimurium strains (Fig. 3B). Differences in the luminal S. Typhimurium population did not affect host pathology: mice in all infected groups experienced moderate to high levels of intestinal inflammation (Fig. 3C), with neutrophil influx into the gut lumen and massive thickening of the mucosa and submucosa (Fig. 3D). In agreement with the observed differences in growth in the intestinal lumen between the napA and narG mutants, we also noted significantly higher expression of the napA gene than of the narG gene in RNAs extracted from the cecal contents of mice infected with wild-type S. Typhimurium (Fig. 3E). No expression of either gene was detected in RNA extracted from the cecal contents of mock-infected control mice (data not shown). Together, our results support the idea that periplasmic nitrate reductase provides the primary benefit during the growth of S. Typhimurium in the host gut by nitrate respiration.
FIG 3.
The periplasmic nitrate reductase NapABC confers a luminal growth benefit during single-infection experiments. (A) Mice were infected with either the wild-type S. Typhimurium strain CAL128 (wt), the napA mutant CAL277, the narG mutant CAL65, or the narZ mutant CAL315. Four days after infection, mice were euthanized, and S. Typhimurium CFU counts in colon contents were determined. *, P < 0.05; **, P < 0.01; ns, not statistically significantly different. (B) Nos2 mRNA levels in the cecal mucosae of the animals used in the experiment for which results are shown in panel A and in those of a control group inoculated with sterile medium (mock). (C) Histopathology scores for the mice used in the experiments for which results are shown in panels A and B. Each data point represents an individual mouse. (D) Representative images of H/E-stained cecal sections scored in panel C. All images were taken at the same magnification. m, mucosa; s, submucosa; ml, muscle layer; lu, lumen. Arrowheads indicate luminal neutrophils. (E) Levels of narG and napA mRNA transcripts were determined in RNAs isolated from the cecal contents of mice 4 days after infection with wild-type S. Typhimurium.
The murine colon is a low-nitrate environment.
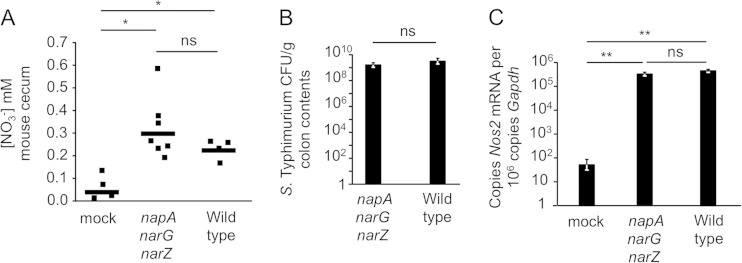
Salmonella nitrate reductase regulation is largely dependent on nitrate concentrations, and on the basis of our results showing that NapABC-deficient S. Typhimurium exhibits a growth defect in vivo, we predicted that the intestine is a nitrate-limited environment. To test this prediction, we determined cecal nitrate concentrations 3 days after infection. This time point was chosen because it coincided with the peak of Nos2 expression observed in a previous study using this model (11). Mice either were mock treated (streptomycin treatment followed 1 day later by inoculation with a sterile medium) or were infected with wild-type S. Typhimurium or with a nitrate respiration-deficient S. Typhimurium mutant (napA narG narZ mutant strain CAL55). Mock-treated mice produced small but measurable amounts of nitrate (0.03 mM) in the cecal mucus layer. In contrast, nitrate was recovered at a 2- to 3-fold-higher level (0.29 mM) in mice 3 days after infection with the S. Typhimurium napA narG narZ mutant. Nitrate concentrations were also elevated in mice infected with wild-type S. Typhimurium (0.21 mM) (Fig. 4A). The two strains were recovered from the colon contents in equivalent numbers at this time point (Fig. 4B), since nitrate-dependent outgrowth is not observed before day 4 after infection in this model (Fig. 3A). Enhanced nitrate production correlated with increased expression of murine Nos2 in infected mice (Fig. 4C). These data suggest that S. Typhimurium encounters, on average, low nitrate concentrations during colonization of the distal gastrointestinal tract.
FIG 4.
Nitrate levels in the murine cecum. (A) Nitrate concentrations were determined in the cecal mucus layers of mice receiving either streptomycin followed 1 day later by inoculation with a sterile medium (mock), an S. Typhimurium nitrate reductase mutant (napA narG narZ mutant CAL64), or the wild-type S. Typhimurium strain CAL63. Measurements were taken 3 days after mice had received bacteria or sterile medium. Values were normalized to initial sample weights. Each data point represents an individual mouse. *, P < 0.05; **, P < 0.01; ns, not statistically significantly different. (B) S. Typhimurium CFU counts 3 days after infection in the colon contents of the mice for which results are shown in panel A. (C) Cecal Nos2 mRNA expression in the mice for which results are shown in panel A.
DISCUSSION
Like many pathogenic bacteria in the family Enterobacteriaceae, S. Typhimurium encodes a variable and highly regulated respiratory network that provides flexibility in responding to changes in the availability of electron donors and acceptors. We are only beginning to understand the environmental conditions that S. Typhimurium experiences during infection and the mechanism by which the resulting changes in central metabolism contribute to successful intestinal colonization. We previously found nitrate respiration to be an important factor for the growth of S. Typhimurium during acute inflammation (11), and here we examined the individual roles of the inducible nitrate reductases (NarGHI and NapABC) in order to understand their contributions to the intestinal lifestyle of S. Typhimurium.
S. Typhimurium directly detects nitrate with the two-component systems NarXL and NarQP, and we sought to determine if loss of the response regulator led to a fitness defect in vivo. Remarkably, inactivation of narL did not reduce growth in the intestinal lumen, although NarL regulates a variety of metabolic genes, and this regulation includes activation of fdnGHI (encoding formate dehydrogenase) and repression of frdABCD (encoding fumarate reductase), dmsABC (encoding dimethyl sulfoxide [DMSO] reductase), dcuSR (encoding a two-component system), and nrfABCDEFG and nirBDC (encoding two nitrite reductases) (reviewed in reference 33). To the best of our knowledge, no evidence exists supporting a role in S. Typhimurium virulence or luminal growth for formate dehydrogenase-N, DMSO reductase, or nitrite reduction via nirBDC. While conclusive data on the individual contributions of these NarL-regulated genes in vivo are lacking, at this point we cannot discount the possibility that NarL-regulated genes other than the nitrate reductases are important in intestinal colonization by S. Typhimurium.
Mutation of narP resulted in an in vivo growth defect, highlighting the importance of the encoded response regulator for the growth of S. Typhimurium in the intestine. NarP positively regulates the napABC genes, encoding a periplasmic nitrate reductase, as well as nrfABCDEFG and nirBDC, two gene clusters encoding nitrite reductases (34). These three gene clusters are thus oppositely regulated by the NarXL and NarQP two-component systems. A napA mutant exhibited a marked luminal growth defect in the mouse colitis model, suggesting that the phenotype of a narP mutant was, at least in part, due to reduced expression of NapABC. In contrast, strains lacking the cytosolic nitrate reductase A exhibited little difference from the wild-type strain in intestinal growth. The third nitrate reductase, NarZYV, also appeared to contribute to the growth of S. Typhimurium, although the benefit was seen only in competition with wild-type S. Typhimurium, not when mice were infected with individual bacterial strains.
Previous work with E. coli would predict that nitrate reductase A contributes most heavily to nitrate-dependent growth in the gut (24, 35). There are technical reasons that may help explain the apparently contradictory results from previous work with E. coli and our in vivo analysis with S. Typhimurium. Early in vitro experiments that probed the functional characteristics of nitrate reductases used batch cultures in which high nitrate levels (as high as 40 mM) were added to the medium (21, 23). This ensured that ample nitrate was available for respiration and growth, but it also biased expression toward nitrate reductase A, which is synthesized optimally with nitrate concentrations of 10 mM or higher (36). In contrast, when nitrate is provided at low concentrations (<1 mM), a condition that favors expression of the periplasmic nitrate reductase, the electron acceptor is consumed rapidly, before growth phenotypes can be recorded (37). Later work confirmed that continuous cultures with chemostats providing nitrate at a constant concentration were needed in order to detect a contribution of the periplasmic nitrate reductase to in vitro growth (23, 36). The high affinity of the periplasmic nitrate reductase for nitrate, coupled with its maximal expression at nitrate concentrations around 1 mM (21), suggest that during acute inflammation, the niche in which S. Typhimurium resides is nitrate limited. In agreement with the hypothesis that the inflamed gut is a nitrate-limited environment, we detected low average nitrate levels in murine ceca, even after infection with an S. Typhimurium mutant that cannot consume nitrate (0.29 mM). A low nitrate concentration in the gut may also explain why the growth of S. Typhimurium in the intestinal lumen requires energy taxis to seek out spatial niches containing this limited electron acceptor (14). Taken together, our data support a model where pathogen-induced inflammation generates low concentrations of nitrate that support the luminal growth of S. Typhimurium using the periplasmic nitrate reductase NapABC.
S. Typhimurium presents a prime example of a bacterial pathogen well adapted to proliferation in an environment greatly altered during infection. Gut inflammation results in shifts in resident microbial populations, disrupting the relative stability of the normally dominant phyla Firmicutes and Bacteroidetes (38, 39), while the reactive oxygen and nitrogen species released by infiltrating immune cells change the availability of key nutrients, such as tetrathionate (40) and ethanolamine (30). Other catabolized nutrients available for the growth of S. Typhimurium include fructose-asparagine (41), hydrogen (42), fucose (13), and sialic acid (13). This ever-changing environment requires the pathogen to exhibit metabolic flexibility in order to maximize its growth by responding to alterations in nutrient availability. This appears to be true with regard to changes in the availability of host-derived nitrate, since regulated expression of the appropriate nitrate respiratory enzymes can significantly impact the fitness of S. Typhimurium by reinforcing its growth via enhanced ATP production, redox balance, or carbon source utilization.
An important role for the periplasmic nitrate reductase may reveal a common metabolic strategy for intestinal bacterial pathogens. The species S. enterica contains two pathovars, one associated with gastrointestinal disease (to which S. Typhimurium belongs) and the other associated with extraintestinal disease (to which Salmonella enterica serovar Typhi belongs) (43). Comparison of the genomes of serovars belonging to the gastrointestinal pathovar with those belonging to the extraintestinal pathovar identifies genes important for intestinal survival, because they are undergoing decay in the latter group (43). That is, extraintestinal serovars accumulate mutations affecting genes important only for luminal growth, since no selective pressure exists to preserve their function; the result is pseudogene formation (43–45). Analysis of the nitrate reductases and their regulators found that only the narQP two-component system was rendered nonfunctional in extraintestinal serovars due to mutations (43). This suggests that NarQP, or genes regulated by NarQP, such as napABC, are critical for gastrointestinal serovars but not for extraintestinal serovars. This hypothesis is consistent with our results showing that maintenance of the periplasmic nitrate reductase strongly supports the enteropathogenic lifestyle of S. Typhimurium. The benefit provided by the periplasmic nitrate reductase is also relevant to bacteria in lineages outside the Enterobacteriaceae. For instance, Vibrio cholerae, the causative agent in the diarrheal disease cholera, multiplies to large numbers in the intestines during infection. V. cholerae encodes a homologue of the periplasmic nitrate reductase, while homologues of cytosolic nitrate reductases are absent. Interestingly, a study examining genes upregulated by V. cholerae after passage through a human host found markedly increased expression of the periplasmic nitrate reductase complex (46). Similarly, Campylobacter jejuni, a frequent cause of food-borne intestinal illness, respires nitrate during infection (47). C. jejuni encodes a periplasmic nitrate reductase but lacks homologues of the cytosolic nitrate reductase. Therefore, a high-affinity periplasmic nitrate reductase may be a common mechanism to assist pathogen growth in a highly competitive intestinal environment.
Our work highlights an important contribution of the periplasmic nitrate reductase to the growth of S. Typhimurium in the intestines during infection and may indicate a common mechanism to support the fitness of intestinal pathogens in diverse lineages. This adds to the small but growing body of literature deciphering the critical role of metabolism in the survival and proliferation of bacterial pathogens.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI096528 (to A.J.B.) and AI112241 (to C.A.L.). F.R.-C. was supported by Public Health Service Grant AI060555.
We declare no conflicts of interest.
REFERENCES
- 1.Jones BD, Ghori N, Falkow S. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J Exp Med 180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark MA, Jepson MA, Simmons NL, Hirst BH. 1994. Preferential interaction of Salmonella typhimurium with mouse Peyer's patch M cells. Res Microbiol 145:543–552. doi: 10.1016/0923-2508(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 3.Galán JE. 2001. Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol 17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 4.Santos RL, Zhang S, Tsolis RM, Bäumler AJ, Adams LG. 2002. Morphologic and molecular characterization of Salmonella typhimurium infection in neonatal calves. Vet Pathol 39:200–215. doi: 10.1354/vp.39-2-200. [DOI] [PubMed] [Google Scholar]
- 5.Santos RL, Raffatellu M, Bevins CL, Adams LG, Tükel C, Tsolis RM, Bäumler AJ. 2009. Life in the inflamed intestine, Salmonella style. Trends Microbiol 17:498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiennimitr P, Winter SE, Bäumler AJ. 2012. Salmonella, the host and its microbiota. Curr Opin Microbiol 15:108–114. doi: 10.1016/j.mib.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keestra-Gounder AM, Tsolis RM, Bäumler AJ. 2015. Now you see me, now you don't: the interaction of Salmonella with innate immune receptors. Nat Rev Microbiol 13:206–216. doi: 10.1038/nrmicro3428. [DOI] [PubMed] [Google Scholar]
- 8.McCormick BA, Colgan SP, Delp-Archer C, Miller SI, Madara JL. 1993. Salmonella typhimurium attachment to human intestinal epithelial monolayers: transcellular signalling to subepithelial neutrophils. J Cell Biol 123:895–907. doi: 10.1083/jcb.123.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupré-Crochet S, Erard M, Nüße O. 2013. ROS production in phagocytes: why, when, and where? J Leukoc Biol 94:657–670. doi: 10.1189/jlb.1012544. [DOI] [PubMed] [Google Scholar]
- 10.Salzman AL, Eaves-Pyles T, Linn SC, Denenberg AG, Szabo C. 1998. Bacterial induction of inducible nitric oxide synthase in cultured human intestinal epithelial cells. Gastroenterology 114:93–102. doi: 10.1016/S0016-5085(98)70637-7. [DOI] [PubMed] [Google Scholar]
- 11.Lopez CA, Winter SE, Rivera-Chávez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Bäumler AJ. 2012. Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3(3):e00143-12. doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundberg JO, Hellstrom PM, Lundberg JM, Alving K. 1994. Greatly increased luminal nitric oxide in ulcerative colitis. Lancet 344:1673–1674. doi: 10.1016/S0140-6736(94)90460-X. [DOI] [PubMed] [Google Scholar]
- 13.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivera-Chávez F, Winter SE, Lopez CA, Xavier MN, Winter MG, Nuccio SP, Russell JM, Laughlin RC, Lawhon SD, Sterzenbach T, Bevins CL, Tsolis RM, Harshey R, Adams LG, Bäumler AJ. 2013. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog 9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Bäumler AJ. 2013. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, Tsolis RM, Bäumler AJ. 2013. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio 4(4):e00430-13. doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iobbi C, Santini CL, Bonnefoy V, Giordano G. 1987. Biochemical and immunological evidence for a second nitrate reductase in Escherichia coli K12. Eur J Biochem 168:451–459. doi: 10.1111/j.1432-1033.1987.tb13438.x. [DOI] [PubMed] [Google Scholar]
- 18.Unden G, Bongaerts J. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta 1320:217–234. doi: 10.1016/S0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 19.Garland PB, Downie JA, Haddock BA. 1975. Proton translocation and the respiratory nitrate reductase of Escherichia coli. Biochem J 152:547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones RW, Lamont A, Garland PB. 1980. The mechanism of proton translocation driven by the respiratory nitrate reductase complex of Escherichia coli. Biochem J 190:79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart V, Lu Y, Darwin AJ. 2002. Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J Bacteriol 184:1314–1323. doi: 10.1128/JB.184.5.1314-1323.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potter LC, Cole JA. 1999. Essential roles for the products of the napABCD genes, but not napFGH, in periplasmic nitrate reduction by Escherichia coli K-12. Biochem J 344(Part 1):69–76. doi: 10.1042/0264-6021:3440069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potter LC, Millington P, Griffiths L, Thomas GH, Cole JA. 1999. Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem J 344(Part 1):77–84. doi: 10.1042/0264-6021:3440077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones SA, Gibson T, Maltby RC, Chowdhury FZ, Stewart V, Cohen PS, Conway T. 2011. Anaerobic respiration of Escherichia coli in the mouse intestine. Infect Immun 79:4218–4226. doi: 10.1128/IAI.05395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darwin AJ, Stewart V. 1995. Expression of the narX, narL, narP, and narQ genes of Escherichia coli K-12: regulation of the regulators. J Bacteriol 177:3865–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noriega CE, Lin HY, Chen LL, Williams SB, Stewart V. 2010. Asymmetric cross-regulation between the nitrate-responsive NarX-NarL and NarQ-NarP two-component regulatory systems from Escherichia coli K-12. Mol Microbiol 75:394–412. doi: 10.1111/j.1365-2958.2009.06987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wimpenny JW, Cole JA. 1967. The regulation of metabolism in facultative bacteria. 3. The effect of nitrate. Biochim Biophys Acta 148:233–242. doi: 10.1016/0304-4165(67)90298-X. [DOI] [PubMed] [Google Scholar]
- 28.Wallace BJ, Young IG. 1977. Role of quinones in electron transport to oxygen and nitrate in Escherichia coli. Studies with a ubiA− menA− double quinone mutant. Biochim Biophys Acta 461:84–100. [DOI] [PubMed] [Google Scholar]
- 29.Cold Spring Harbor Protocols. 2006. M9 recipe. Cold Spring Harb Protoc doi: 10.1101/pdb.rec8146. [DOI] [Google Scholar]
- 30.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsolis RM, Xavier MN, Santos RL, Bäumler AJ. 2011. How to become a top model: impact of animal experimentation on human Salmonella disease research. Infect Immun 79:1806–1814. doi: 10.1128/IAI.01369-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabin RS, Stewart V. 1993. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J Bacteriol 175:3259–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart V. 1993. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol Microbiol 9:425–434. doi: 10.1111/j.1365-2958.1993.tb01704.x. [DOI] [PubMed] [Google Scholar]
- 35.Jones SA, Chowdhury FZ, Fabich AJ, Anderson A, Schreiner DM, House AL, Autieri SM, Leatham MP, Lins JJ, Jorgensen M, Cohen PS, Conway T. 2007. Respiration of Escherichia coli in the mouse intestine. Infect Immun 75:4891–4899. doi: 10.1128/IAI.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Tseng CP, Gunsalus RP. 1999. The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J Bacteriol 181:5303–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeMoss JA, Hsu PY. 1991. NarK enhances nitrate uptake and nitrite excretion in Escherichia coli. J Bacteriol 173:3303–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ali MM, Newsom DL, Gonzalez JF, Sabag-Daigle A, Stahl C, Steidley B, Dubena J, Dyszel JL, Smith JN, Dieye Y, Arsenescu R, Boyaka PN, Krakowka S, Romeo T, Behrman EJ, White P, Ahmer BM. 2014. Fructose-asparagine is a primary nutrient during growth of Salmonella in the inflamed intestine. PLoS Pathog 10:e1004209. doi: 10.1371/journal.ppat.1004209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maier L, Vyas R, Cordova CD, Lindsay H, Schmidt TS, Brugiroux S, Periaswamy B, Bauer R, Sturm A, Schreiber F, von Mering C, Robinson MD, Stecher B, Hardt WD. 2013. Microbiota-derived hydrogen fuels Salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe 14:641–651. doi: 10.1016/j.chom.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Nuccio SP, Bäumler AJ. 2014. Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. mBio 5(2):e00929-14. doi: 10.1128/mBio.00929-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langridge GC, Fookes M, Connor TR, Feltwell T, Feasey N, Parsons BN, Seth-Smith HM, Barquist L, Stedman A, Humphrey T, Wigley P, Peters SE, Maskell DJ, Corander J, Chabalgoity JA, Barrow P, Parkhill J, Dougan G, Thomson NR. 2015. Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proc Natl Acad Sci U S A 112:863–868. doi: 10.1073/pnas.1416707112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nuccio SP, Bäumler AJ. 2015. Reconstructing pathogen evolution from the ruins. Proc Natl Acad Sci U S A 112:647–648. doi: 10.1073/pnas.1423499112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Calderwood SB, Schoolnik GK, Camilli A. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sellars MJ, Hall SJ, Kelly DJ. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J Bacteriol 184:4187–4196. doi: 10.1128/JB.184.15.4187-4196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rankin JD, Taylor RJ. 1966. The estimation of doses of Salmonella typhimurium suitable for the experimental production of disease in calves. Vet Rec 78:706–707. doi: 10.1136/vr.78.21.706. [DOI] [PubMed] [Google Scholar]
- 49.Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 50.Winter SE, Winter MG, Thiennimitr P, Gerriets VA, Nuccio SP, Russmann H, Bäumler AJ. 2009. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol Microbiol 74:175–193. doi: 10.1111/j.1365-2958.2009.06859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51a.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784–791. [Google Scholar]
- 52.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kingsley RA, Reissbrodt R, Rabsch W, Ketley JM, Tsolis RM, Everest P, Dougan G, Bäumler AJ, Roberts M, Williams PH. 1999. Ferrioxamine-mediated iron(III) utilization by Salmonella enterica. Appl Environ Microbiol 65:1610–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kinder SA, Badger JL, Bryant GO, Pepe JC, Miller VL. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R−M+ mutant. Gene 136:271–275. doi: 10.1016/0378-1119(93)90478-L. [DOI] [PubMed] [Google Scholar]