Abstract
Escherichia coli strains expressing the K1 capsule are a major cause of sepsis and meningitis in human neonates. The development of these diseases is dependent on the expression of a range of virulence factors, many of which remain uncharacterized. Here, we show that all but 1 of 34 E. coli K1 neonatal isolates carried clbA and clbP, genes contained within the pks pathogenicity island and required for the synthesis of colibactin, a polyketide-peptide genotoxin that causes genomic instability in eukaryotic cells by induction of double-strand breaks in DNA. Inactivation of clbA and clbP in E. coli A192PP, a virulent strain of serotype O18:K1 that colonizes the gastrointestinal tract and translocates to the blood compartment with very high frequency in experimental infection of the neonatal rat, significantly reduced the capacity of A192PP to colonize the gut, engender double-strand breaks in DNA, and cause invasive, lethal disease. Mutation of clbA, which encodes a pleiotropic enzyme also involved in siderophore synthesis, impacted virulence to a greater extent than mutation of clbP, encoding an enzyme specific to colibactin synthesis. Restoration of colibactin gene function by complementation reestablished the fully virulent phenotype. We conclude that colibactin contributes to the capacity of E. coli K1 to colonize the neonatal gastrointestinal tract and to cause invasive disease in the susceptible neonate.
INTRODUCTION
Escherichia coli strains belonging to phylogenetic group B2 are found with increasing frequency in the feces of healthy individuals from high-income countries (1) and are responsible for a range of extraintestinal diseases that include urinary tract infections, sepsis, pneumonia, and neonatal meningitis (2). These extraintestinal pathogenic E. coli (ExPEC) strains carry genes encoding a diverse range of virulence determinants that promote colonization, invasion, survival in the blood compartment, and the capacity to evade host defenses and damage the host (3). These virulence-enhancing genes tend to cluster on pathogenicity islands, genomic elements that facilitate dissemination by horizontal gene transfer (4). The pks genomic island is found in 30% to 40% of E. coli B2 strains and codes for the production of colibactin, a polyketide-peptide genotoxin of as-yet-unknown chemical structure that causes double-strand (ds) breaks in DNA, leading to cell cycle arrest, chromosome instability, and increased lymphopenia in septic rodents (5–8). Carriage of the pks island is linked to long-term persistence in the gastrointestinal (GI) tract (9), and pks-bearing strains represent a high-virulence subset within the B2 group (6). The 54-kb pks island encodes nonribosomal peptide synthases, polyketide synthases, and hybrid synthases, in addition to accessory, tailoring, and editing enzymes; the clbA gene encodes a phosphopantetheinyl transferase required for colibactin synthesis, and clbP specifies a d-amino peptidase involved in colibactin maturation (10, 11). The pks island also contributes to the synthesis of siderophores by virtue of the broad substrate specificity of ClbA, impacting iron acquisition and the ability to survive in the blood compartment (12).
The pks island was initially identified in an E. coli isolate from a case of neonatal bacterial meningitis (NBM) (6). E. coli is a leading cause of NBM and neonatal sepsis (13–15); these life-threatening conditions arise following vertical transmission of the bacteria from mother to infant during or shortly after birth, and the large majority express the anti-phagocytic polysialic acid K1 capsule (16). Data from human infections are sparse, but results of studies using rat models of infection indicate that E. coli K1 strains initially colonize the GI tract of the neonatal rat and subsequently enter the blood circulation via the mesenteric lymphatic system, causing severe systemic infection with a high likelihood of central nervous system (CNS) involvement and local inflammation (17–21), much as in human infections. The neonatal rat model adopted for these studies replicates the strong age-dependency characteristic of the human condition; systemic infection appears to be related to the capacity of the colonizing bacteria to translocate across the immature GI tract of the newborn (18, 21). Neuropathogenic strains of E. coli K1 have arisen by clonal expansion (22) and, although they are not generally responsible for systemic infection in postneonatal infants and adults, constitute a group of highly virulent opportunistic pathogens for susceptible neonates. We therefore examined the extent of involvement of colibactin production in the pathogenesis of neonatal E. coli K1 systemic infection by determining the degree of pks carriage in clinical isolates and the impact of mutation of clbA and clbP in the course of infection in a rat model of neonatal infection. The data indicate that colibactin and siderophore synthesis has a substantial impact on GI tract colonization and the capacity of pks-positive E. coli K1 to cause invasive disease.
MATERIALS AND METHODS
Bacterial strains.
E. coli O18:K1 strain A192PP was derived from septicemia isolate E. coli A192 (22) by two rounds of passage through neonatal rat pups, with bacterial recovery from the blood (23). Thirty-four isolates of E. coli K1 from neonatal blood, obtained between 2001 and 2014 at Great Ormond Street Hospital, London, United Kingdom, were kindly provided by Nigel Kline and Elaine Cloutman-Green (Institute of Child Health, University College London [UCL]). The presence of K1 capsule was confirmed with K1-specific bacteriophage K1E (24). Inactivation of E. coli A192PP genes was undertaken using bacteriophage λ Red recombinase as described previously (25). The mutants employed are shown in Table 1. For complementation, clbA was cloned into plasmid pASK75 (26) and clbP into plasmid pBRSK (10) (Table 1). clbA::kan and clbP::kan alleles were amplified by PCR from chromosomal DNA of strains SP15clbA::kan and SP15clbP::kan using primers clbA-F/clbA-R and clbP-F/clbP-R (clbA-F, CAG ATA CAC AGA TAC CAT TCA; clbA-R, CTA GAT TAT CCG TGG CGA TTC; clbP-F, GTG AAC TGA GCG AAA TAT TGG CTA ATC; clbP-R, TTA CTC ATC GTC CCA CTC CTT GTT G), respectively, and were used to transform strain A192PP. The allelic exchanges were confirmed by PCR.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype, phenotype, or descriptiona | Reference or source |
|---|---|---|
| E. coli strains | ||
| A192PP | 018:K1:H7; virulent in rat model of neonatal meningitis | 23 |
| A192PPΔclbA::kan | clbA mutant of A192PP; Kanr | This study |
| A192PPΔclbA::kan+pASK75 | clbA mutant of A192PP carrying pASK75 clbA+; Kanr Ampr | This study |
| A192PPΔclbA::kan+pASK75.clbA | clbA mutant of A192PP carrying pASK75; Kanr Ampr | This study |
| A192PPΔclbP::kan | clbP mutant of A192PP; Kanr | This study |
| A192PPΔclbP::kan+pBRSK | clbP mutant of A192PP carrying pBRSK; Kanr Ampr | This study |
| A192PPΔclbP::kan+pBRSK.clbP | clbP mutant of A192PP carrying pBRSK clbP+; Kanr Ampr | This study |
| Plasmids | ||
| pASK75.clbA | Plasmid carrying clbA gene; Ampr | 25 |
| pBRSK.clbP | Plasmid carrying clbP gene | This study |
Amp, ampicillin; Kan, kanamycin.
Identification of the pks island.
Strains were screened by PCR for the presence of clbA and clbP genes representative of the pks island. PCRs were performed using a HotStart Taq kit (Qiagen, Limburg, Netherlands) in a 25-μl reaction volume containing 1 pmol F and R primers, 1× buffer, 1× HotStart Taq buffer mix, 1 μl DNA, and distilled water (dH2O). Reactions were heated to 95°C for 15 min followed by 35 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 90 s followed by 1 cycle of 72°C for 10 min. PCR products were detected following separation by electrophoresis using 1.5% agarose gels.
Genome sequencing.
Bacteria were grown overnight in Mueller-Hinton broth at 37°C using an orbital incubator (minimum of 200 orbits). Genomic DNA was extracted from 1 ml of culture using a QIAamp DNA minikit (Qiagen). Sequencing library preparation was carried out with Illumina Nextera XT according to the manufacturer's instructions. MiSeq sequencing (2 × 151 bp) was carried out following the standard protocols of the manufacturer (Illumina Inc., USA). Sequencing reads were quality controlled using Trimmomatic (26). pks islands were identified in E. coli genomes by BLAST analysis of the pks element in IHE3034 (AM229678). pks islands and their insertion sites from nine E. coli genomes were compared using the Artemis Comparison Tool (27). Fastq files for A192PP were mapped against the IHE3034 pks element using the Burrows-Wheeler Aligner software package (28).
Colibactin production.
Colibactin induces senescence and consequent megalocytosis in cultured eukaryotic cells, which manifest as a progressive enlargement of the cell body and nucleus and the absence of mitosis. We quantified the extent of colibactin-induced megalocytosis using a methylene blue binding assay (12). E. coli strains were added to HeLa cells cultivated as described above at multiplicities of infection (MOIs) of between 12 and 400, cocultured for 4 h, and washed. Cells were then incubated for 72 h with cell culture medium containing 200 μg/ml gentamicin followed by staining with methylene blue. Dye binding was determined spectrophotometrically by measuring optical density at 600 nm (OD660).
Genotoxicity assay.
The capacity of colibactin to engender ds DNA breaks was determined in HeLa cells by γ-H2AX immunofluorescence analysis (5, 29); this assay monitors the phosphorylation of histone H2AX, a sensitive marker of ds DNA breaks. HeLa cells (1.5 × 104 cells in 200 μl Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 80 μg/ml gentamicin, and 0.1 unit/ml bovine insulin) were dispensed into 96-well cell culture plates. After incubation at 37°C in a 5% CO2 atmosphere for 24 h, the cells were washed and incubated with bacteria at MOIs of 20 to 100 bacteria per cell. After 5 h of infection at 37°C in 5% CO2, cells were washed 3 times with Hanks' balanced salt solution and incubated at 37°C in cell culture medium for 20 h with 200 μg/ml gentamicin. Cells were fixed in the plate with 4% paraformaldehyde and processed as previously described (12). Rabbit monoclonal anti-γ-H2AX antibody 9718 (Cell Signaling Technology Inc., Danvers, MA) was diluted 1:500 in blocking solution and incubated for 2 h at room temperature. IRDye 800CW-conjugated goat anti-rabbit secondary antibody was diluted 1:100 in blocking solution and incubated for 1 h. RedDot2 (Biotium) was used for DNA labeling. DNA and γ-H2AX were visualized using an Odyssey infrared imaging scanner (Li-Cor ScienceTec, Les Ulis, France) and 680-nm and 800-nm channels for RedDot2 and IRDye 800, respectively. Relative fluorescence unit values for γ-H2AX per cell (as determined by division of γ-H2AX values by DNA content values) were divided by vehicle control values to determine percentages of changes in phosphorylation of H2AX relative to control levels. All experiments were carried out in triplicate.
Colonization and infection of neonatal rats.
Animal experiments were approved by the Ethical Committee of the UCL School of Pharmacy and the United Kingdom Home Office (HO) and were conducted under HO Project License PPL 80/2243. The procedure has been described in detail previously (30). Briefly, all members of a litter (12 pups) of 2-day-old (P2) Wistar rat pups (Harlan, United Kingdom) were fed 20 μl of mid-logarithmic-phase E. coli bacteria (2 to 6 × 106 CFU) from an Eppendorf micropipette. GI tract colonization was determined by culture of perianal swabs on MacConkey agar; bacteremia was detected by MacConkey agar culture of blood taken postmortem. Disease progression was determined by daily evaluation of symptoms of systemic infection (28). After sacrifice, samples from the proximal small intestine (SI), mid-SI, distal SI, colon, blood, mesenteric lymph nodes (MLN), liver, kidney, and spleen were excised aseptically, transferred to ice-cold phosphate-buffered saline (PBS), and homogenized. Bacteria were quantified by serial dilution culture on MacConkey agar, and the presence of E. coli K1 was confirmed with bacteriophage K1E.
Immunohistochemical evaluation of tissue sections.
The entire GI tract was recovered immediately after culling of rat pups, and 2-cm segments of the mid-SI were removed as previously described (30). Tissues were placed in 10% formalin for 24 h and embedded in paraffin, and 5-μM-thick sections were cut. Dewaxed sections were subjected to antigen retrieval in 0.1 M trypsin for 6 min at 37°C and then in 10 mM sodium citrate buffer (pH 6.0) for 30 min at 94°C. After peroxidase blocking (Dako S2023) for 5 min, sections were incubated with protein block reagent (2.5% normal goat serum) for 20 min and then with monoclonal rabbit anti-γ-H2AX antibody 9718 (Cell Signaling Technology Inc., Danvers, MA) at a dilution of 1:400 for 1 h. The slides were washed with Envision flex buffer (K8000; Dako) for 5 min and then incubated with a polymer-peroxidase conjugate (K4061; Dako) for 30 min. After a second wash, chromogenic development was performed for 5 min with 3,3′-diaminobenzidine (K3468; Dako). The tissues were counterstained with hematoxylin. Histological images were transformed into digital microscopic images with a Pannoramic 250 Flash II scanning system (3DHISTECH, Budapest, Hungary).
Nucleotide sequence accession number.
The nucleotide sequence of the A192PP genome has been deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/ena/; accession number PRJEB9141).
RESULTS
The pks island is widely distributed among neonatal E. coli K1 isolates.
E. coli A192PP displays a high level of virulence in the rodent model of systemic infection (23); lethal infection in susceptible neonatal pups occurs following translocation across the small intestine, with evidence that the invading bacteria adopt a transcellular route across the epithelium (21). E. coli K1 virulence factors enabling colonization and invasion have not been elucidated in an unambiguous fashion, and we therefore examined A192PP for the presence of genes associated with the pks island; PCR indicated that the strain carried clbA and clbP. The presence of the pks island was confirmed by whole-genome sequencing of A192PP; the strain carries all genes required for colibactin production, and the island contains no single nucleotide polymorphisms that distinguish it from the island found in O18:K1:H7 ExPEC strain IHE3034. Similar observations were made with regard to the other sequences we interrogated. We then examined 34 E. coli K1 neonatal isolates for the presence of clbA and clbP; all but one isolate carried these two genes.
Neonatal E. coli K1 isolates elicit genotoxic effects in vitro.
Seven neonatal E. coli K1 pks-positive (pks+) isolates were randomly selected, and their capacity to elicit genotoxic effects in HeLa cocultures was determined; in all cases, exposure of HeLa cells to neonatal E. coli K1 isolates resulted in megalocytosis, a reliable marker of colibactin-producing group B2 strains (Fig. 1) (31). E. coli A192PP also demonstrated the ability to induce megalocytosis in a HeLa cell coculture (Fig. 2B).
FIG 1.
Production of colibactin by E. coli A192PP and derivatives determined by methylene blue staining (OD660) of HeLa cells exposed to the bacteria indicated. MOI, multiplicity of infection, compared to nonexposed control (C).
FIG 2.
Colibactin production by E. coli A192PP induces genotoxic effects. (A) Growth of E. coli strains A192PP, A192PPΔclbA::kan, and A192PPΔclbP::kan in Mueller-Hinton (MH) broth at 37°C (200 orbits/min) (± 1 standard deviation [SD], n = 3). (B) Production of colibactin by E. coli A192PP and derivatives determined by methylene blue staining (OD600) of HeLa cells exposed to the bacteria indicated. MOI, multiplicity of infection compared to nonexposed control. (C) Quantification of double-strand (ds) DNA breaks induced by E. coli A192PP and derivatives. Strains A192PPΔclbA::kan+pASK75.clbA and A192PPΔclbP::kan+pBRSK.clbP are complemented derivatives of strains A192PPΔclbA::kan and A192PPΔclbP::kan, respectively. HeLa cells were incubated with the concentrations of bacteria indicated, and ds breaks were quantified by determination of γ-H2AX induction levels. The Z test was used to determine significant differences (means and standard errors) between A192PP and A192PPΔclbA, A192PP and A192PPΔclbA+pASK75.clbA, A192PPΔclbA and A192PPΔclbA+pASK75.clbA, A192PP and A192PPΔclbP, A192PPΔclbP and A192PPΔclbA+pBRSK.clbP, and A192PPΔclbA+pASK75.clbA and A192PPΔclbA+pBRSK.clbP (all P < 0.001). There were no significant differences between A192PPΔclbA and A192PPΔclbP or between A192PP and A192PPΔclbP+pBRSK.clbP.
While disruption of clbA and clbP in E. coli A192PP did not affect growth kinetics in batch culture (Fig. 2A), induction of megalocytosis was not evident when HeLa cells were exposed to A192PPΔclbA::kan and A192PPΔclbP::kan derivatives of E. coli A192PP (Fig. 2B). Further, in contrast to the parent strain, strains A192PPΔclbA::kan and A192PPΔclbP::kan did not induce ds DNA breaks in HeLa cells (Fig. 2C). This functionality was restored when clbA and clbP were introduced on plasmid vectors (Fig. 2C), and the capacity of the complemented strains to induce γ-H2AX was comparable to that of the progenitor A192PP strain.
Colibactin contributes to the virulence of E. coli K1 strain A192PP.
E. coli strain A192PP and ΔclbA::kan and ΔclbP::kan mutants and the complemented strains were examined for their capacity to induce lethal effects in a rat model of neonatal E. coli K1 systemic infection. We previously established that 2-day-old (P2) rat pups are exquisitely susceptible to lethal infection following oral introduction of E. coli A192PP (23). Over a period of 1 week (P2 to P9), the pups become progressively more resistant to infection, but not to colonization, as a result of rapid maturation of the neonatal GI tract (21, 32). In susceptible animals, E. coli A192PP cells begin to enter the blood circulation around 24 h after GI tract colonization becomes apparent and, within 7 days, all animals in an infected litter succumb to highly disseminated infection. GI colonization of E. coli A192PP is stable in quantitative aspects over the period of investigation (21, 23). Here, P2 neonates became efficiently colonized in the GI tract by E. coli A192PP and all derivatives within 24 to 48 h of administration and remained colonized for the duration of the experiment. As expected, E. coli A192PP produced lethal infection in all colonized pups (Fig. 3) and although a proportion of pups colonized with the A192PPΔclbA::kan (Fig. 3A) and A192PPΔclbP::kan (Fig. 3B) mutants did not survive, the overall lethal effect of these mutations was significantly attenuated. Thus, disruption of clbA increased survival by 50% and disruption of clbP increased survival by 30%. Introduction of an intact clbA gene restored the lethal effect, while enhanced lethality in comparison to that seen with E. coli A192PP was found when the clbP gene was complemented using plasmid pBRSK.clbP.
FIG 3.
Colibactin genes are required for expression of full virulence in the rat model of neonatal systemic infection. Rates of survival of P2 rats colonized with E. coli K1 A192PP and (A) A192PPΔclbA or (B) A192PPΔclbP are shown. GI tract colonization was established by manual feeding of 2 × 106 to 6 × 106 CFU. For panel A, strain A192PP n = 24, A192PPΔclbA::kan n = 24, A192PPΔclbA::kan+pASK75 n = 12, and A192PPΔclbA::kan +pASK75.clbA n = 12. For panel B, strain A192PP n = 36, A192PPΔclbP::kan n = 36, A192PPΔclbP::kan+pBRSK n = 12, and A192PPΔclbP::kan+pBRSK.clbP n = 12 (log-rank [Mantel-Cox] test; ns, nonsignificant; *, P < 0.05; **, P < 0.01).
Colibactin influences colonization of, and induces genotoxic effects in, the neonatal GI tract.
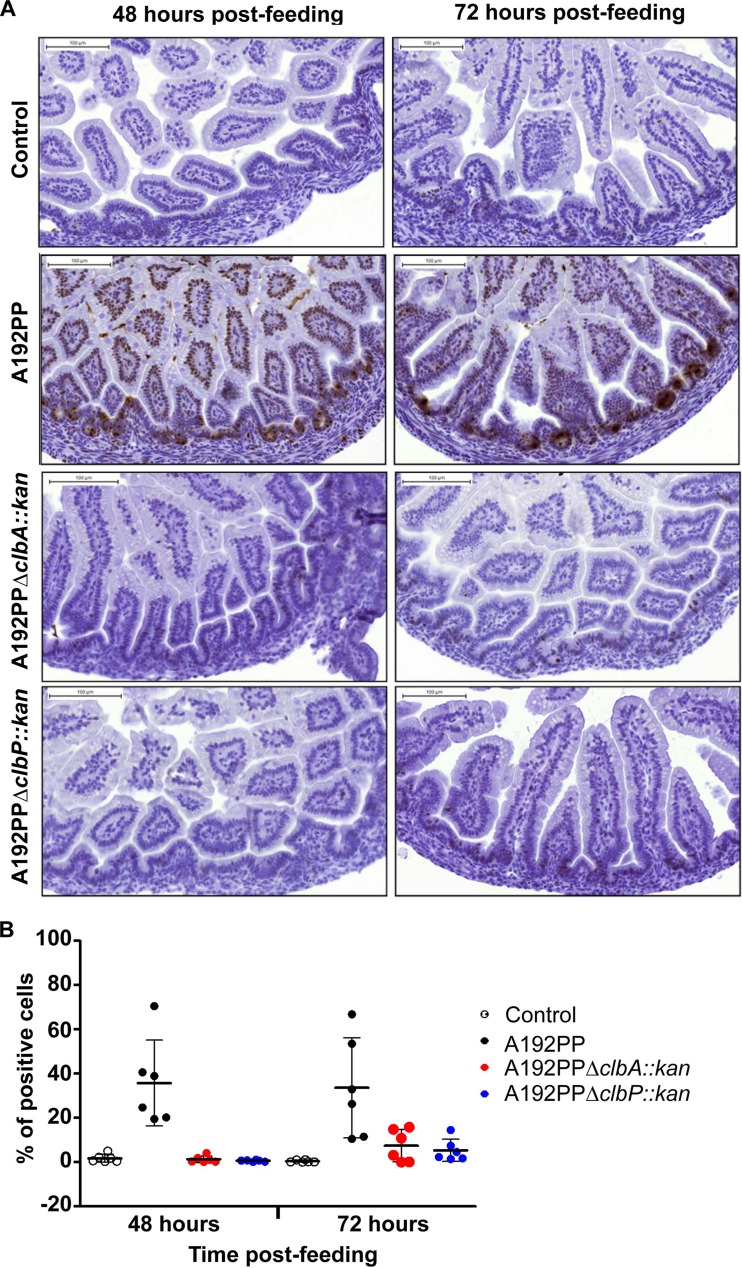
Current evidence suggests that the likely site of translocation of E. coli K1 from the GI lumen to the blood compartment is the small intestine (21), with the proximal SI and the midsection of this region being the most likely loci of colonization preceding systemic invasion. At both sites, inactivation of clbA and clbP in E. coli A192PP resulted in a significantly lower population of colonizing bacteria (Fig. 4). With strains A192PPΔclbA::kan and A192PPΔclbP::kan, significantly fewer viable bacteria were recovered from the blood compared to the parent strain and fewer bacteria were recovered from the major organs of bacterial clearance but, with the exception of the spleen, following infection with the clbP mutant, these did not reach levels of statistical significance (Fig. 4). We also examined the possibility that GI-colonizing E. coli A192PP engendered ds DNA breaks in enterocytes lining the mid-SI. We therefore obtained mid-SI sections from P2 pups 48 h and 72 h after oral administration of E. coli A192PP strains and assessed DNA damage by monitoring phosphorylation of histone γ-H2AX, a sensitive marker of ds DNA breaks (33), with monoclonal rabbit anti-γ-H2AX antibody. There was evidence of extensive ds DNA breaks in cells lining the mid-SI lumen from pups exposed to A192PP but not in those exposed to A192PPΔclbA::kan and A192PPΔclbP::kan mutants or the negative control (Fig. 5A). At 48 h and 72 h after colonization, dsDNA breaks were found in, respectively, 35.7% and 33.6% of cells lining the mid-SI lumen of pups exposed to E. coli A192PP, significantly more than in the cells from pups exposed to A192PPΔclbA::kan and A192PPΔclbP::kan mutants (P < 0.01 and P < 0.001 at 48 and 72 h, respectively; analysis of variance [ANOVA]) (Fig. 5B).
FIG 4.
Colibactin genes influence the degree of GI tract colonization and blood and organ dissemination of E. coli A192PP in the susceptible P2 neonatal rat. Tissues were removed 24 h after colonization and CFU/g of tissue determined by plating on MacConkey agar. E. coli A192PP colonies were detected using K1-specific bacteriophage; mutants were selected on medium containing 50 μg/ml kanamycin. SI, small intestine. For panel A, strain A192PP n = 18 and strain A192PPΔclbA::kan n = 18. For panel B, strain A192PP n = 17 and strain A192PPΔclbP::kan n = 17 (Student's t test; ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
FIG 5.
E. coli A192PP induces double-strand (ds) DNA breaks in epithelial cells of the mid-SI. P2 rats were colonized with E. coli A192PP, A192PPΔclbA::kan, or A192PPΔclbP::kan by feeding from an Eppendorf pipette; controls (uninfected group) were fed MH broth. Animals were sacrificed at 48 h (n = 6) or 72 h (n = 6) after administration of the colonizing bacteria or broth. Sections of the mid-SI were embedded in paraffin and stained for γ-H2AX (brown), a biomarker for breaks in ds DNA, and counterstained with hematoxylin. (A) Representative mid-SI sections. Bar, 100 μm. (B) Quantification of ds DNA breaks in mid-SI sections. Groups of 6 rats were analyzed. Data represent means ± SD (one-way ANOVA with Tukey's multiple-comparison test; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
The blood isolate E. coli A192 efficiently (100%) colonized the GI tract of neonatal rats but elicited bacteremia in only 25% to 35% of colonized animals (18, 32), whereas the passaged derivative A192PP caused invasive disease in essentially all pups colonized at P2 (23, 32). GI tract colonization of P2 pups by E. coli A192PP was accompanied by extensive replication of A192PP to yield steady-state populations of 107 to 108/g within 24 h (21). A similar degree of clonal expansion was noted at 24 h in this study, particularly in the proximal and mid-SI; inactivation of both clbA and clbP gave rise to a statistically significant, 5- to 10-fold reduction in the size of the colonizing population in these regions of the GI tract (Fig. 4). It is likely that the capacity of potential neuropathogens to colonize the neonate in numbers sufficient to permit their translocation to the blood compartment during the first few days of life represents a key determinant of outcome, although very little experimental or epidemiological data are available to assess the relative importance of quantitative aspects of early gastrointestinal colonization in either naturally occurring or experimental infections. However, these reductions in colonization are concomitant with a reduced capacity to cause bacteremia and death (Fig. 3). In this context, it is known that colonization with A192PP induces massive downregulation of tff2 encoding Trefoil factor 2, a compact protein involved in assembly and maturation of the mucin layer that keeps luminal bacteria away from the enterocyte surface (21, 33).
Colibactin production has a substantial impact on the capacity of A192PP to cause lethal infection (Fig. 3) following invasion of the blood compartment (Fig. 4), providing a practical demonstration that the pks island is associated with bacteremia (6, 9, 12). In line with previous observations (5, 7), we found that pks carriage elicits ds DNA breaks in cultured cells. DNA ds cleavage is also manifest in gut epithelial cells (Fig. 5) during the period (48 to 72 h) of translocation from the GI lumen to the blood compartment (21). The blood bioburden of E. coli A192PPΔclbA::kan was significantly lower than that achieved by strain A192PPΔclbP::kan 24 h after colonization (Fig. 4). ClbA impacts colibactin and siderophore synthesis, while ClbP is involved only in colibactin synthesis (12), suggesting that low-molecular-weight iron chelators may also play a significant role in the invasive potential of E. coli K1 infection in the neonatal rat. We have previously shown that mutation of clbA impacts siderophore biosynthesis when the entD gene (present in E. coli A192PP) is inactivated (12), emphasizing that the clbA mutation has the potential to engender pleiotropic effects on siderophore and colibactin biosynthesis, even in the presence of a functional entD gene. We therefore hypothesize that strain A192PPΔclbA::kan, compromised with regard to colibactin biosynthesis, also displays altered siderophore production, which could contribute to the decreased virulence of the clbA mutant compared to the clbP mutant. It should be borne in mind, however, that siderphore production in vitro is not informative with respect to the levels of synthesis of the chelator during colonization and invasion of infected animals.
Colibactin requires cell-bacterium contact in order to elicit genotoxic effects (5). We have recently obtained as-yet-unpublished histological and immunohistochemical evidence that A192PP comes into close proximity to enterocytes lining the GI tract, adopts a transcellular route across the epithelial barrier, and gains access to the circulation via the mesenteric lymphatic system. The observations reported here provide additional evidence for this mode of translocation, as it is likely that DNA strand breakage occurs predominantly either during close-proximity colonization with or after internalization of pks-carrying bacteria. Colibactin-mediated DNA damage leads to activation of DNA damage repair pathways that promote the accumulation of mutations with long-term health consequences for the host if exposure to the genotoxin persists (7). More-transient exposure leads to formation of anaphase bridges, chromosomal abnormalities, and senescence in dividing cells (7, 34). Recently, it has been demonstrated, using a mother-to-offspring transmission model, that colibactin-producing E. coli strains impair intestinal permeability by low-molecular-weight molecules and impact oral tolerance (35). We will determine if A192PP colonization impacts the permeability of the gut by micromolecules, macromolecules, and particulates in order to reconcile transient or long-term DNA damage with an enhanced capacity to translocate across the gut epithelium. Additional processes associated with colibactin production remain to be established.
In summary, this is the first report to show that colibactin production contributes to the capacity of E. coli to cause systemic lethal infections in experimental models that follow that natural pathway to infection. Moreover, our findings indicate that the pks island is widely distributed among neonatal E. coli K1 isolates, suggesting that this virulence factor has an important role to play in invasive disease in susceptible neonates. Additional processes associated with colibactin production remain to be established.
ACKNOWLEDGMENTS
This work was supported by research grant MR/K018396/1 from the Medical Research Council. This work was also supported by research grant ANR-13-BSV1-0028-01 from the Agence Nationale de la Recherche and the Aninfimip platform, an EquipEx (Equipement d'Excellence) platform supported by the French government through the Investments for the Future program (ANR-11-EQPX-0003). The National Institute for Health Research University College London Hospitals Biomedical Research Centre provided further support.
REFERENCES
- 1.Escobar-Páramo P, Grenet K, Le Menac'h A, Rode L, Salgado E, Amorin C, Gouriou S, Picard B, Rahimy MC, Andremont A, Denamur E, Ruimy R. 2004. Large-scale population structure of human commensal Escherichia coli isolates. Appl Environ Microbiol 70:5698–5700. doi: 10.1128/AEM.70.9.5698-5700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 5:449–456. doi: 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 3.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt H, Hensel M. 2004. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev 17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 6.Johnson JR, Johnston B, Kuskowski MA, Nougayrede JP, Oswald E. 2008. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J Clin Microbiol 46:3906–3911. doi: 10.1128/JCM.00949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. 2010. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A 107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcq I, Martin P, Payros D, Cuevas-Ramos G, Boury M, Watrin C, Nougayrède JP, Olier M, Oswald E. 2014. The genotoxin colibactin exacerbates lymphopenia and decreases survival rate in mice infected with septicemic Escherichia coli. J Infect Dis 210:285–294. doi: 10.1093/infdis/jiu071. [DOI] [PubMed] [Google Scholar]
- 9.Nowrouzian FL, Oswald E. 2012. Escherichia coli strains with the capacity for long-term persistence in the bowel microbiota carry the potentially genotoxic pks island. Microb Pathog 53:180–182. doi: 10.1016/j.micpath.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Dubois D, Baron O, Cougnoux A, Delmas J, Pradel N, Boury M, Bouchon B, Bringer MA, Nougayrède JP, Oswald E, Bonnet R. 2011. ClbP is a prototype of a peptidase subgroup involved in biosynthesis of nonribosomal peptides. J Biol Chem 286:35562–35570. doi: 10.1074/jbc.M111.221960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brotherton CA, Balskus EP. 2013. A prodrug resistance mechanism is involved in colibactin biosynthesis and cytotoxicity. J Am Chem Soc 135:3359–3362. doi: 10.1021/ja312154m. [DOI] [PubMed] [Google Scholar]
- 12.Martin P, Marcq I, Magistro G, Penary M, Garcie C, Payros D, Boury M, Olier M, Nougayrède JP, Audebert M, Chalut C, Schubert S, Oswald E. 2013. Interplay between siderophores and colibactin genotoxin biosynthetic pathways in Escherichia coli. PLoS Pathog 9:e1003437. doi: 10.1371/journal.ppat.1003437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson JR, Oswald E, O'Bryan TT, Kuskowski MA, Spanjaard L. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J Infect Dis 185:774–784. doi: 10.1086/339343. [DOI] [PubMed] [Google Scholar]
- 14.Polin RA, Harris MC. 2001. Neonatal bacterial meningitis. Semin Neonatol 6:157–172. doi: 10.1053/siny.2001.0045. [DOI] [PubMed] [Google Scholar]
- 15.Simonsen KA, Anderson-Berry AL, Delair SF, Davies HD. 2014. Early-onset neonatal sepsis. Clin Microbiol Rev 27:21–47. doi: 10.1128/CMR.00031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarff LD, McCracken GH, Schiffer MS, Glode MP, Robbins JB, Ørskov I, Ørskov F. 1975. Epidemiology of Escherichia coli K1 in healthy and diseased newborns. Lancet i:1099–1104. [DOI] [PubMed] [Google Scholar]
- 17.Glode MP, Sutton A, Moxon ER, Robbins JB. 1977. Pathogenesis of neonatal Escherichia coli meningitis: induction of bacteremia and meningitis in infant rats fed E. coli K1. Infect Immun 16:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pluschke G, Mercer A, Kusećek B, Pohl A, Achtman M. 1983. Induction of bacteremia in newborn rats by Escherichia coli K1 is correlated with only certain O (lipopolysaccharide) antigen types. Infect Immun 39:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zelmer A, Bowen M, Jokilammi A, Finne J, Luzio JP, Taylor PW. 2008. Differential expression of the polysialyl capsule during blood-to-brain transit of neuropathogenic Escherichia coli K1. Microbiology 154:2522–2532. doi: 10.1099/mic.0.2008/017988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zelmer A, Martin M, Gundogdu O, Birchenough G, Lever R, Wren BW, Luzio JP, Taylor PW. 2010. Administration of capsule-selective endosialidase E minimizes changes in organ gene expression induced by experimental systemic infection with Escherichia coli K1. Microbiology 156:2205–2215. doi: 10.1099/mic.0.036145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birchenough GMH, Johannson MEV, Stabler RA, Dalgakiran F, Hansson GC, Wren BW, Luzio JP, Taylor PW. 2013. Altered innate defenses in the neonatal gastrointestinal tract in response to colonization by neuropathogenic Escherichia coli. Infect Immun 81:3264–3275. doi: 10.1128/IAI.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Achtman M, Mercer A, Kusećek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver RP. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun 39:315–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mushtaq N, Redpath MB, Luzio JP, Taylor PW. 2004. Prevention and cure of systemic Escherichia coli K1 infection by modification of the bacterial phenotype. Antimicrob Agents Chemother 48:1503–1508. doi: 10.1128/AAC.48.5.1503-1508.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross RJ, Cheasty T, Rowe B. 1977. Isolation of bacteriophages specific for the K1 polysaccharide antigen of Escherichia coli. J Clin Microbiol 6:548–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olier M, Marcq I, Salvador-Cartier C, Secher T, Dobrindt U, Boury M, Bacquié V, Pénary M, Gaultier E, Nougayrède JP, Fioramonti J, Oswald E. 2012. Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes 3:501–509. doi: 10.4161/gmic.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalgakiran F, Witcomb L, McCarthy A, Birchenough GMH, Taylor PW. 2014. Non-invasive model of neuropathogenic Escherichia coli infection in the neonatal rat. J Vis Exp 92:e52018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mushtaq N, Redpath MB, Luzio JP, Taylor PW. 2005. Treatment of experimental Escherichia coli infection with recombinant bacteriophage-derived capsule depolymerase. J Antimicrob Chemother 56:160–165. doi: 10.1093/jac/dki177. [DOI] [PubMed] [Google Scholar]
- 32.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 33.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Secher T, Samba-Louaka A, Oswald E, Nougayrède JP. 2013. Escherichia coli producing colibactin triggers premature and transmissible senescence in mammalian cells. PLoS One 8:e77157. doi: 10.1371/journal.pone.0077157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Secher T, Payros D, Brehin C, Boury M, Watrin C, Gillet M, Bernard-Cadenat I, Menard S, Theodorou V, Saoudi A, Olier M, Oswald E. 2015. Oral tolerance failure upon neonatal gut colonization with Escherichia coli producing the genotoxin colibactin. Infect Immun 83:2420–2429. doi: 10.1128/IAI.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]