Abstract
The Lyme disease spirochete, Borrelia burgdorferi, expresses RevA and numerous outer surface lipoproteins during mammalian infection. As an adhesin that promotes bacterial interaction with fibronectin, RevA is poised to interact with the extracellular matrix of the host. To further define the role(s) of RevA during mammalian infection, we created a mutant that is unable to produce RevA. The mutant was still infectious to mice, although it was significantly less well able to infect cardiac tissues. Complementation of the mutant with a wild-type revA gene restored heart infectivity to wild-type levels. Additionally, revA mutants led to increased evidence of arthritis, with increased fibrotic collagen deposition in tibiotarsal joints. The mutants also induced increased levels of the chemokine CCL2, a monocyte chemoattractant, in serum, and this increase was abolished in the complemented strain. Therefore, while revA is not absolutely essential for infection, deletion of revA had distinct effects on dissemination, arthritis severity, and host response.
INTRODUCTION
Borrelia burgdorferi, the causative agent of Lyme disease, is a vector-borne pathogen that successfully colonizes both ticks and a variety of vertebrate hosts. In mammals, B. burgdorferi is frequently found associated with connective tissues (1–13), and the bacterium is often detected in cartilaginous or collagen-rich tissues, such as skin and joints (6, 7, 9, 11–18). Adhesins expressed by B. burgdorferi facilitate interactions with these tissues. B. burgdorferi expresses a plethora of outer surface proteins that interact with numerous components of the extracellular matrix (ECM), such as fibronectin, decorin, and integrins (19–23). The attachment of B. burgdorferi to the host ECM is likely critical for pathogenesis and persistence in mammals. Indeed, for many of these adhesins, deletion mutants have significant defects in infectivity, fail to disseminate widely in the host, or have other effects on disease, such as alterations in the severity of arthritis (24–27).
The interactions of B. burgdorferi with fibronectin are facilitated by several distinct bacterial surface proteins, including BBK32, RevA, BB0347, and CspA (BbCRASP-1) (28–31). The redundancy in adhesins makes it difficult to dissect the role of each B. burgdorferi fibronectin binding protein in the pathogenesis of Lyme disease. For example, real-time microscopic imaging in live mice revealed a significant role for BBK32 in interactions with host vasculature, yet bbk32 mutants are still able to infect mammals (32).
RevA is a 19-kDa surface protein encoded on the circular plasmid 32 (cp32) family of plasmids (33). RevA expression is elevated during mammalian infection over its expression in the tick vector, suggesting a role in pathogenesis (30, 34–36). RevA binds to fibronectin, and anti-RevA antibodies block the binding of whole B. burgdorferi bacteria to fibronectin (30). RevA is antigenic, as evidenced by the fact that both mice and human Lyme disease patients produce antibodies against RevA (36, 37). Antibodies against RevA are bactericidal in vitro, and passive immunization with anti-RevA antibodies prevents infection (38). However, mice vaccinated with recombinant RevA protein were not protected when challenged with B. burgdorferi by needle or tick bite (38).
Analysis of RevA has been complicated by the fact that two separate revA genes are present in the B31 type strain, revA1 on cp32-1 and revA6 on cp32-6. The mature amino acid sequences of the two encoded RevA proteins are identical, however. Intriguingly, a revA1 mutant uncovered in a transposon mutagenesis study demonstrated an infectivity deficit in dissemination (39). To further elucidate the function of RevA and its role in the pathogenesis of B. burgdorferi, we created a double revA deletion mutant and characterized its infectious properties.
MATERIALS AND METHODS
Bacteria.
B. burgdorferi strain B31-A3 is an infectious clone of the sequenced type strain (40, 41) that contains all parental plasmids except cp9 (42). Bacteria were grown at 34°C to densities of approximately 1 × 107 cells/ml in modified Barbour-Stoenner-Kelly (BSK-II) medium supplemented with 6% rabbit serum (43). Total DNA (genomic and plasmid DNA) was isolated using a DNeasy blood and tissue kit (Qiagen, Valencia, CA). Plasmid contents were determined by multiplex PCR as described by Bunikis et al. (44).
Generation of revA deletion mutant and complemented mutant clones.
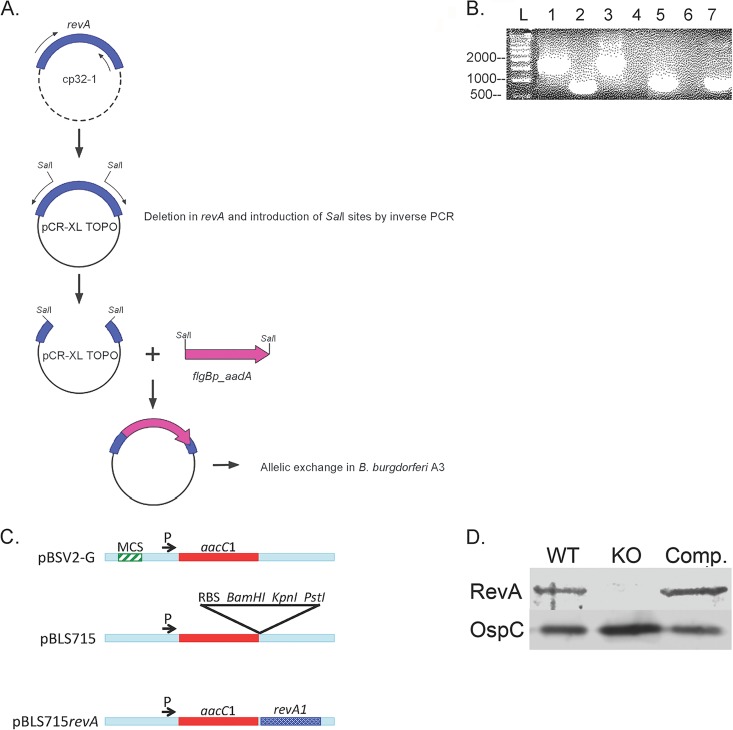
B. burgdorferi strain B31-A3 has 2 copies of the revA gene: revA1 on plasmid cp32-1 (open reading frame [ORF] bbp27) and revA6 (ORF bbm27) on plasmid cp32-6. To create a doubly deleted mutant, the revA1 region was first PCR amplified with the primers listed in Table 1 and then cloned into the TOPO XL vector (Fig. 1A). Restriction enzyme sites (SalI) were created, and the revA open reading frame was deleted, by inverse PCR. A streptomycin resistance cassette was first ligated into the SalI site and then introduced into B. burgdorferi B31-A3 by electroporation (45). The revA6 locus was deleted in a similar matter (using a kanamycin cassette), and the final construct was introduced into the ΔrevA1 mutant. Deletion of both revA loci was confirmed by PCR (primers listed in Table 1) and bidirectional Sanger sequencing, and the double deletion mutant was designated B31-A3ΔrevA1ΔrevA6.
TABLE 1.
Oligonucleotides used in this study
| Primer name | Sequence (5′ → 3′) | Purpose | Source or reference |
|---|---|---|---|
| BBP27flkF | TATTTAGTAGTAGTAAAAATAAACAAAATAATATG | Cloning of revA + flank | This study |
| BBP27flkR | CTAATTTATGATCAAATCGGCTTTTGC | Cloning of revA + flank | This study |
| Pflg_aadAF | GTCGACTACCCGAGCTTCAAGGAAG | Cloning; addition of SalI sites; PCR confirmationa | This study |
| Pflg_aadAR | CAGCTGTTATTTGCCGACTACCTTGGTGATC | Cloning; addition of SalI sites; PCR confirmationa | This study |
| Inv_PCR_BBP27F | TAATGGCTTGTAAAGCATATGTAGAAGCAAAATTAGGTCGAC | Overlap PCR for revA disruption | This study |
| Inv_PCR_BBP27R | CAGCTGCAGTTACTCCAACTAATTTTGCTTCTACATATG | Overlap PCR | This study |
| BBM27flkF | GGGTTAAATCACGCTTTCCCCTGC | Cloning of revA + flank; PCR confirmationa | This study |
| BBM27flkR | CATTGTCTACTAATTGCTTTGCTGCTAT | Cloning of revA + flank; PCR confirmationa | This study |
| Pflg_kanF | CTCGAGTACCCGAGCTTCAAGGAAG | Cloning; addition of XhoI sites | This study |
| Pflg_kanR | GAGCTCTTAGAAAAACTCATCGAGCATC | Cloning; addition of XhoI sites | This study |
| Inv_PCR_BBM27F | AGCAGCGTTAAAAGCAGCTAAAAATACTTAATTGGGCTCGAG | Overlap PCR | This study |
| Inv_PCR_BBM27R | GAGCTCATAATAAATATTCCCAATTAAGTATTTTTAGCT | Overlap PCR | This study |
| AadF | TACCCGAGCTTCAAGGAAG | Confirmation of aad | This study |
| AadR | TTATTTGCCGACTACCTTGGTGATC | Confirmation of aad | This study |
| KanF | TACCCGAGCTTCAAGGAAG | Confirmation of kan; Southern blotting | This study |
| KanR | TTAGAAAAACTCATCGAGCATC | Confirmation of kan | This study |
| BamHIRevF | GGATCCATGAGAAATAAAAACATATTTAAATTATTTTTTG | Cloning of revA for complementation | This study |
| PstIRevR | CTGCAGTTAATTAGTGCCCTCTCC | Cloning of revA for complementation | This study |
| RevAF | ATGAGAAATAAAAACATATTTAAATTATTTTTTG | Confirmation of revA for complementation | This study |
| RevAR | TTAATTAGTGCCCTCTCC | Confirmation of revA for complementation | This study |
| nTM17F | GTGGATCTATTGTATTAGATGAGGCTCTCG | qPCR | 49 |
| nTM17R | GCCAAAGTTCTGCAACATTAACACCTAAAG | qPCR | 49 |
| nidoF | CCAGCCACAGAATACCATCC | qPCR | 38 |
| nidoR | GGACATACTCTGCTGCCATC | qPCR | 38 |
| Fla3 | GGGTCTCAAGCGTCTTGG | qPCR | 48 |
| Fla4 | GAACCGGTGCAGCCTGAG | qPCR | 48 |
| Iscap16sF | CGGTCTGAACTCAGATCAAG | qPCR | 48 |
| Iscap16sR | GGGACAAGAAGACCCTATG | qPCR | 48 |
See Fig. 1B.
FIG 1.
Construction of the revA-deficient mutant. (A) Schematic of mutant construction. See Materials and Methods for details of cloning. (B) PCR confirmation of revA deletions by use of flanking primers for the revA6 locus (lanes 1 to 4) and primers for aad (lanes 5 to 7). Lanes 1 and 5, B31-A3ΔrevA1ΔrevA6; lanes 2 and 6, the parental strain, B31-A3; lane 3, cloning plasmid control (with the whole flanking region plus revA and the kan cassette); lane 4, no template; lane 7, cloning plasmid control (with the whole flanking region plus revA and the aad cassette). L, ladder. (C) Schematic of construction of the revA complement vector. MCS, multiple cloning site; RBS, ribosome-binding site. (D) Immunoblotting for RevA protein in whole-cell lysates. Lane 1, B31-A3 (wild type [WT]); lane 2, overloaded B31-A3ΔrevA1ΔrevA6 (knockout [KO]); lane 3, complemented strain. OspC serves as a loading control.
For complementation, the wild-type revA1 gene was cloned into a B. burgdorferi-Escherichia coli shuttle plasmid under the control of the constitutive promoter PflgB. Briefly, pBSV2-G (46) was modified by overlap extension PCR mutagenesis to delete the existing multiple cloning site and then add a ribosome-binding site and BamHI, PstI, and KpnI cleavage sites (Fig. 1C). The revA1 gene, without its native promoter, was PCR amplified using primers with added BamHI and PstI sites and was then cloned into pBLS715. This insertion was verified by PCR and bidirectional Sanger sequencing (Davis Sequencing). The new construct, pBLS715revA, was introduced into B31-A3ΔrevA1ΔrevA6 (the revA double deletion mutant) by electroporation (45). Several individual clones were isolated by pour plating in the presence of gentamicin. The presence of pBLS715revA was verified by PCR (primers listed in Table 1) and sequencing. The plasmid content of the resulting complemented strain, B31-A3ΔrevA1ΔrevA6/pBLS715revA, was assessed by multiplex PCR (44).
Immunoblot analysis.
Whole-cell lysates were separated on 12.5% SDS-PAGE gels and were transferred to nitrocellulose membranes. The membranes were blocked overnight at 4°C with 5% (wt/vol) bovine serum albumin (BSA) in Tris-buffered saline–Tween 20 (TBS-T), consisting of 20 mM Tris (pH 7.5), 150 mM NaCl, and 0.05% (vol/vol) Tween 20. The membranes were washed with TBS-T and were incubated for 2 h at room temperature with purified anti-RevA or anti-OspC (loading control) diluted 1:500 in TBS-T (30). After 3 washes with TBS-T for 5 min each time, the membranes were incubated for 1 h at room temperature with a horseradish peroxidase-conjugated donkey anti-rabbit IgG antibody (GE Healthcare) diluted 1:5,000 in TBS-T. After 5 washes with TBS-T for 10 min each time, bound antibodies were detected using the SuperSignal West Pico enhanced chemiluminescence substrate (Pierce). Blots were visualized on a Li-Cor bioimaging system by using the associated imaging software.
Infection of mice and ticks.
For studies of 50% infective doses (ID50) and tissue dissemination, female C3H/HeN mice (4 to 6 weeks old) were infected by subcutaneous injection with 1 × 105, 1 × 104, 1 × 103, or 1 × 102 bacteria of strain B31-A3, the B31-A3ΔrevA1ΔrevA6 mutant, or the complemented strain, B31-A3ΔrevA1ΔrevA6/pBLS715revA, from mid-exponential-phase cultures grown at 34°C. Cultures were harvested at 1 × 107 bacteria/ml. Infection of mice was confirmed by analysis of serum samples by enzyme-linked immunosorbent assays (ELISAs) for antibodies directed against B. burgdorferi whole-cell lysates as described previously (30, 38). Two weeks after infection, ear pinnae, hearts, bladders, skin from the inoculation site, and tibiotarsal joints were collected and were either frozen for DNA extraction and quantitative PCR (qPCR) or cultured in BSK-II medium plus 6% rabbit serum and 50 μg/ml rifampin.
To initiate tick infection studies, female C3H/HeN mice (4 to 6 weeks old) were infected by subcutaneous injection of 1 × 106 bacteria of strain B31-A3 or the revA-deficient mutant from mid-exponential-phase cultures grown at 34°C. These mice then served to infect Ixodes scapularis larvae as follows. Egg masses laid by pathogen-free I. scapularis ticks were obtained from the Department of Entomology, Oklahoma State University—Stillwater, Stillwater, OK, and were held in a humidified chamber until they hatched. For B. burgdorferi acquisition studies, approximately 200 naïve larvae were placed on each of the B. burgdorferi-infected mice. After 96 h, the ticks had fully engorged and naturally dropped off the mice. Immediately after the completion of feeding, cohorts of approximately 30 engorged larvae from each mouse were analyzed by qPCR (primers are given in Table 1) for the acquisition of B. burgdorferi. The remainder of the ticks were returned to the humidified chamber and were allowed to molt to the nymphal stage. Approximately 3 weeks after ecdysis, the infected nymphs were allowed to feed on naïve female C3H/HeN mice. Mice infected through feeding by infected nymphs were killed 2 weeks after the completion of tick feeding. Ear pinnae, hearts, bladders, and tibiotarsal joints were processed for DNA extraction and culture. For arthritis development, female C3H/HeN mice (3 to 4 weeks old) were infected by injection into the left footpad of 1 × 103 or 1 × 104 bacteria of strain B31-A3, the B31-A3ΔrevA1ΔrevA6 mutant, or the complemented strain from mid-exponential-phase cultures grown at 34°C.
Measurement of tibiotarsal joints.
Joint measurements were taken for both ankles with a digital metric caliper (General Tools, Montreal, QC, Canada) at 0, 2, and 4 weeks postinfection. Measurements were taken in the anterior-to-posterior position with the knee extended, through the thickest portion of the ankle (47). Three measurements were taken per time point and were averaged. A researcher blinded to the strain of B. burgdorferi took the measurements, while a second researcher recorded the results. The percentage of change in ankle diameter was determined by subtracting the preinfection joint diameter from the measurements taken at weeks 2 and 4 [e.g., (individual ankle measurement at 2 weeks − ankle measurement at baseline) ×100].
Histology of tibiotarsal joints.
At 4 weeks postinfection, mice were sacrificed and the tibiotarsal joints collected for histopathology. The joints were first decalcified and then fixed in 10% neutral buffered formalin. Sections from the decalcified joints were embedded in paraffin and were stained with hematoxylin and eosin (H&E; AML Laboratories, Baltimore, MD). Sections were scored blindly by a veterinary pathologist (North Dakota State University [NDSU], Fargo, ND) from 0 to 5, as follows: 0, no inflammation; 1, minimal change; 2, mild change (1 to 25% of the area infiltrated with leukocytes); 3, moderate change (25 to 50% of the area infiltrated with leukocytes, often with synovial hyperplasia and/or fibrosis); 4, severe change (including more than 50% infiltrating leukocytes, synovial hyperplasia, and fibrosis); 5, most severe change (characteristics of score 4 plus exudates within the joint or tendon sheath). For collagen staining, paraffin-embedded H&E-stained slides were deparaffinized and placed in phosphate-buffered saline (PBS). The slides were then stained with Masson's trichrome stains (Sigma-Aldrich) according to the manufacturer's standard procedure. Slides were first dehydrated with increasing concentrations of ethanol-water solutions and then equilibrated in a xylene-ethanol solution (1:1, by volume) followed by 100% xylene. Coverslips were then placed on the slides by using Permount (Thermo Fisher). Sections from at least 3 mice per infection strain were examined.
ELISA.
Mouse blood was drawn from the saphenous vein and was collected in heparin-coated tubes. Blood samples were centrifuged (6,000 × g) to remove red blood cells, and the serum was stored at −20°C. To measure mouse IgM or IgG against B. burgdorferi, 96-well plates were coated overnight with 100 μl/well of 10 μg/ml B. burgdorferi lysate (mid-log-phase B. burgdorferi cultures, pelleted and washed 3 times in PBS) in carbonate coating buffer (0.32 g Na2CO3 and 0.586 g NaHCO3 per 200 ml [pH 9.6]) at 4°C. Room temperature plates were washed three times with PBS containing 0.05% Tween 20 (by volume) (PBS-T). Wells were first blocked for 2 h at room temperature with PBS containing 10% fetal bovine serum and then washed three times with PBS-T. At the time of the assay, a 1:100 dilution of serum was placed on the plate and was incubated for 2 h at 37°C. Wells were first washed three times with PBS-T and then incubated for 1 h at room temperature with a horseradish peroxidase (HRP)-conjugated goat antiserum against mouse IgM (Pierce) or IgG (GE Healthcare, Piscataway, NJ) diluted to 1:5,000 in PBS. Color development was performed by adding a tetramethylbenzidine substrate (TMB; Thermo Fisher Scientific, Waltham, MA) for 15 min and was stopped by the addition of an equal volume of 2 N sulfuric acid. Commercial ELISA kits were used to measure mouse chemokines according to the manufacturer's instructions (R&D Systems, Minneapolis, MN).
Analysis of B. burgdorferi loads in mouse tissues.
Total DNA was extracted from tissue samples by using a DNeasy kit according to the manufacturer's instructions (Qiagen). Frozen mouse tissue samples (20 mg) were first minced with sterile single-use razor blades on a DNA/DNase-free glass surface and were resuspended in Buffer ATL with proteinase K for overnight digestion at 56°C as recommended by the manufacturer (Qiagen). For ticks, cohorts of 30 fed larvae were processed according to the method of Jutras et al. (48). qPCR was performed using a Bio-Rad MyiQ2 thermal cycler and Bio-Rad SYBR green Supermix. All DNA samples were analyzed in triplicate. Each run included a sample that lacked a template in order to test for DNA contamination of reagents. The oligonucleotide primers used for amplification of B. burgdorferi recA (nTM17F and nTM17R) (49), Ixodes scapularis 16S rRNA, B. burgdorferi flaB (48), and mouse nidogen are given in Table 1. Reaction conditions consisted of a 10-min initial denaturation step at 95°C; 40 cycles of 95°C for 15 s and 55°C (for recA) or 60°C (for nidogen) for 1 min; 95°C for 1 min; 60°C for 1 min; and melting analysis starting at 60°C and increasing by increments of 0.5°C, with a hold at each temperature for 10 s. Tenfold serial dilutions of B. burgdorferi genomic DNA, mouse genomic DNA, or Ixodes genomic DNA were included in every assay for each primer set. This enabled the generation of standard curves from which the amount of DNA present in each sample could be calculated by using Bio-Rad MyiQ2 software. The same software package was also used for melting-curve analyses. To verify amplicon sizes and purities, all products were separated by agarose gel electrophoresis, and DNA was visualized with ethidium bromide. Average values obtained from triplicate runs of each DNA sample for B. burgdorferi recA copies or flaB copies were calculated relative to the average triplicate value for the mouse nidogen or I. scapularis 16S rRNA housekeeping gene from the same DNA preparation. Statistical analyses of data were performed using Student's t test and assuming unequal variances.
RESULTS
Generation of B. burgdorferi revA double deletion and complemented strains.
The B. burgdorferi type strain, B31, naturally carries 2 nearly identical copies of the revA gene: revA1 on plasmid cp32-1 and revA6 on plasmid cp32-6. The amino acid sequences of the mature RevA1 and RevA6 proteins are identical. The revA loci were sequentially deleted by allelic exchange, as diagramed in Fig. 1A, creating strain B31-A3ΔrevA1ΔrevA6. A schematic of the construction of the mutant is shown in Fig. 1A. The deletion of both copies of revA through allelic exchange was confirmed by PCR (Fig. 1B).
To demonstrate that any observed phenotypes of the mutant strain were due to revA deletion only, a revA trans-complemented strain was also constructed (B31-A3ΔrevA1ΔrevA6/pBLS715revA). The relevant sequences of both strains were confirmed by PCR and sequencing. The inability of the mutant to produce RevA and the production of RevA by the complemented strain were confirmed by immunoblotting (Fig. 1D).
Effect of RevA deletion on infectivity.
To assess the contribution of RevA to the pathogenic process, we tested the ability of the mutant bacterium to complete each stage of the natural infection cycle. B31-A3ΔrevA1ΔrevA6 mutant bacteria were first assessed for their ability to infect mammals. Cohorts of mice were inoculated with serially diluted bacteria, ranging from 10 to 105 spirochetes per animal (50). Wild-type B. burgdorferi generally exhibits ID50 of 10 to 100 bacteria in mice (51–53). We found no differences in the ID50 between the wild-type and revA-deficient B. burgdorferi strains (Table 2).
TABLE 2.
ID50 of wild-type and revA-deficient B. burgdorferi strainsa
| Strain and inoculum | No. culture positive/total no. |
|
|---|---|---|
| Mice | Organsb | |
| B31-A3 | ||
| 105 | 6/6 | 18/18 |
| 104 | 10/10 | 29/34 |
| 103 | 10/10 | 20/34 |
| 102 | 0/6 | 0/24 |
| B31-A3ΔrevA1ΔrevA6 | ||
| 105 | 6/6 | 15/15 |
| 104 | 8/10 | 26/34 |
| 103 | 8/10 | 17/34 |
| 102 | 0/6 | 0/24 |
Mice were infected subcutaneously with increasing inocula of the B. burgdorferi parental strain B31-A3 or the B31-A3ΔrevA1ΔrevA6 mutant. Mice were sacrificed 2 weeks postinfection, and organs were cultured.
Including bladders, hearts, ears, and tibiotarsal joints.
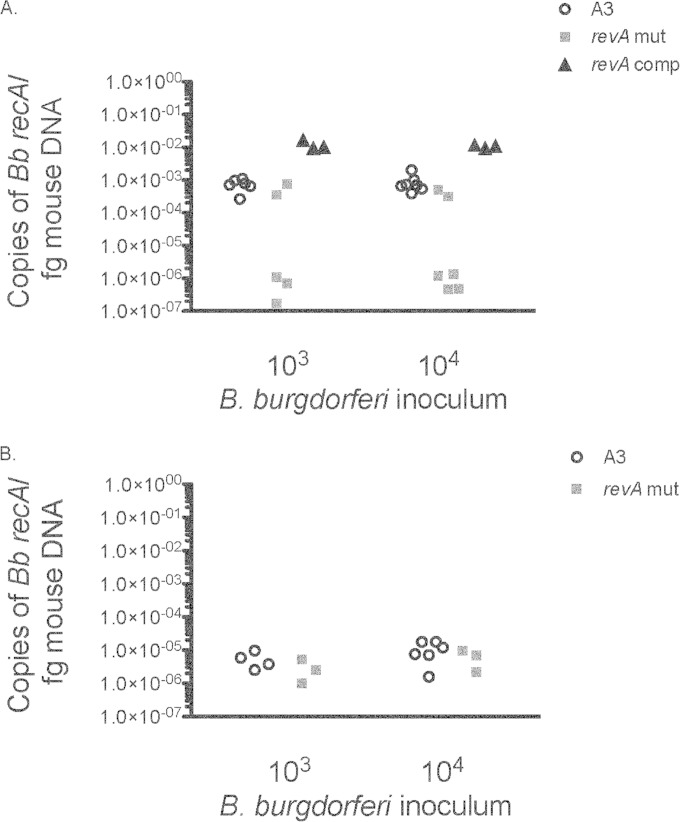
Experience with other B. burgdorferi mutants has demonstrated subtle defects in infectivity, such as impaired dissemination or differences in tissue pathology (see, e.g., references 25 and 54). Therefore, we also determined the ability of revA-deficient bacteria to disseminate to distal organs. We found that the B31-A3ΔrevA1ΔrevA6 mutant was significantly less able to colonize heart tissues (P = 0.003) than the wild-type strain (Fig. 2A). This defect was restored upon complementation. No significant defects were observed for tibiotarsal joint colonization (Fig. 2B).
FIG 2.
revA-deficient mutants are impaired in colonization of the heart but not the joint. One month after infection, hearts (A) and joints (B) from mice infected with either B31-A3ΔrevA1ΔrevA6 (revA mut), the parental strain, B31-A3 (A3), or the complemented strain (revA comp) were collected, DNA was extracted, and levels of B. burgdorferi DNA were determined by qPCR. Data are expressed as copies of B. burgdorferi recA per femtogram of mouse nidogen DNA. Samples were analyzed in triplicate, and each data point represents an individual animal. Two-way analysis of variance was used to calculate statistical differences between the groups infected with the parental strain or the B31-A3ΔrevA1ΔrevA6 mutant. The difference in the ability to colonize heart tissues (A) was statistically significant (P = 0.003).
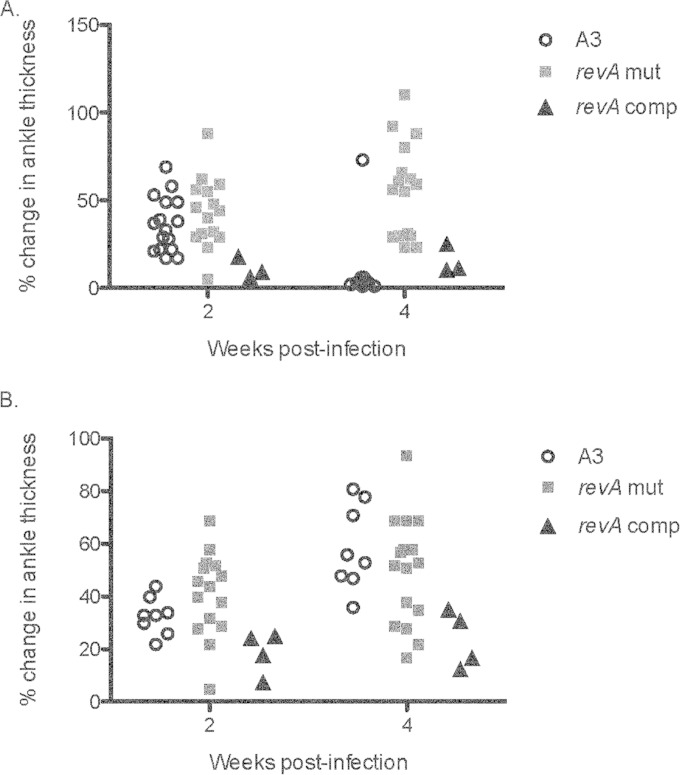
Noting that other borrelial adhesins have been found to have an impact on tissue-specific pathology, we tested the effects of RevA deficiency on arthritis severity. Cohorts of C3H/HeN mice were injected subcutaneously or in the left footpad with 103 or 104 B. burgdorferi B31-A3 or B31-A3ΔrevA1ΔrevA6 mutant bacteria. Tibiotarsal joint (ankle) thickness was measured before and after infection. Mice that were injected in the footpad with 103 B31-A3ΔrevA1ΔrevA6 bacteria showed significantly greater ankle thickness at 4 weeks postinfection (P < 0.001) (Fig. 3A). No significant differences were apparent following inoculation with the higher dose (104 bacteria) (Fig. 3B). No significant swelling of the right ankle was noted for the footpad-injected mice (data not shown). For all mice injected subcutaneously, ankle thickness increased over time, but there were no differences between B31-A3, the revA-deficient mutant, and the complemented strain (data not shown).
FIG 3.
Effects of RevA deficiency on ankle thickness. Three- to 4-week-old female C3H/HeN mice were injected in the left footpad with 103 (A) or 104 (B) bacteria of strain B31-A3, the B31-A3ΔrevA1ΔrevA6 mutant, or the complemented strain. Ankle thickness was measured with calipers (triplicate measurements) at baseline and at 2 and 4 weeks postinfection. Two-way analysis of variance was used to calculate statistical differences between the groups injected with the parental strain or the B31-A3ΔrevA1ΔrevA6 mutant. For mice injected with 103 bacteria (A), the difference was significant (P < 0.0001) at 4 weeks postinfection.
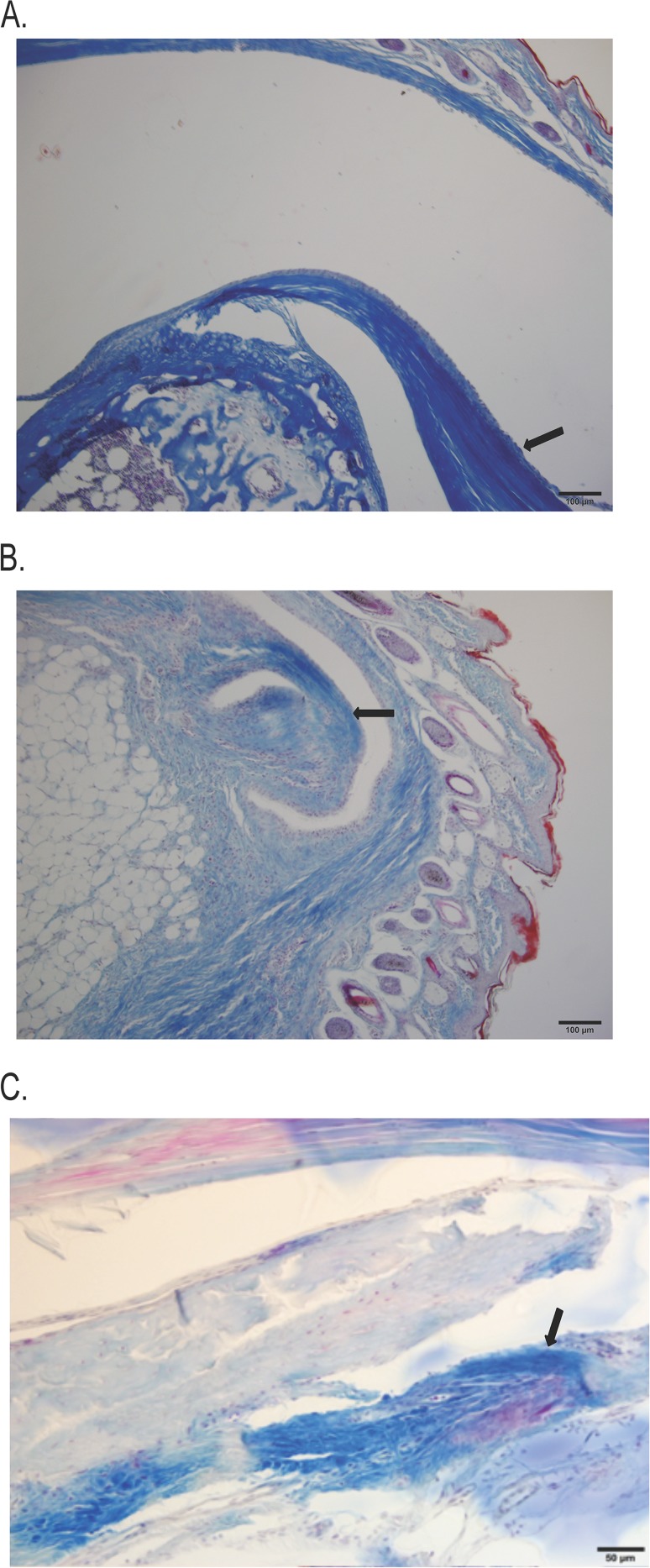
Histological examination of tibiotarsal joints was then performed, for more precise evaluation of arthritis. Tibiotarsal joints were collected after 4 weeks of infection, sectioned, and stained with hematoxylin and eosin. Each section was blindly examined by a veterinary pathologist and was given a score ranging from 0 to 5, with 0 indicating normal tissue and 5 representing the most severe lesions. In mice infected with either the parental strain or the revA-deficient mutant, all sections showed chronic active inflammation with infiltration of plasma cells, lymphocytes, and macrophages, as well as a proliferative response in the periarticular connective tissue (Table 3). However, joints from mice infected with the revA-deficient mutant had lower average histopathology scores than joints from mice infected with the parental strain, B31-A3 (Table 3). Yet Masson's trichome stain for collagen revealed more fibrotic connective tissue deposition in the joints of mice infected with the revA-deficient mutant, extending into and obliterating the entire joint space (Fig. 4B). In contrast, collagen staining was dense and regular, with little fibrosis inside the joint space, in mice infected with either B31-A3 or the revA-complemented strain (Fig. 4A and C). Mice infected with the complemented strain had no demonstrable pathology in the ankle joint. These data suggest that revA deletion results in increased levels of edema and remodeling in the affected joint.
TABLE 3.
Histology scores for tibiotarsal joints
| Strain and inoculuma | No. of mice | Histopathology scoreb |
|---|---|---|
| PBS | 8 | 0.57 ± 0.27 |
| B31-A3 | ||
| 103 | 8 | 4.50 ± 0.26 |
| 104 | 4 | 4.00 ± 0.58 |
| revA mutant | ||
| 103 | 5 | 3.10 ± 0.50 |
| 104 | 7 | 3.30 ± 0.42 |
| revA-complemented strain | ||
| 103 | 3 | 0.33 ± 0.10 |
| 104 | 3 | 0.33 ± 0.10 |
The left footpad of each mouse was injected with B. burgdorferi or the control (PBS).
Each section was given a score ranging from 0 to 5, with 0 indicating normal tissue and 5 representing the most severe lesions. Each data point represents the average score per section ± the standard error of the mean.
FIG 4.
Proliferative response in periarticular connective tissue. Tibiotarsal sections from infected mice were stained for collagen. (A) Section from a B31-A3-infected mouse with patent synovial spaces and clear demarcations of bone and dense, regular connective tissue. (B) Section from a B31-A3ΔrevA1ΔrevA6 mutant-infected mouse with apparent degeneration of bone and infiltration of connective tissue into the joint space. (C) Section from a mouse infected with the complemented strain. Three individual sections from 3 infected animals were observed per infection strain; representative slides are shown. Arrows indicate collagen staining at the edge of the joint space.
Effect of RevA deletion on systemic chemokine production.
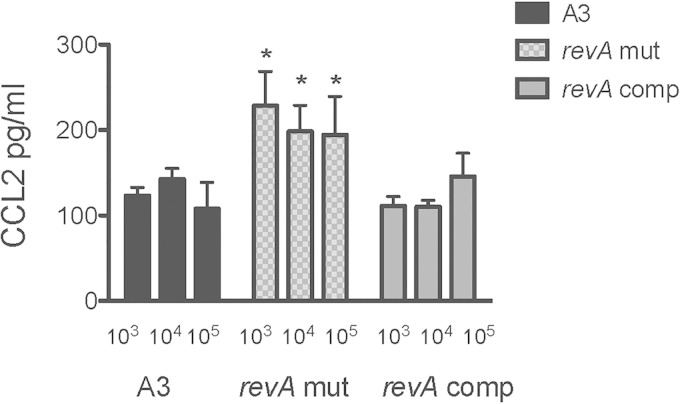
To determine whether the systemic levels of cytokines or chemokines elicited in revA-deficient mutant- and B31-A3-infected mice differed, serum was collected from all revA-deficient mutant- and B31-A3 infected mice at the time of euthanasia. An initial multianalyte screen for cytokines and chemokines suggested differences between the levels of tumor necrosis factor alpha (TNF-α), interleukin 17 (IL-17), and CCL-2 in the sera of animals infected with the RevA-deficient strain and those infected with the wild-type strain (data not shown). ELISAs indicated that the revA-deficient mutant induced significantly higher systemic CCL-2 levels at all inocula than did either the wild-type B31-A3 or the complemented mutant (P = 0.014) (Fig. 5). There were no significant differences in TNF-α or IL-17 induction for any strain.
FIG 5.
The revA-deficient mutant induces higher systemic CCL-2 levels than the wild-type strain. Blood was collected at the time of sacrifice from mice infected with B. burgdorferi (B31-A3, the B31-A3ΔrevA1ΔrevA6 mutant, or the complemented strain; subcutaneous dose of 103, 104, or 105 bacteria). Red blood cells were pelleted, and serum was frozen at −80°C. A commercial sandwich ELISA was used to assay CCL-2 levels according to the manufacturer's instructions (R&D Systems). Samples from 6 to 8 animals per inoculum were analyzed in triplicate. Two-way analysis of variance was used to calculate statistical differences between the groups infected with the parental strain or the B31-A3ΔrevA1ΔrevA6 mutant (P = 0.014).
Acquisition and transmission by ticks.
The potential impacts of RevA deficiency throughout the remainder of the mouse-tick infectious cycle were also examined. First, mice infected with either B31-A3 or B31-A3ΔrevA1ΔrevA6 were fed upon by I. scapularis tick larvae. Immediately after the completion of feeding, cohorts of approximately 30 engorged larvae from each mouse were analyzed by qPCR, according to established procedures (30, 42, 55, 56), for the acquisition of B. burgdorferi. No significant differences in acquisition were detected (Table 4). We then tested the ability of those bacteria to colonize ticks and to be transmitted to mice. Three weeks after molting, ticks were allowed to feed on naïve mice. All naïve mice (6 each for the wild-type strain and B31-A3ΔrevA1ΔrevA6) became infected, as determined by detection of anti-B. burgdorferi IgG by ELISA and by positive culture of hearts, ears, and joints (Table 4). Taken together, these data demonstrate that revA is not required for the acquisition or transmission of B. burgdorferi by ticks.
TABLE 4.
Acquisition and transmission of revA-deficient mutant B. burgdorferi by tick feedinga
| Strain | Acquisition by larvaeb as determined by: |
Transmission from nymphs (no. of culture-positive mouse organs/total no. of organs) | |
|---|---|---|---|
| PCR | ELISA | ||
| B31-A3 | + | 6/6 | 16/18 |
| revA-deficient mutant | + | 6/6 | 15/18 |
B31-A3 and revA-deficient mutants were assessed for their abilities to be transmitted from infected mice to feeding tick larvae. Immediately after the completion of feeding, cohorts of approximately 30 engorged larvae from each mouse were analyzed by qPCR for acquisition of B. burgdorferi. Cohorts of larvae from B31-A3- and B31-A3ΔrevA1ΔrevA6 mutant-infected mice were allowed to molt. Three weeks after the molt, those ticks were allowed to feed on naïve mice. Infection was determined by detection of anti-B. burgdorferi IgG by ELISA and by positive culture of hearts, ears, and joints.
PCR results are shown as positive (+) or negative (−). ELISA results are shown as the number of infected mice/total number of mice.
DISCUSSION
RevA was initially identified as a surface-exposed, immunogenic protein with differential expression patterns that indicated a potential role in mammalian infection (34–36, 55, 57). Indeed, the Rrp2-RpoN-RpoS pathway, involved in the modulation of many mammalian infection-associated genes, may play a role in the upregulation of revA, since both copies in strain B31 were upregulated 16-fold in wild-type B. burgdorferi over expression in an isogenic rrp2 mutant, and the orthologous gene of B. burgdorferi strain 297 was upregulated in wild-type B. burgdorferi over expression in an isogenic rpoS mutant within dialysis membrane chambers (58, 59). Although the function of RevA was unknown, Carroll et al. suggested the possibility that RevA functions as an adhesin (34). We subsequently determined that RevA is a relatively strong fibronectin-binding protein with a calculated Kd (dissociation constant) of 12.5 nM, which also exhibits weak binding affinity for other ECM substrates, including laminin (30). Taken together, the evidence suggests that RevA is poised to interact with the mammalian host and facilitate infection.
The present study determined that the RevA-deficient mutant was less able to colonize heart tissues than was the wild type or the complemented mutant. In fact, complementation of revA, under the control of a strong constitutive promoter, enhanced colonization of the heart. In contrast, no differences were detected in bacterial loads in tibiotarsal joints. These results suggest that interactions between RevA and components of cardiac tissue are beneficial to bacterial infection. Moreover, this apparently tissue-specific effect of RevA echoes the subtle phenotypes seen with mutants of other borrelial adhesins, such as DbpA and BBK32 (22, 24, 25, 60).
There was a pronounced difference in ankle swelling between mice infected with the parental strain and mice infected with the revA-deficient mutant at the lower inoculum. Perhaps paradoxically, the revA-deficient mutant led to lower average histopathology scores for the ankle joint but more collagen deposition. An intriguing possibility is that lack of RevA alters the expression of other adhesins, thus altering the bacterium's tropism to and interaction with the joint tissues. Histopathology scores were based on the numbers of infiltrating cells and the degree of tissue damage. There may be differences in the types of infiltrating cells; indeed, the blinded histology reports indicated the presence of purulent exudates in the joints of revA-deficient mutant-infected mice but not in those of mice infected with the parental strain (data not shown). The composition of the cellular infiltrate and its effect on swelling and collagen deposition remain to be determined.
We also detected differences in the levels of the chemokine CCL-2 in serum, which were higher in mice infected with the revA-deficient mutant than in those infected with the parental strain. This increase was resolved by trans-complementation of revA. CCL-2 (also known as monocyte chemoattractant protein 1 [MCP-1]) is a chemokine that recruits monocytes and dendritic cells to sites of infection or tissue damage, and several studies have demonstrated a role for CCL-2 in Lyme arthritis (61–64). We did not examine chemokine levels in specific tissues, but increased monocyte attraction to joints could account for the differences observed in swelling and fibrosis.
The relative subtlety of the effect of RevA deficiency on infection may be due to the redundancy in adhesins. To date, at least five fibronectin-binding proteins have been identified in B. burgdorferi (28–31), and different adhesins are likely involved in different aspects of the infection process, such as initial attachment or dissemination. For example, overexpression of BBK32 in a nonadherent B. burgdorferi strain enhanced bacterial binding to vascular endothelium in vivo, whereas similar expression of RevA did not measurably alter vascular adhesion, yet BBK32-deficient B. burgdorferi strains exhibit only a slight impairment in mammalian infection (24, 25, 54). The borrelial model is one of redundancy in its host-interacting proteins (e.g., adhesins and complement-regulator binding proteins [19, 65]). Only a few adhesins have been identified as absolutely essential for B. burgdorferi infectivity (66, 67).
It is possible that RevA has additional/other functions in vivo, such as immune evasion. Indeed, the increased CCL-2 levels and the defect in heart colonization seen in B31-A3ΔrevA1ΔrevA6-infected mice could point to differential clearance of the mutant from certain tissues early in the infection process. In contrast, there were no differences in bacterial loads in the tibiotarsal joints of mice infected with the parental strain or the revA-deficient strain. However, the revA-deficient mutant caused increased edema and fibrotic connective tissue deposition in the joint space. Here again, immune evasion could be key; the revA-deficient mutant may be inducing a more pronounced, but tissue-specific, inflammatory response that causes damage to the host while failing to clear the bacteria. Studies of the establishment and early dissemination of bacteria deficient in other adhesins, such as DbpA and BBK32, have hinted at roles in avoiding immune clearance (24, 68). Further research employing bioluminescent whole-body imaging, combined with comprehensive examination of tissue-specific pathology and immune responses, could provide insights into the role of RevA in both dissemination and colonization in the early stages of infection.
ACKNOWLEDGMENTS
This work was supported by NIH/NIAID grant K22AI093671 to C.A.B.
We thank John Lee of the UND School of Medicine and Health Sciences, Medical Media, for expert assistance with graphics. We also thank Ann Flower and John Watt for helpful discussions.
REFERENCES
- 1.Häupl T, Hahn G, Rittig M, Krause A, Schoerner C, Schonherr U, Kalden JR, Burmester GR. 1993. Persistence of Borrelia burgdorferi in ligamentous tissue from a patient with chronic Lyme borreliosis. Arthritis Rheum 36:1621–1626. doi: 10.1002/art.1780361118. [DOI] [PubMed] [Google Scholar]
- 2.Cabello FC, Godfrey HP, Newman SA. 2007. Hidden in plain sight: Borrelia burgdorferi and the extracellular matrix. Trends Microbiol 15:350–354. doi: 10.1016/j.tim.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Franz JK, Fritze O, Rittig M, Keysser G, Priem S, Zacher J, Burmester GR, Krause A. 2001. Insights from a novel three-dimensional in vitro model of Lyme arthritis: standardized analysis of cellular and molecular interactions between Borrelia burgdorferi and synovial explants and fibroblasts. Arthritis Rheum 44:151–162. doi:. [DOI] [PubMed] [Google Scholar]
- 4.Barthold SW, de Souza MS, Janotka JL, Smith AL, Persing DH. 1993. Chronic Lyme borreliosis in the laboratory mouse. Am J Pathol 143:959–972. [PMC free article] [PubMed] [Google Scholar]
- 5.De Koning J, Bosma RB, Hoogkamp-Korstanje JA. 1987. Demonstration of spirochaetes in patients with Lyme disease with a modified silver stain. J Med Microbiol 23:261–267. doi: 10.1099/00222615-23-3-261. [DOI] [PubMed] [Google Scholar]
- 6.Pachner AR, Basta J, Delaney E, Hulinska D. 1995. Localization of Borrelia burgdorferi in murine Lyme borreliosis by electron microscopy. Am J Trop Med Hyg 52:128–133. [DOI] [PubMed] [Google Scholar]
- 7.Barthold SW, Persing DH, Armstrong AL, Peeples RA. 1991. Kinetics of Borrelia burgdorferi dissemination and evolution of disease after intradermal inoculation of mice. Am J Pathol 139:263–273. [PMC free article] [PubMed] [Google Scholar]
- 8.Barthold SW, de Souza M, Fikrig E, Persing DH. 1992. Lyme borreliosis in the laboratory mouse, p 223–242. In Schutzer SE. (ed), Lyme disease: molecular and immunologic approaches. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 9.Barthold SW, Sidman CL, Smith AL. 1992. Lyme borreliosis in genetically resistant and susceptible mice with severe combined immunodeficiency. Am J Trop Med Hyg 47:605–613. [DOI] [PubMed] [Google Scholar]
- 10.Cadavid D, Bai Y, Dail D, Hurd M, Narayan K, Hodzic E, Barthold SW, Pachner AR. 2003. Infection and inflammation in skeletal muscle from nonhuman primates infected with different genospecies of the Lyme disease spirochete Borrelia burgdorferi. Infect Immun 71:7087–7098. doi: 10.1128/IAI.71.12.7087-7098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Defosse DL, Duray PH, Johnson RC. 1992. The NIH-3 immunodeficient mouse is a model for Lyme borreliosis myositis and carditis. Am J Pathol 141:3–10. [PMC free article] [PubMed] [Google Scholar]
- 12.Kornblatt AN, Steere AC, Brownstein DG. 1984. Experimental Lyme disease in rabbits: spirochetes found in erythema migrans and blood. Infect Immun 46:220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coburn J, Medrano M, Cugini C. 2002. Borrelia burgdorferi and its tropisms for adhesion molecules in the joint. Curr Opin Rheumatol 14:394–398. doi: 10.1097/00002281-200207000-00010. [DOI] [PubMed] [Google Scholar]
- 14.Bykowski T, Woodman ME, Cooley AE, Brissette CA, Brade V, Wallich R, Kraiczy P, Stevenson B. 2007. Coordinated expression of Borrelia burgdorferi complement regulator-acquiring surface proteins during the Lyme disease spirochete's mammal-tick infection cycle. Infect Immun 75:4227–4236. doi: 10.1128/IAI.00604-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwan TG, Burgdorfer W, Schrumpf ME, Karstens RH. 1988. The urinary bladder, a consistent source of Borrelia burgdorferi in experimentally infected white-footed mice (Peromyscus leucopus). J Clin Microbiol 26:893–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller JC, von Lackum K, Woodman ME, Stevenson B. 2006. Detection of Borrelia burgdorferi gene expression during mammalian infection using transcriptional fusions that produce green fluorescent protein. Microb Pathog 41:43–47. doi: 10.1016/j.micpath.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Shih C-M, Pollack RJ, Telford SR, Spielman A. 1992. Delayed dissemination of Lyme disease spirochetes from the site of deposition in the skin of mice. J Infect Dis 166:827–831. doi: 10.1093/infdis/166.4.827. [DOI] [PubMed] [Google Scholar]
- 18.Sinsky RJ, Piesman J. 1989. Ear punch biopsy method for detection and isolation of Borrelia burgdorferi from rodents. J Clin Microbiol 27:1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brissette CA, Gaultney RA. 2014. That's my story, and I'm sticking to it—an update on B. burgdorferi adhesins. Front Cell Infect Microbiol 4:41. doi: 10.3389/fcimb.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonara S, Ristow L, Coburn J. 2011. Adhesion mechanisms of Borrelia burgdorferi. Adv Exp Med Biol 715:35–49. doi: 10.1007/978-94-007-0940-9_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ristow LC, Bonde M, Lin YP, Sato H, Curtis M, Geissler E, Hahn BL, Fang J, Wilcox DA, Leong JM, Bergstrom S, Coburn J. 2015. Integrin binding by Borrelia burgdorferi P66 facilitates dissemination but is not required for infectivity. Cell Microbiol doi: 10.1111/cmi.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin YP, Chen Q, Ritchie JA, Dufour NP, Fischer JR, Coburn J, Leong JM. 2015. Glycosaminoglycan binding by Borrelia burgdorferi adhesin BBK32 specifically and uniquely promotes joint colonization. Cell Microbiol doi: 10.1111/cmi.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coburn J, Leong J, Chaconas G. 2013. Illuminating the roles of the Borrelia burgdorferi adhesins. Trends Microbiol 21:372–379. doi: 10.1016/j.tim.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hyde JA, Weening EH, Chang M, Trzeciakowski JP, Hook M, Cirillo JD, Skare JT. 2011. Bioluminescent imaging of Borrelia burgdorferi in vivo demonstrates that the fibronectin-binding protein BBK32 is required for optimal infectivity. Mol Microbiol 82:99–113. doi: 10.1111/j.1365-2958.2011.07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seshu J, Esteve-Gassent MD, Labandeira-Rey M, Kim JH, Trzeciakowski JP, Hook M, Skare JT. 2006. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol Microbiol 59:1591–1601. doi: 10.1111/j.1365-2958.2005.05042.x. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y, Xu Q, McShan K, Liang FT. 2008. Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infect Immun 76:1239–1246. doi: 10.1128/IAI.00897-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortune DE, Lin YP, Deka RK, Groshong AM, Moore BP, Hagman KE, Leong JM, Tomchick DR, Blevins JS. 2014. Identification of lysine residues in the Borrelia burgdorferi DbpA adhesin required for murine infection. Infect Immun 82:3186–3198. doi: 10.1128/IAI.02036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaultney RA, Gonzalez T, Floden AM, Brissette CA. 2013. BB0347, from the Lyme disease spirochete Borrelia burgdorferi, is surface exposed and interacts with the CS1 heparin-binding domain of human fibronectin. PLoS One 8:e75643. doi: 10.1371/journal.pone.0075643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallström T, Haupt K, Kraiczy P, Hortschansky P, Wallich R, Skerka C, Zipfel PF. 2010. Complement regulator-acquiring surface protein 1 of Borrelia burgdorferi binds to human bone morphogenic protein 2, several extracellular matrix proteins, and plasminogen. J Infect Dis 202:490–498. doi: 10.1086/653825. [DOI] [PubMed] [Google Scholar]
- 30.Brissette CA, Bykowski T, Cooley AE, Bowman A, Stevenson B. 2009. Borrelia burgdorferi RevA antigen binds host fibronectin. Infect Immun 77:2802–2812. doi: 10.1128/IAI.00227-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Probert WS, Johnson BJB. 1998. Identification of a 47 kDA fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol Microbiol 30:1003–1015. doi: 10.1046/j.1365-2958.1998.01127.x. [DOI] [PubMed] [Google Scholar]
- 32.Moriarty TJ, Shi M, Lin YP, Ebady R, Zhou H, Odisho T, Hardy PO, Salman-Dilgimen A, Wu J, Weening EH, Skare JT, Kubes P, Leong J, Chaconas G. 2012. Vascular binding of a pathogen under shear force through mechanistically distinct sequential interactions with host macromolecules. Mol Microbiol 86:1116–1131. doi: 10.1111/mmi.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porcella SF, Popova TG, Akins DR, Li M, Radolf JD, Norgard MV. 1996. Borrelia burgdorferi supercoiled plasmids encode multi-copy tandem open reading frames and a lipoprotein gene family. J Bacteriol 178:3293–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carroll JA, El-Hage N, Miller JC, Babb K, Stevenson B. 2001. Borrelia burgdorferi RevA antigen is a surface-exposed outer membrane protein whose expression is regulated in response to environmental temperature and pH. Infect Immun 69:5286–5293. doi: 10.1128/IAI.69.9.5286-5293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilmore RD Jr, Mbow ML. 1998. A monoclonal antibody generated by antigen inoculation via tick bite is reactive to the Borrelia burgdorferi Rev protein, a member of the 2.9 gene family locus. Infect Immun 66:980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mbow ML, Gilmore RD Jr, Stevenson B, Golde WT, Piesman J, Johnson BJB. 2002. Borrelia burgdorferi-specific monoclonal antibodies derived from mice primed with Lyme disease spirochete-infected Ixodes scapularis ticks. Hybrid Hybridomics 21:179–182. doi: 10.1089/153685902760173890. [DOI] [PubMed] [Google Scholar]
- 37.Brissette CA, Rossmann E, Bowman A, Cooley AE, Riley SP, Hunfeld KP, Bechtel M, Kraiczy P, Stevenson B. 2010. The borrelial fibronectin-binding protein RevA is an early antigen of human Lyme disease. Clin Vaccine Immunol 17:274–280. doi: 10.1128/CVI.00437-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Floden AM, Gonzalez T, Gaultney RA, Brissette CA. 2013. Evaluation of RevA, a fibronectin-binding protein of Borrelia burgdorferi, as a potential vaccine candidate for Lyme disease. Clin Vaccine Immunol 20:892–899. doi: 10.1128/CVI.00758-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin T, Gao L, Zhang C, Odeh E, Jacobs MB, Coutte L, Chaconas G, Philipp MT, Norris SJ. 2012. Analysis of an ordered, comprehensive STM mutant library in infectious Borrelia burgdorferi: insights into the genes required for mouse infectivity. PLoS One 7:e47532. doi: 10.1371/journal.pone.0047532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser C. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 41.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb J-F, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidmann J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 42.Miller JC, von Lackum K, Babb K, McAlister JD, Stevenson B. 2003. Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle. Infect Immun 71:6943–6952. doi: 10.1128/IAI.71.12.6943-6952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zückert WR. 2007. Laboratory maintenance of Borrelia burgdorferi. Curr Protoc Microbiol; Chapter 12:Unit 12C.1. doi: 10.1002/9780471729259.mc12c01s4. [DOI] [PubMed] [Google Scholar]
- 44.Bunikis I, Kutschan-Bunikis S, Bonde M, Bergstrom S. 2011. Multiplex PCR as a tool for validating plasmid content of Borrelia burgdorferi. J Microbiol Methods 86:243–247. doi: 10.1016/j.mimet.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Tilly K, Elias AF, Bono JL, Stewart P, Rosa P. 2000. DNA exchange and insertional inactivation in spirochetes. J Mol Microbiol Biotechnol 2:433–442. [PubMed] [Google Scholar]
- 46.Stewart PE, Thalken R, Bono JL, Rosa P. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol 39:714–721. doi: 10.1046/j.1365-2958.2001.02256.x. [DOI] [PubMed] [Google Scholar]
- 47.Ma Y, Seiler KP, Eichwald EJ, Weis JH, Teuscher C, Weis JJ. 1998. Distinct characteristics of resistance to Borrelia burgdorferi-induced arthritis in C57BL/6N mice. Infect Immun 66:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jutras BL, Liu Z, Brissette CA. 2010. Simultaneous isolation of Ixodidae and bacterial (Borrelia spp.) genomic DNA. Curr Protoc Microbiol Chapter 1:Unit 1E.2. doi: 10.1002/9780471729259.mc01e02s19. [DOI] [PubMed] [Google Scholar]
- 49.Ornstein K, Barbour AG. 2006. A reverse transcriptase-polymerase chain reaction assay of Borrelia burgdorferi 16S rRNA for highly sensitive quantification of pathogen load in a vector. Vector Borne Zoonotic Dis 6:103–112. doi: 10.1089/vbz.2006.6.103. [DOI] [PubMed] [Google Scholar]
- 50.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg (Lond) 27:493–497. [Google Scholar]
- 51.Purser JE, Norris SJ. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A 97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purser JE, Lawrenz MB, Caimano MJ, Howell JK, Radolf JD, Norris SJ. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol 48:753–764. doi: 10.1046/j.1365-2958.2003.03452.x. [DOI] [PubMed] [Google Scholar]
- 53.Norris SJ, Howell JK, Garza SA, Ferdows MS, Barbour AG. 1995. High- and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect Immun 63:2206–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X, Liu X, Beck DS, Kantor FS, Fikrig E. 2006. Borrelia burgdorferi lacking BBK32, a fibronectin-binding protein, retains full pathogenicity. Infect Immun 74:3305–3313. doi: 10.1128/IAI.02035-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilmore RD Jr, Mbow ML, Stevenson B. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect 3:799–808. doi: 10.1016/S1286-4579(01)01435-6. [DOI] [PubMed] [Google Scholar]
- 56.Morrison TB, Ma Y, Weis JH, Weis JJ. 1999. Rapid and sensitive quantification of Borrelia burgdorferi-infected mouse tissues by continuous fluorescent monitoring of PCR. J Clin Microbiol 37:987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nowalk AJ, Gilmore RD Jr, Carroll JA. 2006. Serologic proteome analysis of Borrelia burgdorferi membrane-associated proteins. Infect Immun 74:3864–3873. doi: 10.1128/IAI.00189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boardman BK, He M, Ouyang Z, Xu H, Pang X, Yang XF. 2008. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect Immun 76:3844–3853. doi: 10.1128/IAI.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol 65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weening EH, Parveen N, Trzeciakowski JP, Leong JM, Hook M, Skare JT. 2008. Borrelia burgdorferi lacking DbpBA exhibits an early survival defect during experimental infection. Infect Immun 76:5694–5705. doi: 10.1128/IAI.00690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strle K, Jones KL, Drouin EE, Li X, Steere AC. 2011. Borrelia burgdorferi RST1 (OspC type A) genotype is associated with greater inflammation and more severe Lyme disease. Am J Pathol 178:2726–2739. doi: 10.1016/j.ajpath.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strle K, Shin JJ, Glickstein LJ, Steere AC. 2012. Association of a Toll-like receptor 1 polymorphism with heightened Th1 inflammatory responses and antibiotic-refractory Lyme arthritis. Arthritis Rheum 64:1497–1507. doi: 10.1002/art.34383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Codolo G, Bossi F, Durigutto P, Bella CD, Fischetti F, Amedei A, Tedesco F, D'Elios S, Cimmino M, Micheletti A, Cassatella MA, D'Elios MM, de Bernard M. 2013. Orchestration of inflammation and adaptive immunity in Borrelia burgdorferi-induced arthritis by neutrophil-activating protein A. Arthritis Rheum 65:1232–1242. doi: 10.1002/art.37875. [DOI] [PubMed] [Google Scholar]
- 64.Brown CR, Blaho VA, Loiacono CM. 2003. Susceptibility to experimental Lyme arthritis correlates with KC and monocyte chemoattractant protein-1 production in joints and requires neutrophil recruitment via CXCR2. J Immunol 171:893–901. doi: 10.4049/jimmunol.171.2.893. [DOI] [PubMed] [Google Scholar]
- 65.Kraiczy P, Stevenson B. 2013. Complement regulator-acquiring surface proteins of Borrelia burgdorferi: structure, function and regulation of gene expression. Ticks Tick Borne Dis 4:26–34. doi: 10.1016/j.ttbdis.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhi H, Weening EH, Barbu EM, Hyde JA, Hook M, Skare JT. 2015. The BBA33 lipoprotein binds collagen and impacts Borrelia burgdorferi pathogenesis. Mol Microbiol doi: 10.1111/mmi.12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ristow LC, Miller HE, Padmore LJ, Chettri R, Salzman N, Caimano MJ, Rosa PA, Coburn J. 2012. The β3-integrin ligand of Borrelia burgdorferi is critical for infection of mice but not ticks. Mol Microbiol 85:1105–1118. doi: 10.1111/j.1365-2958.2012.08160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Imai DM, Samuels DS, Feng S, Hodzic E, Olsen K, Barthold SW. 2013. The early dissemination defect attributed to disruption of decorin-binding proteins is abolished in chronic murine Lyme borreliosis. Infect Immun 81:1663–1673. doi: 10.1128/IAI.01359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]