Abstract
Avian pathogenic Escherichia coli (APEC) strains cause one of the three most significant infectious diseases in the poultry industry and are also potential food-borne pathogens threating human health. In this study, we showed that ArcA (aerobic respiratory control), a global regulator important for E. coli's adaptation from anaerobic to aerobic conditions and control of that bacterium's enzymatic defenses against reactive oxygen species (ROS), is involved in the virulence of APEC. Deletion of arcA significantly attenuates the virulence of APEC in the duck model. Transcriptome sequencing (RNA-Seq) analyses comparing the APEC wild type and the arcA mutant indicate that ArcA regulates the expression of 129 genes, including genes involved in citrate transport and metabolism, flagellum synthesis, and chemotaxis. Further investigations revealed that citCEFXG contributed to APEC's microaerobic growth at the lag and log phases when cultured in duck serum and that ArcA played a dual role in the control of citrate metabolism and transportation. In addition, deletion of flagellar genes motA and motB and chemotaxis gene cheA significantly attenuated the virulence of APEC, and ArcA was shown to directly regulate the expression of motA, motB, and cheA. The combined results indicate that ArcA controls metabolism, chemotaxis, and motility contributing to the pathogenicity of APEC.
INTRODUCTION
Commensal and pathogenic Escherichia coli strains can grow under both aerobic and anaerobic conditions. The enzymes required for catabolism under aerobic versus anaerobic conditions are substantially different, and E. coli has the ability to switch among anaerobic, anaerobic respiratory, and fermentative pathways (1). The expression of genes involved in cellular functions such as nutrient uptake and/or excretion systems, biosynthetic pathways, and macromolecular synthesis are also adjusted in response to oxygen availability (2). The Arc two-component signal transduction system, comprised of the kinase sensor ArcB and its cognate response regulator ArcA, is one of the mechanisms that enable E. coli adaptation to changing oxygen availability (3). Once an environmental signal is received, ArcB undergoes autophosphorylation and catalyzes the transphosphorylation of ArcA, which then promotes or represses expression of Arc-regulated genes. Rather than directly detecting environmental O2, ArcB probably senses the redox state of the cell through detection of a reduced electron transport component (4). Under aerobic conditions (5), oxidized forms of quinone electron carriers in the membrane inhibit the autophosphorylation of ArcB, while under anaerobic and microaerobic conditions, ArcB undergoes autophosphorylation. The net activity of ArcB as a kinase for ArcA is expected to progressively increase during the transition from aerobic to anaerobic growth.
The ArcA regulon of E. coli K-12 strains has been extensively studied under both aerobic and anaerobic conditions (6, 7). A previous study indicated that about 1,139 genes in the E. coli K-12 genome are regulated either directly or indirectly by the ArcA protein (6). Under anaerobic conditions, ArcA inhibits the expression of genes required for aerobic metabolism, energy generation, amino acid transport, and fatty acid transport. In particular, the genes involved in the tricarboxylic acid cycle and cytochrome o ubiquinol oxidation were repressed by ArcA under anaerobic conditions (8). ArcA also functions as an activator. Transcription of about 93 genes was positively regulated by ArcA in the absence of oxygen: most of these genes contribute to the anaerobic respiratory and metabolism pathways (6).
A second transcription factor involved in controlling anaerobic gene expression and facilitating bacterial adaptation to anaerobic conditions is FNR (fumarate and nitrate respiration) (9). A comparison of the ArcA and FNR regulons showed that 303 genes were regulated by both proteins (6). Our recent study showed that FNR is a key global regulator of ExPEC virulence, controlling expression of several important virulence traits. These include motility, adherence, invasion, modulation of hemolysin expression, and expression of a novel pathogenicity island involved in α-ketoglutarate metabolism under anaerobic conditions (10).
Based on the similar roles of ArcA and FNR in bacterial adaptation to anaerobic conditions, we propose that ArcA should also be important for pathogenic E. coli's virulence. However, the regulon of ArcA in pathogenic E. coli and its role in pathogenesis have never been explored. To address these questions, we constructed and characterized an arcA mutant and a complemented strain of an avian pathogenic E. coli (APEC) strain. The genes regulated by ArcA in duck serum were identified by use of transcriptome sequencing (RNA-Seq) analyses. A total of 129 genes were significantly differentially expressed between the wild type (WT) and the arcA mutant, and ArcA selectively regulated genes required for the metabolism, motility, and chemotaxis contributing to APEC virulence.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
The APEC strain XM (O2:K1:H7), belonging to the phylogenetic E. coli reference (ECOR) group B2, was isolated from the brain of a duck which had septicemia and neurological symptoms (11). The strain was also found to cause serious meningitis in a neonatal rat model of human disease. The strains and plasmids used in this study are listed in Table 1. Microaerobic growth was achieved by static culture at 37°C with 5% oxygen. For genetic manipulations, all E. coli strains were routinely grown in Luria-Bertani (LB) broth medium. Selective antibiotics and IPTG (isopropyl-β-d-thiogalactopyranoside) were added when necessary at the following concentrations: ampicillin (Amp), 100 μg · ml−1; kanamycin (Kan), 50 μg · ml−1; chloramphenicol (Chl), 25 μg · ml−1; and IPTG, 0.1 mM (11).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | Plasmid propagation strain | Invitrogen |
| APEC XM | Brain isolate from a duck with septicemia and neurological symptoms | 11 |
| arcA mutant | APEC XM ΔarcA | This study |
| cheA mutant | APEC XM ΔcheA | This study |
| cheAB mutant | APEC XM ΔcheAW | This study |
| motAB mutant | APEC XM ΔmotAB | This study |
| citCEFXG mutant | APEC XM ΔcitCEFXG | This study |
| flgBCDEFGHIJKL mutant | APEC XM ΔflgBCDEFGHIJKL | This study |
| fliCD mutant | APEC XM ΔfliCD | This study |
| Plasmids | ||
| pET28a-arcA | Expression plasmid for ArcA protein | 46 |
| pGEN-MCS | Low-copy-no. plasmid for complementation | 17 |
| pGEM-arcA | pGEN-MCS carrying arcA coding region and 500-bp upstream promoter region | This study |
| pKD3 | Template for λ-Red Chlr cassette | 13 |
| pCP20 | Encodes FLP recombinase for removal of resistance cassette | 13 |
| pKD46 | λ-Red recombinase expression | 13 |
Recombinant DNA techniques.
PCR, DNA ligation, electroporation, and DNA gel electrophoresis were performed as described by Sambrook and Russell (12), unless otherwise indicated. All oligonucleotide primers were purchased from BGI Tech Solutions Co., Ltd. (BGI, Guangzhou, China), and are listed in Table S1 in the supplemental material. All restriction and DNA-modifying enzymes were purchased from TaKaRa Biotechnology Co., Ltd. (Dalian, China), and used on the basis of the supplier's recommendations. Recombinant plasmids, PCR products, and restriction fragments were purified using TaKaRa MiniBEST plasmid purification kits or agarose gel extraction kits (Shiga, Japan) as recommended by the supplier. DNA sequencing was performed at the DNA facility, Shanghai Sunny Biotechnology Co., Ltd.
Deletion mutants were constructed using the bacteriophage lambda red recombinase system described by Datsenko and Wanner (13–15), and primers used for mutagenesis are listed in Table S1 in the supplemental material. Chromosomal transcriptional arcA fusions were performed using the XM strain. The homologous recombination constructions used PCR-purified products with a selectable antibiotic resistance gene and 60-nucleotide (nt) homology extensions. The mutants were confirmed by PCR and sequencing. For complementation, the coding sequences of genes plus their putative promoter regions were amplified (the primers used are listed in Table S1 in the supplemental material) from the XM strain and independently cloned into pGEN-MCS (16, 17) using NdeI and BamHI restriction sites.
To construct the plasmid overproducing pET28a(+)-arcA-His6 fusion protein, a 717-bp fragment containing the coding region of arcA was obtained by PCR from genomic DNA using arcAHistag-F and arcAHistag-R (see Table S1 in the supplemental material), carrying codons for 6×His, and subsequently cloned into pET28a(+) vectors (Novagen, Madison, WI, USA) using BamHI and HindIII sites. The resultant plasmid contains pet28a-arcA-His6 under the control of the T7 promoter.
RNA isolation, rRNA removal, sequencing library preparation, and sequencing.
RNAs from the WT and arcA mutant cultured in duck serum and RNAs from the WT and arcA mutant grown in LB broth were extracted using an RNeasy minikit (Promega) with a 15-min on-column DNase digestion (Promega) according to the manufacturer's instructions. Three biological replicates were used for each combination of genotype (WT or arcA mutant) and medium (duck serum or LB broth). The extracted RNA was quantified by the use of a Qubit 2.0 spectrophotometer (Life Technologies, Carlsbad, CA) to estimate concentrations and then stored at −80°C until use. The cDNA libraries were constructed from 1,000 ng of total RNA, and rRNA was removed using the MICROBExpress bacterial mRNA enrichment kit (Life Technologies, Carlsbad, CA). The sequencing libraries were prepared using a TruSeq Stranded total RNA sample preparation kit (Illumina, San Diego, CA) following the guidelines of the manufacturer. Multiplex libraries were prepared using barcoded primers and a median insert size of 250 bp. Libraries were analyzed for size distribution using a Bioanalyzer and quantified by quantitative reverse transcription-PCR (RT-PCR) using a Kapa library quantification kit (Kapa Biosystems, Boston, MA), and relative volumes were pooled accordingly. The pooled libraries were sequenced on an Illumina HiSeq platform with 100-bp end reads following standard Illumina protocols at BGI.
Data analyses.
Since the genome of APEC XM was not fully sequenced, the RNA sequencing data were mapped to the genome sequences of APEC O1 (1) using Bowtie (18). The mapped reads were counted by eXpress (http://bio.math.berkeley.edu/eXpress/). Genes with an average of at least one uniquely mapped read across samples and at least four nonzero read counts were tested for differential expression between genotypes within each growth medium, using the R package QuasiSeq (http://cran.r-project.org/web/packages/QuasiSeq). The negative binomial QLShrink method implemented in the QuasiSeq package and described previously (19) was used to compute a P value for each gene and each genotype comparison. The log of each count mean was modeled as the sum of an intercept term, a genotype effect, a growth medium effect, an interaction between genotype and growth medium, and an offset normalization factor, determined for each sample by the log of the TMM normalization factor (20). Estimates of the fold change between the different growth media and between genotypes were computed by evaluating the exponential function at estimates of effect differences. Using the P values for each comparison, the approach of Nettleton et al. (21) was used to estimate the number of genes with true null hypotheses among all genes tested, and this estimate was used to convert the P values to q values (22).To control the false-discovery rate at 10%, comparisons with q values no larger than 0.10 and estimated fold changes of at least 2 were declared significant.
RT-PCR and quantitative real-time RT-PCR.
For the cotranscription test, one microgram of total RNA was reverse transcribed in triplicate using random hexamers and ImProm-II reverse transcriptase (Promega). Reactions without reverse transcriptase were used as a DNA contamination control. cDNA was then used as the template for subsequent PCRs.
Real-time quantitative RT-PCR (qPCR) was used to validate the expression levels for selected genes. Primers are listed in Table S1 in the supplemental material. Total RNAs from the APEC XM WT and arcA mutant, cultured in LB broth and serum, were extracted as described above. Treatment of total RNA with DNase was performed, followed by purification using the RNeasy kit. RNA samples without reverse transcription served as PCR templates to confirm that they were free of contaminating DNA. One microgram of total RNA was reverse transcribed in triplicate using random hexamers and Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega). The primer pairs used are listed in Table S1 in the supplemental material, and real-time PCR was performed as previously described (23, 24). Melting-curve analyses were performed after each reaction to ensure amplification specificity. Differences (n-fold) in transcripts were calculated using the relative comparison method, and amplification efficacies of each primer set were verified as described by Schmittgen et al. (25). RNA levels were normalized using the housekeeping gene for the replication terminator protein Tus as a control (23, 24).
EMSA.
E. coli BL21 with pET28a-arcA was grown in 200 ml of LB medium for 16 h at 28°C, and protein expression was induced by adding 0.1 mM IPTG. The ArcA-His6 fusion protein was purified to homogeneity using Ni-nitrilotriacetic acid spin columns and dialyzed in buffer (20 mM Tris, 50 mM NaCl, 40 mM EDTA, 4 mM dithiothreitol [DTT], 10% glycerol, pH 7.4) at 4°C overnight with three buffer changes. To study the binding of ArcA to the DNA probe, electrophoretic mobility shift assays (EMSAs) were performed as described previously. Briefly, DNA probes were amplified using specific primers and purified using a TaKaRa MiniBEST gel extraction kit. EMSAs were performed by adding increasing amounts of purified ArcA-His6 fusion protein (00 to 3,000 ng) to the DNA probe (50 ng) in binding buffer (10 mM Tris-HCl, 7 mM MgCl2, 50% glycerol, 40 μg/ml bovine serum albumin, pH 8.0) for 45 min at room temperature. The reaction mixtures were then subjected to electrophoresis on a 6% polyacrylamide gel in 0.5× TBE buffer (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA, pH 8.0) at 200 V for 45 min. The gel was stained in 0.5× TBE buffer containing 1× SYBR Gold nucleic acid staining solution (Life Technologies, Grand Island, NY) for 30 min, and then the image was recorded.
Animal tests.
Healthy 7-day-old ducks testing negative for antibodies to APEC XM were used in this study. All the animals were allowed free access to sterile water and animal feed and were handled according to the guidelines of the Laboratory Animal Monitoring Committee of Jiangsu Province. Seven-day-old ducks were divided into different groups according to the experiment, and mortalities were recorded by intraperitoneal challenge with various doses of viable bacteria. Strains were grown in LB medium at 37°C to the logarithmic phase, diluted as required in sterile phosphate-buffered saline (PBS), and given 200 μl of various doses of inoculum by the intraperitoneal route. Eight ducks were used per dose. The ducks were observed for 7 days, and mortality rates were calculated. Fisher's exact tests at a significance level of 0.05 were used to identify statistically significant differences between estimated mortality rates.
RNA-Seq data accession number.
The RNA-Seq data are available at the European Bioinformatics Institute ArrayExpress-functional genomics database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-3396.
RESULTS
Deletion of arcA significantly attenuates virulence of APEC in the duck model.
To address the role of ArcA in APEC virulence, we compared the APEC XM wild type with its isogenic arcA deletion mutant strain for its ability to grow in vitro (LB) under aerobic and anaerobic conditions. The arcA mutant grew only slightly worse than the WT under aerobic and anaerobic conditions (see Fig. S1 in the supplemental material). The virulences of the WT and mutant strains were then compared using a 7-day-old duck model. The WT and mutant strains were inoculated into eight ducks with inoculation doses of 5 × 106, 1 × 106, 5 × 105, and 1 × 105 CFU. No significant difference (P > 0.05) in mortality between the WT and mutant strains was observed in the groups with higher inoculation doses. However, the WT strain killed significantly (P < 0.05) more ducks than the arcA mutant when the inoculation doses were lower (Table 2), and deletion of arcA resulted significant decrease (P < 0.05) of the bacterial burden in the brain tissue (see Fig. S2 in the supplemental material). Specifically, for inoculation doses of 5 × 105 and 1 × 105 CFU, the WT strain killed 7 and 6 ducks, respectively, while no death of ducks was observed in the groups infected with the arcA mutant.
TABLE 2.
Mortality rates among ducks inoculated with different strains
| Challenge dose (CFU) | No. of dead animals/total no. tested after inoculation with: |
||
|---|---|---|---|
| WT | arcA mutant | arcA mutant complementation strain | |
| 5 × 106 | 8/8 | 7/8 | 8/8 |
| 1 × 106 | 8/8 | 4/8 | 6/8 |
| 5 × 105 | 7/8 | 0/8a | 6/8 |
| 1 × 105 | 6/8 | 0/8a | 5/8 |
Estimated mortality rate significantly (P < 0.05 by Fisher's test) lower than other estimated mortality rates within a dose.
To verify that this impact on mortality was not due to an unwanted mutation, a stable low-copy-number plasmid, pGEN-MCS, which had been successfully used for an in vivo complementation assay (10, 16, 17), was used to construct a mutant complementation strain. To do this, the coding region of arcA plus its predicted promoter region was cloned into pGEN-MCS, and the resultant plasmid was transformed into the arcA mutant. As shown in Table 1, the reintroduction of arcA back into the mutants restored duck mortality to WT levels (Table 2). These results demonstrate that the arcA mutation significantly attenuates virulence in the duck model.
Identification of genes differentially expressed between WT APEC and arcA mutants.
To further understand the mechanisms by which ArcA contributes to APEC pathogenicity, we used RNA-Seq to compare the transcriptomes of the APEC XM WT strain and its arcA mutant in duck serum and in LB broth. Results are reported here for the WT versus arcA mutant comparison in duck serum. Results for this same genotype comparison in LB broth will be reported elsewhere. The normal values of pO2 in the microenvironments of various tissues range from 1 to 10% oxygen (26). We used culture conditions of 5% oxygen and static growth in duck serum to imitate in vivo oxygen tension. Overall, 129 genes were significantly differentially expressed between the WT and the arcA mutant, using a threshold false-discovery rate of <10%, comparisons with q values of <0.10, and estimated fold changes of at least two. Eighty-one genes were significantly upregulated and 48 significantly downregulated in the arcA mutant compared to the WT (see Table S2 in the supplemental material). In contrast to the effects of deletion of the FNR global regulator in uropathogenic E. coli (UPEC) strain CFT073, which regulates the expression of several important virulence factors of UPEC (10), we did not find many well-known virulence factors of APEC regulated by ArcA, such as iron acquisition systems (aerobactin, SitABCD, salmochelin, yersiniabactin, and ChuA system), temperature-sensitive hemagglutinin (Tsh), vacuolating autotransporter toxin (Vat), and virulence traits involved in resistance to the bactericidal effects of serum and phagocytosis (outer membrane protein A, Iss protein, lipopolysaccharide [LPS], and capsule).
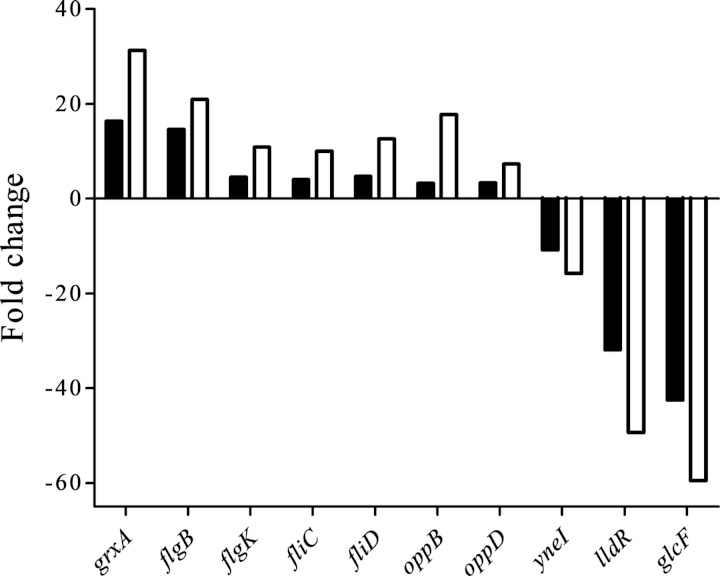
To confirm the results from the RNA sequencing analyses, real-time quantitative reverse transcription-PCR was performed on the 10 genes listed in Table S2 in the supplemental material. The expression of 7 of these genes was upregulated, and that of the other 3 was downregulated, according to RNA sequencing analyses. The qPCR results were consistent with the results obtained from the RNA-Seq analysis (Fig. 1). However, for all genes, the qPCR results showed fold changes greater than those seen with the corresponding RNA-Seq analysis. Therefore, results of the qPCR analysis provided evidence that the transcriptional data obtained from RNA-Seq were reliable.
FIG 1.
Comparison of gene regulation analyzed by RNA sequencing analysis or real-time quantitative RT-PCR. Real-time quantitative RT-PCR (open bars) was used to validate the expression level change for 10 selected genes, including 7 upregulated genes and 3 downregulated genes, revealed by RNA-Seq analysis (filled bars).
Genes involved in flagellum synthesis contribute to APEC virulence.
The E. coli flagellum consists of six components, i.e., a basal body, a motor, a switch, a hook, a filament, and an export apparatus (27), and we found in this study that several categories of genes involved in flagellum synthesis were significantly downregulated in the arcA mutant compared to the WT strain. These downregulated genes in the arcA mutant include (see Table S2 in the supplemental material) (i) the motA and motB genes, which are involved in the synthesis of the motor; (ii) flgB, flgC, and flgG, which contribute to the synthesis of the rod (basal body); (iii) flgE and flgK, which encode genes that specify proteins to form the hook and the hook-filament junction; and (iv) fliC and fliD, which encode the filament and hook-capping protein, respectively. Interestingly, all genes that had lower transcript levels in the arcA mutant than in the WT strain belong to the middle (class 2) and late (class 3) flagellar genes, while there was no significant difference in the transcript levels of the early flagellar genes (class 1) flhD and flhC, whose gene products FlhD/FlhC are the master regulators of flagellar biosynthesis.
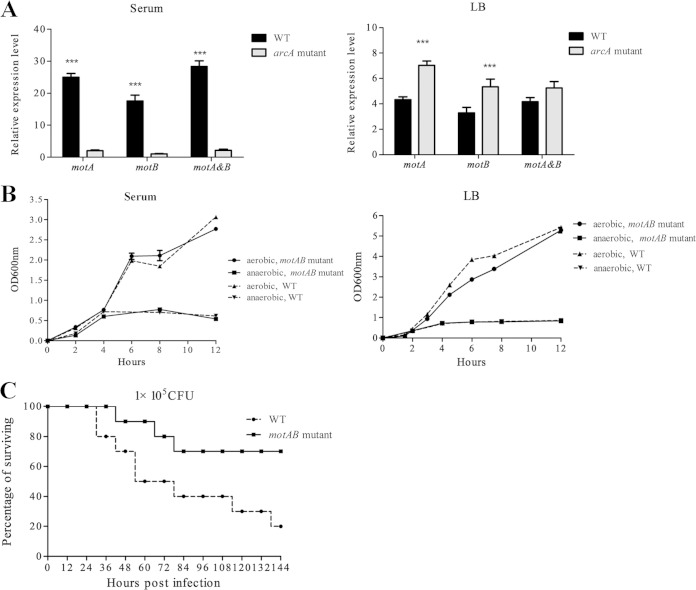
Since the role of motA and motB in APEC is unknown, we further compared the virulence of a mutant with these double mutations to that of the WT. This mutant grew as well as the WT strain (no significant difference) in LB and serum under both aerobic and microaerobic conditions, and it demonstrated significant attenuation (P < 0.05) of virulence compared to the WT, with significantly lower mortality and longer survival time for those ducks that eventually died due to treatment, suggesting that these genes contributes to the pathogenicity of APEC (Fig. 2).
FIG 2.
MotA and MotB contribute to APEC virulence. (A) The expression of motA and motB was significantly downregulated in the arcA mutant in response to duck serum under microaerobic (5% oxygen) conditions. (B) Deletion of motAB did not affect the growth of APEC XM in either LB or serum under either aerobic or microaerobic conditions. Bacterial strains were grown in LB and serum under both aerobic and microaerobic conditions, and the optical density at 600 nm (OD600) was monitored every 2 h. The experiments were performed at least three times in quadruplicate. (C) Deletion of motAB resulted in attenuation of APEC XM. The motAB mutant and the WT (1 × 105 CFU) were inoculated into 10 7-day-old ducks per group, and the mortality was monitored for 1 week. The virulence of the motAB mutant was significant attenuated compared to that of the WT strain (P < 0.05 by Fisher's exact test.).
citCEFXG contributes to APEC's microaerobic growth at the lag and log phases when cultured in duck serum.
The expression of genes citE and citF was significantly downregulated in the arcA mutant compared to the WT when cultured in serum under microaerobic conditions. Bioinformatics analysis suggested that these genes very likely form one operon, citCEFXG, that contributes to E. coli's utilization of citrate under anoxic conditions. Our reverse transcription-qPCR analyses further confirmed that the operon of cit was downregulated in the arcA mutant. The citC gene encodes a ligase required for the acetylation of the 2-(50-phosphoribosyl)-39-dephosphocoenzyme-A prosthetic group, and the genes citE and citF encode the three subunits of citrate lyase, which splits the citrate to acetate and oxaloacetate. CitX was confirmed to be a citrate transporter (28, 29). The function of CitG is currently unknown, and it presumably is involved in the biosynthesis or the covalent attachment of the prosthetic group.
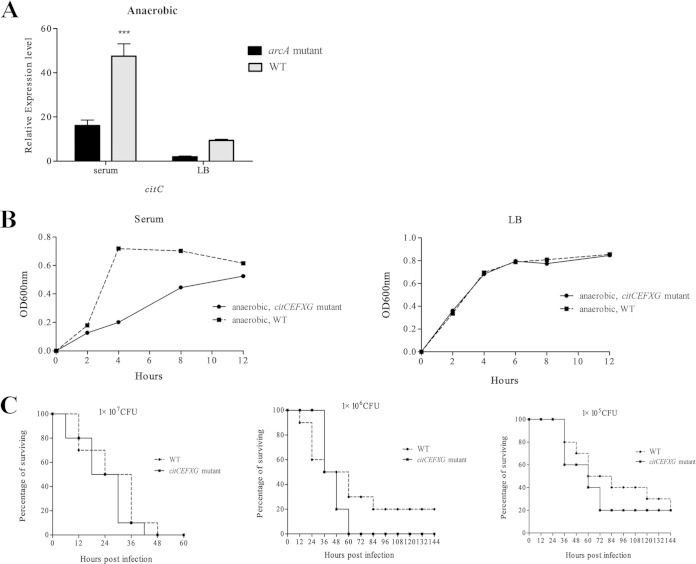
It was reported that the genes citC, citE, citF, citX, and citG were induced in LB culture under anaerobic conditions (29). We deleted these genes from APEC XM to generate the ΔcitCEFXG mutant and compared the growth of the ΔcitCEFXG strain in LB and serum under microaerobic conditions with that of the WT. The ΔcitCEFXG mutant grew slower than the WT during the lag and log phases when cultured in serum, but no significant difference was found when it was cultured in LB. However, the mutant and WT reached the same growth level in the stationary phase when cultured both in LB and in serum. To determine if the genes citCEFXG contribute to the pathogenicity of APEC in the duck model, we inoculated ducks with the ΔcitCEFXG and WT strains; no significant difference in mortality was found between the citCEFXG mutant and the WT (P > 0.05) (Fig. 3). Thus, citCEFXG increases APEC's growth in duck serum under microaerobic conditions, but it is not important for APEC's growth in normal LB medium.
FIG 3.
The expression of the cit operon is regulated by ArcA, and deletion of cit genes decreases the APEC growth at the lag and log phases when cultured in duck serum. (A) Levels of citC expression in the arcA mutant and WT strains. citC is the first gene of the cit operon, and thus its expression was used to represent the expression level of the whole cit operon. Bacteria were grown in serum and LB under microaerobic (5% oxygen) conditions before being subjected to RNA extraction. qRT-PCR expression values are means plus standard deviations (error bars) from at least three independent experiments. Analysis of statistical significance was performed using two-way analysis of variance (ANOVA) (***, P < 0.01). (B) Deletion of citCEFXG retards the growth of APEC XM at the lag and log phases when cultured in duck serum but not in LB under microaerobic conditions. The OD600 was monitored every 2 h, and the experiments were performed at least three times in quadruplicate. (C) Deletion of citCEFXG affects the mortality of APEC XM. Groups of 10 7-day-old ducks were inoculated with 1 × 107, 1 × 106, or 1 × 105 CFU/duck, and the mortality was monitored for 1 week.
Deletion of arcA results in downregulation of the chemosensor CheA.
CheA is the sensor histidine kinase that is necessary for the chemotactic behavior of E. coli (30, 31). Transmembrane methyl-accepting chemoreceptor proteins (MCPs) bind to the chemical attractant and then interact with CheA and its coupling protein, CheW. These interactions result in autophosphorylation of CheA and transphosphorylation of the response regulator CheY. Phospho-CheY interacts with flagellar motor proteins, thus prolonging cell travel in the preferred direction (32). The expression of CheA and its coupling protein CheW is decreased in the ArcA deletion mutant in duck serum under microaerobic conditions. Specifically, CheA was downregulated by 3.86-fold and CheW by 3.66-fold in the arcA mutant compared to the WT strain when these strains were cultured in duck serum under microaerobic conditions.
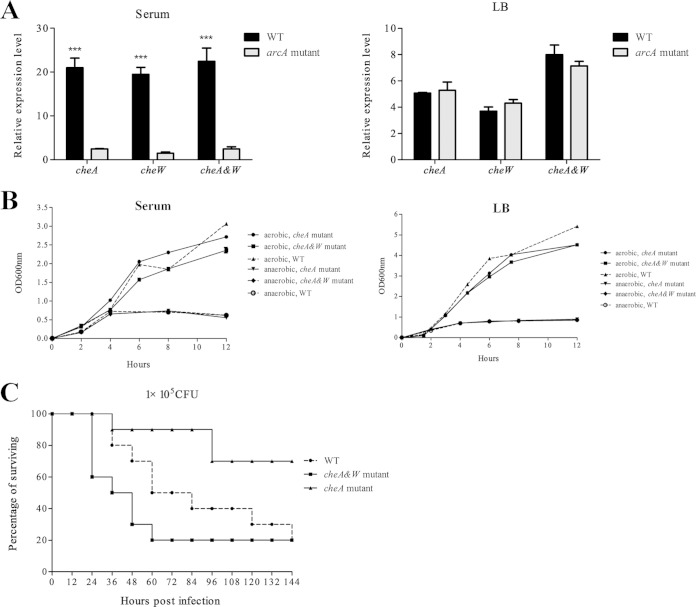
We examined the expression of CheA and CheW in more detail using reverse transcription-qPCR. CheA and CheW expression was evaluated in the arcA and WT strains under aerobic and anaerobic conditions (Fig. 4). Expression of CheA and CheW was much lower in the arcA mutant than in the WT strain when cultured in duck serum under anaerobic conditions. In contrast, if the strains were cultured in LB under aerobic and anaerobic conditions or in duck serum under aerobic conditions, there was no significant difference in expression of the cheA and cheW genes between the arcA mutant and WT strains. Upregulation of cheA and cheW under culture conditions simulating the in vivo environment suggests that CheA and Chew should be important for APEC's virulence.
FIG 4.
The expression of cheA is regulated by ArcA, and cheA contributes to APEC virulence. (A) The expression of cheA and cheW is significantly downregulated in the arcA mutant in response to duck serum but not in response to LB under microaerobic (5% oxygen) conditions. Bacteria were grown in serum and LB under microaerobic conditions before being subjected to RNA extraction. qRT-PCR expression values are means plus standard deviations (error bars) from three independent experiments. Analysis of statistical significance was performed using two-way ANOVA (***, P < 0.01). (B) Deletion of cheA and cheW does not affect the growth of APEC XM in LB or duck serum under aerobic or microaerobic conditions. the OD600 was monitored every 2 h. The experiments were performed at least three times in quadruplicate. (C) Deletion of cheA significantly attenuates the virulence of APEC XM. The cheA mutant, the cheA and cheW double deletion mutant, and the WT strain were inoculated into 10 7-day-old ducks using a dose of 1 × 105 CFU of per bird, and the mortality was monitored for 1 week. The virulence of the cheA mutant is significant attenuated compared to that of the WT strain (P < 0.05 by Fisher's exact test). Deletion of both cheA and cheW recovers virulence to the WT level.
To investigate the role of cheA and cheW in virulence, we generated a mutant with a cheA deletion and a mutant with deletions of cheA and cheW. Both mutants grew similarly to the WT strain in LB medium and serum under aerobic and microaerobic conditions. However, when the three strains were inoculated in the duck model, the virulence of the cheA mutant was significantly attenuated (P < 0.05). With an inoculation dose of 1 × 105 CFU, eight out of 10 ducks inoculated with the WT strain died, while only two of the 10 ducks inoculated with the cheA mutant strain died (P < 0.05), suggesting that cheA increase the virulence of APEC. Surprisingly, the mutant with a double deletion of cheA and cheW was as virulent as the wild type (Fig. 4). Since the complementation test of this mutant was not finished, at this time we do not know if this effect was due to an unwanted mutation.
The regulator ArcA binds to the promoter regions of genes motA, motB, and cheA.
Prediction by bioinformatics tools (Softberry, Mount Kisco, NY) followed by reverse transcription-PCR indicates that motA, motB, and cheA form one operon (Fig. 5A) and thus share one promoter. To determine if ArcA directly regulates the expression of motAB and cheA, an electrophoretic mobility shift assay was performed. The promoter region was predicted by the BProm program (SoftBerry). The potential binding site of ArcA was identified with Patser software (version 3d) (8). DNA fragments containing the potential binding site were then PCR amplified for use as probes. Fragments amplified from the coding region of motA were used as negative controls. As shown in Fig. 5B, the ArcA fusion protein was able to shift the promoter fragment of motA, motB, and cheA but not the control fragment. These results suggest that ArcA directly regulates the expression of these three genes.
FIG 5.
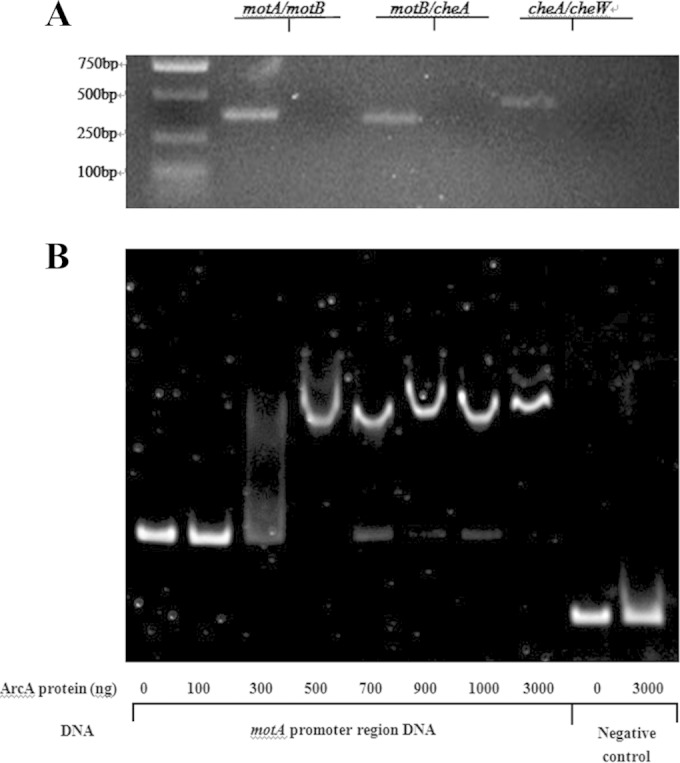
ArcA directly regulates the expression of motA, motB, and cheA. (A) The motA, motB, and cheA genes form one operon, as represented by RT-PCR results using cDNA and a negative control without reverse transcriptase. Primers were designed to span open reading frames (ORFs) motA and motB, motB and cheA, and cheA and cheW. RNA was purified and reverse transcribed to cDNA. The RNA that was not reverse transcribed served as a negative control. (B) Nonradioactive EMSA of binding of ArcA-His6 to the promoter regions of motA, motB, and cheA. PCR products of the motA promoter region were used as probes at 50 ng per reaction mixture. Purified ArcA-His6 fusion protein was added to each reaction mixture at different concentrations, as indicated; the coding-region DNA probes with and without the ArcA protein were used as negative controls. DNA fragments were stained with SYBR green.
DISCUSSION
Bacterial pathogens coordinate the expression of virulence determinants in response to the local microenvironment stimuli of the host, and availability of oxygen has been reported to affect the expression of virulence factors of several microorganisms (33–35). The two-component regulator ArcA is one of the main transcriptional regulators that facilitates the metabolic shift from anaerobic to aerobic conditions and activates the enzymatic defenses of bacteria against reactive oxygen species (ROS) (2, 8). Although the ArcA regulon had been extensively analyzed in nonvirulent E. coli K-12 strains (5–7), the virulence role of ArcA in pathogenic E. coli was still unexplored. Here, we demonstrated that deletion of arcA significantly attenuates the virulence of APEC in a duck model and began to define the virulence-associated genes that are affected by ArcA. To our best knowledge, this is the first report of the virulence mechanisms by which ArcA contributes to APEC pathogenicity.
The expression of over one-third of the genes expressed during growth under aerobic conditions is altered when E. coli cells are transitioned to an anaerobic growth state (6). To identify genes targeted by ArcA during host in vivo infection, we imitated the microenvironment that the APEC would encounter in the host. This included the use of duck serum and of microaerobic conditions (5% oxygen) in order to replicate the oxygen tension thought to occur during host infection (26). Previous studies of genes targeted by the ArcA regulon in E. coli K-12 strain MC4100 have used either aerobic (19.95% oxygen) or completely anaerobic conditions (6, 7). Compared to previous reports, we identified few genes (129 genes, compared to 1,139 in E. coli K-12 strain MC4100) (6) regulated by ArcA in response to duck serum, likely due to the microaerobic conditions that we used or to the different E. coli strain used. We believe that our results more accurately reflect the regulon of ArcA during host infection.
An interesting finding is that not many “well-known” virulence factors were targeted by ArcA under the in vivo imitation conditions. This contrasts to the targets of FNR, another factor that mediates bacterial adaptation to anaerobic conditions. Studies of uropathogenic E. coli (UPEC) indicate that FNR regulates genes controlling motility and multiple virulence factors (including expression of type I and P fimbriae), modulation of hemolysin expression, and expression of a novel pathogenicity island involved in α-ketoglutarate metabolism (10). This may or may not be due to the facts that (i) the FNR study targeted UPEC strain CFT073 (serotype O6:K2:H1), a distinct clade within B2 strains, and (ii) the conditions used for transcriptome examinations differed.
Citrate, like the other tricarboxylic acid (TCA) cycle intermediates α-ketoglutarate, succinate, and fumarate, is a metabolic pivot for catabolic and anabolic processes that supply key metabolic intermediates and are used to generate energy (36, 37). Under anaerobic and microaerobic conditions, TCA cycle intermediates contribute more significantly to cellular biosynthesis than to energy generation (17, 36). Expression of most enzymes of the TCA cycle is inhibited in the absence of oxygen, and to maintain carbon flux, bacteria have evolved alternative enzymes at certain steps of the TCA cycle and/or possess pathways that bypass the TCA cycle to metabolically link oxidative and reductive branches of TCA cycles (17). Citrate can be synthesized by the enzyme GltA; however, expression of the gene encoding this citrate synthase, like genes encoding most other enzymes of the TCA cycle, is significantly inhibited by ArcA in the absence of oxygen (37). Here, we found that under microaerobic conditions, ArcA significantly activates the expression of the citCEFTG gene cluster. citT encodes the citrate/succinate antiporter CitT, which imports citrate from the environment, and citCEF encode citrate lyase, which can cleave the citrate to acetate and oxaloacetate (OAA) (28). Therefore, the contribution of citCEFTG to APEC's growth under microaerobic conditions in duck serum is likely due to the supply of key metabolic intermediates that maintain carbon flux under microaerobic conditions. Indeed, oxaloacetate can be further reduced to malate, malate subsequently can be converted to fumarate, and fumarate can be finally reduced to succinate, all of which contribute to the microaerobic and anaerobic growth of bacteria (38). Thus, our data identify citCEFTG as a regulon of ArcA and reveal the dual role of ArcA in the control of citrate metabolism and transportation: under microaerobic and anaerobic conditions, ArcA inhibits citrate synthesis through inhibition of gltA expression in the TCA cycle pathway (37), while it promotes citrate import and citrate metabolism through citCEFTG, enhancing the cellular biosynthesis pathway.
One of the most provocative findings of this study is that ArcA may contribute to APEC virulence by affecting the expression of genes involved in motility and chemotaxis. Deletion of arcA significantly downregulates the expression of the middle (class 2) and late (class 3) flagellar genes. More importantly, strains of APEC XM with deletions in the motA and motB genes show severely decreased virulence, confirming that motility is important for the pathogenesis of APEC. Deletion of arcA also leads to significant downregulation of the chemotaxis-signaling histidine kinase gene cheA and the coupling protein gene cheW, and further deletions of cheA significantly attenuated APEC virulence. The results indicate that ArcA's contribution to APEC pathogenicity is at least partially through regulation of the expression of motility and chemotaxis genes. Furthermore, ArcA was shown to bind to the promoter regions of the motA, motB, and cheA genes, suggesting that ArcA directly regulates the expression these three genes.
This finding is consistent with studies of the role of flagella in uropathogenic E. coli (UPEC) and the chemotactic response of Salmonella. Expression of flagellar genes in UPEC is coincident with that pathogen's ascension to the upper urinary tract and significantly enhances the degree of colonization of the urinary tract (39). In addition, replacing the flagellin gene fliC by an antibiotic gene reduced APEC's adherence, invasion, and colonization, indicating that flagella may contribute to the virulence of extraintestinal pathogenic E. coli (40). Chemotaxis enables the Salmonella pathogen to seek out favorable metabolic niches and thus contributes to virulence in the early stages of host invasion and colonization (41). In our study, the expression of cheA was upregulated in duck serum compared to LB under microaerobic conditions, consistent with the notion that chemotaxis toward a favorable attractant(s) may also contribute to virulence in APEC. The mechanism by which chemotaxis may contributes to virulence remains unknown.
In summary, our data indicate that ArcA contributes to the virulence of APEC by affecting E. coli metabolism, motility, and chemotaxis. The two-component signal transduction systems PhoP-PhoQ (24), BarA-UvrY (42), and PhoB-PhoR (43) have each been reported to contribute to the virulence of APEC. Now, ArcA and its target CheA can be included in the growing list of signal transduction networks that control APEC pathogenicity. Most of the metabolic and motility and chemotaxis pathways that are affected by ArcA are highly conserved across bacterial pathogens, while deletion of arcA does not confer attenuation in all microbial pathogens. Salmonella arcA mutants show regulation of Salmonella pathogenicity island 1 (SPI1) and SPI2 and dysregulation of metabolic pathways but do not display virulence attenuation (44, 45). These findings suggest that studies are needed to determine the breadth of the effect of arcA deletion in the attenuation of virulence in bacterial pathogens and to understand the mechanisms by which arcA contributes to virulence.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Natural Science Foundation of China (31470243) and the Fundamental Research Funds for the Central Universities. The Center for Metabolic Biology at Iowa State University contributed to funding of data analysis.
The transcriptome sequencing was conducted in conjunction with BGI. We thank BGI for contributing its expertise in genomic sequencing and bioinformatics analysis to provide processed sequencing data.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00312-15.
REFERENCES
- 1.Gunsalus RP, Park SJ. 1994. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol 145:437–450. doi: 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 2.Iuchi S, Lin EC. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci U S A 85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch AS, Lin EC. 1996. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol 178:6238–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iuchi S, Chepuri V, Fu HA, Gennis RB, Lin EC. 1990. Requirement for terminal cytochromes in generation of the aerobic signal for the arc regulatory system in Escherichia coli: study utilizing deletions and lac fusions of cyo and cyd. J Bacteriol 172:6020–6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, De Wulf P. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem 279:12588–12597. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- 6.Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP. 2005. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J Biol Chem 280:15084–15096. doi: 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- 7.Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol 46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- 8.Nystrom T, Larsson C, Gustafsson L. 1996. Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. EMBO J 15:3219–3228. [PMC free article] [PubMed] [Google Scholar]
- 9.Myers KS, Yan H, Ong IM, Chung D, Liang K, Tran F, Keles S, Landick R, Kiley PJ. 2013. Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet 9:e1003565. doi: 10.1371/journal.pgen.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbieri NL, Nicholson B, Hussein A, Cai W, Wannemuehler YM, Dell'Anna G, Logue CM, Horn F, Nolan LK, Li G. 2014. FNR regulates expression of important virulence factors contributing to pathogenicity of uropathogenic Escherichia coli. Infect Immun 82:5086–5098. doi: 10.1128/IAI.02315-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma J, Bao Y, Sun M, Dong W, Pan Z, Zhang W, Lu C, Yao H. 2014. Two functional type VI secretion systems in avian pathogenic Escherichia coli are involved in different pathogenic pathways. Infect Immun 82:3867–3879. doi: 10.1128/IAI.01769-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 13.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G, Cai W, Hussein A, Wannemuehler YM, Logue CM, Nolan LK. 2012. Proteome response of an extraintestinal pathogenic Escherichia coli strain with zoonotic potential to human and chicken sera. J Proteomics 75:4853–4862. doi: 10.1016/j.jprot.2012.05.044. [DOI] [PubMed] [Google Scholar]
- 15.Li G, Feng Y, Kariyawasam S, Tivendale KA, Wannemuehler Y, Zhou F, Logue CM, Miller CL, Nolan LK. 2010. AatA is a novel autotransporter and virulence factor of avian pathogenic Escherichia coli. Infect Immun 78:898–906. doi: 10.1128/IAI.00513-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alteri CJ, Smith SN, Mobley HL. 2009. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog 5:e1000448. doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai W, Wannemuehler Y, Dell'anna G, Nicholson B, Barbieri NL, Kariyawasam S, Feng Y, Logue CM, Nolan LK, Li G. 2013. A novel two-component signaling system facilitates uropathogenic Escherichia coli's ability to exploit abundant host metabolites. PLoS Pathog 9:e1003428. doi: 10.1371/journal.ppat.1003428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lund SP, Nettleton D, McCarthy DJ, Smyth GK. 2012. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat Appl Genet Mol Biol 11. doi: 10.1515/1544-6115.1826. [DOI] [PubMed] [Google Scholar]
- 20.Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nettleton DHJ, Caldo RA, Wise RP. 2006. Estimating the number of true null hypotheses from a histogram of p values. J Agric Biol Environ Stat 11:337–356. doi: 10.1198/108571106X129135. [DOI] [Google Scholar]
- 22.Storey JD. 2000. A direct approach to false discovery rates. J R Stat Soc Ser B 64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 23.Li G, Ewers C, Laturnus C, Diehl I, Alt K, Dai J, Antao EM, Schnetz K, Wieler LH. 2008. Characterization of a yjjQ mutant of avian pathogenic Escherichia coli (APEC). Microbiology 154:1082–1093. doi: 10.1099/mic.0.2007/015784-0. [DOI] [PubMed] [Google Scholar]
- 24.Li G, Tivendale KA, Liu P, Feng Y, Wannemuehler Y, Cai W, Mangiamele P, Johnson TJ, Constantinidou C, Penn CW, Nolan LK. 2011. Transcriptome analysis of avian pathogenic Escherichia coli O1 in chicken serum reveals adaptive responses to systemic infection. Infect Immun 79:1951–1960. doi: 10.1128/IAI.01230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. 2000. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem 285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 26.Carreau A, El Hafny-Rahbi B, Matejuk A, Grillon C, Kieda C. 2011. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J Cell Mol Med 15:1239–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev 64:694–708. doi: 10.1128/MMBR.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pos KM, Dimroth P, Bott M. 1998. The Escherichia coli citrate carrier CitT: a member of a novel eubacterial transporter family related to the 2-oxoglutarate/malate translocator from spinach chloroplasts. J Bacteriol 180:4160–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheu PD, Witan J, Rauschmeier M, Graf S, Liao YF, Ebert-Jung A, Basche T, Erker W, Unden G. 2012. CitA/CitB two-component system regulating citrate fermentation in Escherichia coli and its relation to the DcuS/DcuR system in vivo. J Bacteriol 194:636–645. doi: 10.1128/JB.06345-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrera Seitz MK, Frank V, Massazza DA, Vaknin A, Studdert CA. 2014. Bacterial chemoreceptors of different length classes signal independently. Mol Microbiol 93:814–822. doi: 10.1111/mmi.12700. [DOI] [PubMed] [Google Scholar]
- 31.Ringgaard S, Zepeda-Rivera M, Wu X, Schirner K, Davis BM, Waldor MK. 2014. ParP prevents dissociation of CheA from chemotactic signaling arrays and tethers them to a polar anchor. Proc Natl Acad Sci U S A 111:E255–E264. doi: 10.1073/pnas.1315722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazelbauer GL. 2012. Bacterial chemotaxis: the early years of molecular studies. Annu Rev Microbiol 66:285–303. doi: 10.1146/annurev-micro-092611-150120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, Palm F, Mounier J, Prevost MC, Sansonetti P, Tang CM. 2010. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane MC, Li X, Pearson MM, Simms AN, Mobley HL. 2009. Oxygen-limiting conditions enrich for fimbriate cells of uropathogenic Proteus mirabilis and Escherichia coli. J Bacteriol 191:1382–1392. doi: 10.1128/JB.01550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marteyn B, Scorza FB, Sansonetti PJ, Tang C. 2011. Breathing life into pathogens: the influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell Microbiol 13:171–176. doi: 10.1111/j.1462-5822.2010.01549.x. [DOI] [PubMed] [Google Scholar]
- 36.Kornberg HL. 1970. The role and maintenance of the tricarboxylic acid cycle in Escherichia coli. Biochem Soc Symp 30:155–171. [PubMed] [Google Scholar]
- 37.Park SJ, McCabe J, Turna J, Gunsalus RP. 1994. Regulation of the citrate synthase (gltA) gene of Escherichia coli in response to anaerobiosis and carbon supply: role of the arcA gene product. J Bacteriol 176:5086–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimroth P, Schink B. 1998. Energy conservation in the decarboxylation of dicarboxylic acids by fermenting bacteria. Arch Microbiol 170:69–77. doi: 10.1007/s002030050616. [DOI] [PubMed] [Google Scholar]
- 39.Lane MC, Alteri CJ, Smith SN, Mobley HL. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104:16669–16674. doi: 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.La Ragione RM, Sayers AR, Woodward MJ. 2000. The role of fimbriae and flagella in the colonization, invasion and persistence of Escherichia coli O78:K80 in the day-old-chick model. Epidemiol Infect 124:351–363. doi: 10.1017/S0950268899004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rivera-Chavez F, Winter SE, Lopez CA, Xavier MN, Winter MG, Nuccio SP, Russell JM, Laughlin RC, Lawhon SD, Sterzenbach T, Bevins CL, Tsolis RM, Harshey R, Adams LG, Baumler AJ. 2013. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog 9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herren CD, Mitra A, Palaniyandi SK, Coleman A, Elankumaran S, Mukhopadhyay S. 2006. The BarA-UvrY two-component system regulates virulence in avian pathogenic Escherichia coli O78:K80:H9. Infect Immun 74:4900–4909. doi: 10.1128/IAI.00412-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertrand N, Houle S, LeBihan G, Poirier E, Dozois CM, Harel J. 2010. Increased Pho regulon activation correlates with decreased virulence of an avian pathogenic Escherichia coli O78 strain. Infect Immun 78:5324–5331. doi: 10.1128/IAI.00452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim S, Yoon H, Kim M, Han A, Choi J, Choi J, Ryu S. 2013. Hfq and ArcA are involved in the stationary phase-dependent activation of Salmonella pathogenicity island 1 (SPI1) under shaking culture conditions. J Microbiol Biotechnol 23:1664–1672. doi: 10.4014/jmb.1305.05022. [DOI] [PubMed] [Google Scholar]
- 45.Evans MR, Fink RC, Vazquez-Torres A, Porwollik S, Jones-Carson J, McClelland M, Hassan HM. 2011. Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiol 11:58. doi: 10.1186/1471-2180-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shan Y, Pan Q, Liu J, Huang F, Sun H, Nishino K, Yan A. 2012. Covalently linking the Escherichia coli global anaerobic regulator FNR in tandem allows it to function as an oxygen stable dimer. Biochem Biophys Res Commun 419:43–48. doi: 10.1016/j.bbrc.2012.01.121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.