Abstract
Intracellular pathogens are differentially sensed by the compartmentalized host immune system. Nevertheless, gene expression studies of infected cells commonly average the immune responses, neglecting the precise pathogen localization. To overcome this limitation, we dissected the transcriptional immune response to Shigella flexneri across different infection stages in bulk and single cells. This identified six distinct transcriptional profiles characterizing the dynamic, multilayered host response in both bystander and infected cells. These profiles were regulated by external and internal danger signals, as well as whether bacteria were membrane bound or cytosolic. We found that bacterial internalization triggers a complex, effector-independent response in bystander cells, possibly to compensate for the undermined host gene expression in infected cells caused by bacterial effectors, particularly OspF. Single-cell analysis revealed an important bacterial strategy to subvert host responses in infected cells, demonstrating that OspF disrupts concomitant gene expression of proinflammatory, apoptosis, and stress pathways within cells. This study points to novel mechanisms through which bacterial internalization, localization, and injected effectors orchestrate immune response transcriptional signatures.
INTRODUCTION
The host immune system discriminates nonpathogenic and pathogenic bacteria through sensing of so-called patterns of pathogenesis (1). Differentiation between extra- and intracellular bacteria includes sensing of microbe- and danger-associated molecular patterns (MAMPs and DAMPs) through a dual system of pattern recognition molecules (PRMs) with different subcellular localizations. The specific sensing of patterns with different localizations has been proposed to be “compartmentalization” of cellular self-defense (2). PRMs include Toll-like receptors (TLRs) at the cell surface and in some membrane-enclosed compartments and Nod-like receptors (NLR) or nucleic acid receptors in the cytosolic compartment. They detect a range of microbial components, including parts of the bacterial cell wall (e.g., lipopolysaccharide [LPS], peptidoglycan [PG], or lipopetides [LP]) or nucleic acids, as well as DAMPs (3). MAMP/DAMP sensing leads to activation of distinct signaling pathways that converge at a few transcription factors, such as nuclear factor of κB (NF-κB) or interferon (IFN)-regulatory factor 3 (IRF3), and mitogen-activated protein kinases (MAPKs). Ultimately, this culminates in induction of cytokines or chemokines (e.g., interleukin 8 [IL-8], CXCL-2, or type I IFNs) and of antimicrobial peptides (e.g., human β defensin 3 [HBD3]). Other patterns of pathogenesis are microbial invasion, viability, and growth, as well as perturbations of core internal processes by bacterial effectors, all of which can lead to transcriptional and posttranslational changes in the host. Perturbed processes include cytoskeletal disruptions, membrane damage, and concomitant cytosolic access of microbial material or changes of nutrient levels (1, 4).
One major human pathogen traversing the membrane-bound and the cytosolic subcellular compartments is the intracellular Gram-negative bacterium Shigella flexneri, the main cause of bacterial dysentery. Shigella uses a virulence plasmid-encoded type 3 secretion system (T3SS) that injects sets of primary and secondary effectors into the host cytosol, which are directly induced by the bacterial regulators/effectors VirB and MxiE, respectively, and which interfere with diverse host cell signaling pathways (5, 6). It attaches to the epithelial cell membrane, where it induces actin focus formation and subsequent internalization into a pathogen-containing vacuole (7). Then, within 5 to 15 min, Shigella triggers vacuolar rupture and invades the host cytosolic compartment as a niche for replication and intracellular spread (8). During invasion, it subverts the activated host immune responses through injected bacterial effectors. The primary effector, IpgD, is an inositol 4-phosphatase that alters multiple host signaling pathways, including the release of extracellular ATP (eATP), which activates host inflammation, and the onset of the phosphatidylinositol 3-kinase (PI3K)/Akt anti-apoptotic signaling pathways (9, 10). The secondary effector, OspG, has ubiquitin-binding properties that attenuate IκB degradation and NF-κB-dependent immune signaling (11, 12). The VirB- and MxiE-dependent effector OspF is a dual-specificity phospholyase that irreversibly dephosphorylates the MAPKs p38 and ERK, thereby reducing levels of proinflammatory cytokines (13, 14).
Measurements of complex host immune responses are routinely simplified. They are averaged from large, heterogeneous cell populations, neglecting the onset of specific immune response pathways depending on the bacterial subcellular localization, and the impact of noninfected neighboring, so-called bystander, cells has only recently been considered (15, 16). However, the study of the localization-specific impact caused by pathogen traversal from one subcellular compartment to another requires direct assessment in single cells. To examine this, we established an experimental and analytical pipeline using fluorescent pathogen localization reporters and gene expression profiling. We used this pipeline to determine that the epithelial cell host immune response during Shigella contact segregates into distinguishable bystander, vacuolar, and cytosolic portions. Computational analysis of complex, dynamic, pathogen-induced expression signatures revealed six distinct profiles of coregulated gene sets that readily fit specific signaling pathways. The host response of bystander cells was activated solely through bacterial internalization in infected cells and was very minimally affected by bacterial effector activities. Transcriptional signatures in infected cells were shaped through bacterial localization, T3SS effectors, or both. Among them, OspF disrupted concomitant host gene expression of apoptosis, inflammatory, and stress response pathways as a major infection strategy of the invading pathogen.
MATERIALS AND METHODS
Bacterial strains and bacterial preparation.
S. flexneri M90T wild-type (WT) and mutant strains deficient in the bacterial effectors OspF (ΔospF) (13), OspG (ΔospG) (17), MxiE (ΔmxiE) (18), IpgD (ΔipgD) (19), and MxiD (ΔmxiD) (20) were transformed by electroporation with the pGG2(Ampr)-dsRed.T3_S4T plasmid (21). For infection experiments, bacteria were grown overnight (ON) in tryptic casein soy broth (TCSB) supplemented with ampicillin (50 μg/ml) at 37°C with shaking at 220 rpm and subsequently subcultured at a 1/100 dilution for 2.25 h to an optical density at 600 nm (OD600) of >0.3.
Cell culture, transfection procedures, fluorescence resonance energy transfer (FRET) assay, infections, and stimulations.
Human epithelial HeLa cells (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% decomplemented fetal calf serum (FCS) at 37°C, 5% CO2. Transfection of HeLa cells with p-mOrange-empty (7, 8), p-mOrange-bla, p-mOrange-Actin, or p-YFP-p65 (a kind gift of R. Weil) was performed using Fugene 6 transfection reagent (Promega) according to the manufacturer's instructions 48 h prior to experimentation.
All experiments using the FRET assay were carried out in EM medium (120 mM NaCl, 7 mM KCl, 1.8 mM CaCl2, 0.8 mM MgCl2, 5 mM glucose, and 25 mM HEPES at pH 7.3) as described previously (8), and all buffers and media were supplemented with 2.5 μM probenecid until the final measurements. Bacteria were washed with phosphate-buffered saline (PBS) and coated for 10 min at room temperature with 10 μg/ml poly-l-lysine (Sigma) and 10 μg/ml soluble β-lactamase (Sigma). Infection was carried out at a multiplicity of infection (MOI) of 25. To synchronize infection, bacteria were allowed to settle for 15 min at room temperature, followed by incubation for 30 min at 37°C to promote T3SS-dependent bacterial invasion. Subsequently, the cells were extensively washed to remove bacteria from the medium and further incubated for 2.5 h at 37°C. Infection was stopped by maintaining the cells at 4°C until they were sorted. Cytochalasin D treatment (2 μM; Sigma) was performed 30 min prior to and during the course of infection.
Live imaging of Shigella infection.
HeLa cells were washed with PBS, infected at an MOI of 20, and imaged on a Leica DM inverted microscope equipped with a heated stage using a 20× N-Plan objective with excitation at 465 to 500 nm (for yellow fluorescent protein [YFP]) and 532 to 554 nm (for mOrange), and emission was detected with 516- to 556-nm (YFP) and 573- to 613-nm (mOrange) filters. Images were acquired in the two fluorescent channels and in trans every 5 min using a CoolSnap2 camera (Roeper Scientific). Time-lapse series were analyzed using Fiji (http://fiji.sc/Fiji).
Fluorescence-activated cell sorter (FACS)-based cell sorting, sample processing, and multiplex qPCR.
Cells were trypsinized for 5 min at 37°C, centrifuged for 5 min at 200 × g and 4°C, and sorted on a FACS Aria III cell sorter (BD Bioscience) into 96-well plates containing 9 μl lysis–reverse transcription-specific target amplification (RT-STA) mixture per well (5 μl Cells Direct 2× Reaction Mix, 0.2 μl SuperScript III RT PlatinumR Taq Mix [CellsDirect One-Step qRT-PCR kit; Life Technologies], 2 U Superase-In RNase inhibitor [Life Technologies], 2.5 μl assay mixture of 96 pooled Solaris qPCR gene expression assays [ThermoFisher] at a final concentration of 0.2×, and 1.2 μl 0.1 mM EDTA-10 mM Tris buffer). After sorting, samples were subjected to one-step RT-STA using the following program: 50°C for 15 min, 95°C for 2 min (for RT), and 23 cycles of 95°C for 15 s and 60°C for 4 min (for STA). For multiplex quantitative PCR (qPCR) on a BioMark System (Fluidigm), cDNA was diluted 1/5 with 1 mM EDTA-10 mM Tris buffer, and 2.9 μl cDNA was mixed with 3.25 μl 2× Solaris Universal qPCR MasterMix (ThermoFisher) and 0.32 μl sample-loading reagent (Fluidigm). Ninety-six samples and 96 individual gene expression assays were loaded on a 96-by-96 dynamic array. The gene expression curves obtained were analyzed using the manufacturer's software.
Bulk cell data processing, statistical analysis, and visualization.
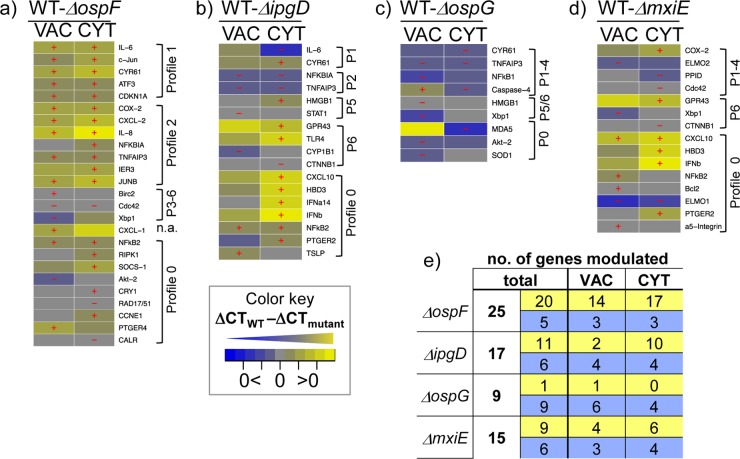
Further analysis was performed using different custom and commercial computational analysis programs. Samples that gave less than six threshold cycle (CT) values were removed from the analysis. ΔCT values were calculated using the average CT of the two reference genes chosen for each cell line, i.e., B2M and PGK1 genes for HeLa cells and PGK1 and ALDOA genes for Caco-2 cells. Principal-component analysis (PCA) plots were generated using R (see Fig. S3d and e in the supplemental material) (http://www.r-project.org) or the Qlucore software (all other PCA plots), which provides tools to apply P and q value (false-discovery rate) cutoffs based on analysis of variance (ANOVA) and nearest-neighbor connections (by Euclidean distance) to indicate highly similar samples. Heat maps, box plots, and representations of gene expression profiles were generated by custom software implemented in Matlab and R. To generate gene expression profiles, we developed a computational profiling procedure in order to classify each gene (see the flowchart in Fig. S5a in the supplemental material). To this end, we computed scores for each expression change at each transition to the next indicated stage (i.e., CO→BY→VAC→CYT) as follows: −1 for a significant (P < 0.05) decrease, +1 for a significant increase, and 0 if no significant change was detected. Subtractive heat maps (see Fig. 4a to d) were generated by subtracting the median ΔCT values of mutant-infected cells from those of WT-infected cells (ΔCTWT − ΔCTmutant) at the indicated infection stages. Only the genes that showed significantly different expression (P < 0.05) for at least one of the stages are presented in the heat maps.
FIG 4.
OspF regulates apoptosis and the host inflammatory and stress response. (a to d) Heat maps showing subtracted ΔCTs and indicating if the relative difference in gene expression was higher (yellow) or lower (blue) in mutant- compared to WT-infected cells. All the genes for which this difference was significant (P < 0.05) for at least one of the indicated stages upon infection with the ΔospF (a), ΔipgD (b), ΔospG (c), or ΔmxiE (d) mutant are shown. Subtracted median values of eight samples (quadruplicates of bulk cells from two independent experiments) are shown. Significant differences (P < 0.05 by Mann-Whitney U test) are marked with a plus for significantly higher expression and a minus for significantly lower expression. (e) Overview table of the total numbers of tested genes whose expression was significantly higher (yellow) or lower (blue) at vacuolar (VAC) or cytosolic (CYT) bacterial localization in cells infected with the respective mutant than in WT Shigella-infected cells.
Single-cell data processing and analysis.
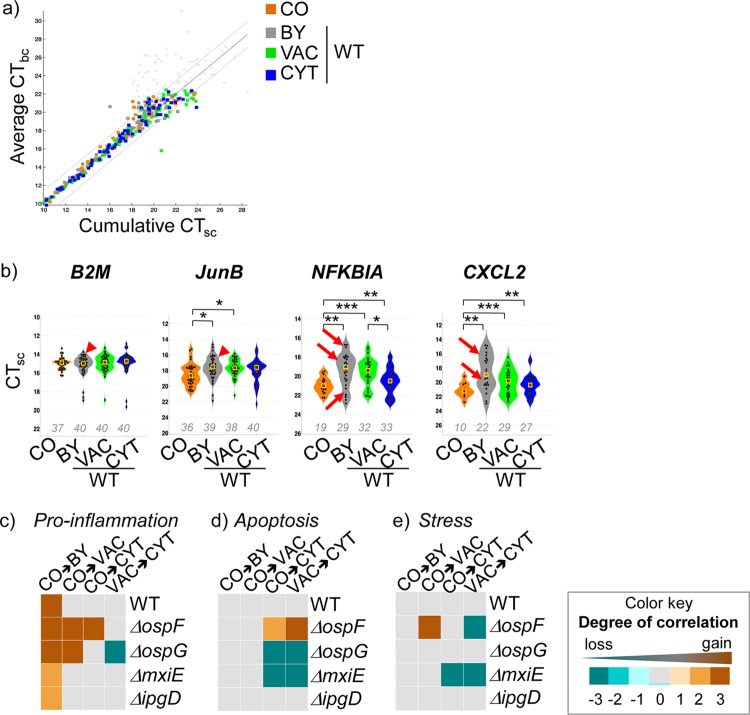
Samples with less than 6 out of 96 valid expression values were removed from the analysis. Concordance between single- and bulk cell expression values was assessed by comparing the average CT value to the bulk cell samples of each respective experimental condition (CTbc) to the sum of an in silico pool of single-cell expression values, which is referred to as the cumulative single-cell CT value (cCTsc) (for a more detailed description of single-cell data processing, see the supplemental material). Statistical significance was determined using a likelihood ratio test (LRT). Single-cell expression data were analyzed using box and violin plots. For pathway correlation analysis, we assessed concomitant expression of gene pairs assigned to common functional pathways (e.g., proinflammation, apoptosis, and stress) (see the supplemental material) using Spearman's rank correlation coefficients. To this end, we defined a score that measures the degree of concomitant expression between genes assigned to a given pathway. To detect differences in concomitant expression upon a change from one stage of infection to another, we developed a statistical analysis that computes a significance score, t, on a scale of +3 to −3 for gain (t > 0) or loss (t < 0) of pairwise gene correlations of the pathway (further details are provided in the supplemental material). Significance scores are shown as heat maps (see Fig. 5c to e).
FIG 5.
Single-cell analysis revealed the targeted disruption of concomitant inflammatory gene expression by the bacterial effector OspF. (a) Concordance of single-cell (sc) and bulk cell (bc) measurements. The cumulative CTsc and the average CTbc from two independent experiments for the indicated stages of bacterial infection (BY, VAC, and CYT) or uninfected (CO) control cells are shown. The gray lines indicate a deviation of ±1 CT from absolute concordance (black line). The gray data points indicate values eliminated after applying a cutoff at a CT of 23 (CTmax). (b) Violin plots showing CTsc values (black dots) and the means (red crosses) and medians (yellow squares) for the indicated genes per condition measured from 40 cells from two independent experiments. The numbers show the proportions of the 40 cells that expressed the respective genes with a CTsc greater than or equal to the CTmax. The P values were calculated using the log likelihood ratio test. *, P < 0.05; **, P < 0.005; ***, P < 0.001; not significant if nothing is indicated. The arrows indicate a multimodal distribution of the CTsc values; the arrowheads indicate that the CTsc values were log-normally distributed around the mean. (c to e) Heat maps showing the significance scores for concomitant expression of genes that were classified as proinflammation (c), apoptosis (d), or stress (e) genes. They indicate the degree of significance of gene pair correlation changes at the change of infection stage. The color gradients represent significance scores ranging from +3 for a significant gain of correlation (orange) to −3 for a significant loss of correlation (blue) and 0 if no change of correlation was detected (gray).
RESULTS
Identification of host immune response signatures for distinct stages of Shigella invasion.
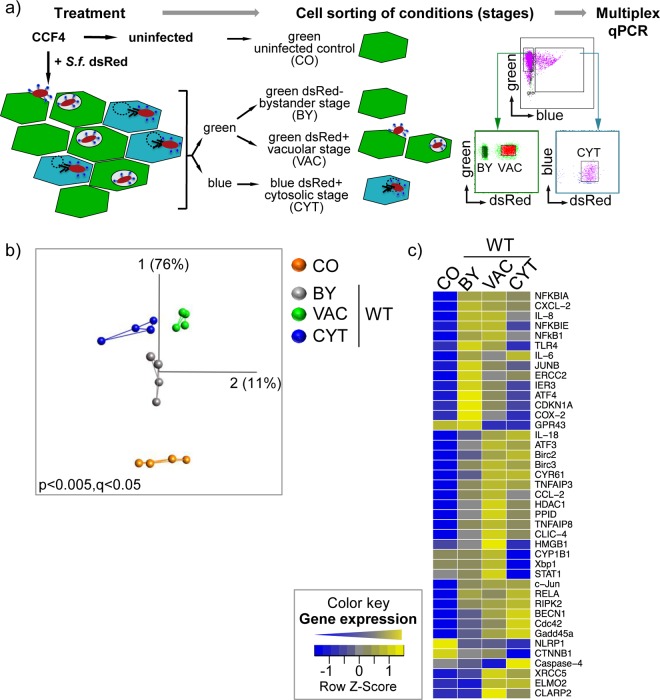
We hypothesized that three stages during S. flexneri infection lead to the activation of distinct host response signaling pathways: (i) the bystander stage of noninfected neighboring cells (BY) and two stages of infected cells with (ii) membrane- or vacuole-bound bacteria (VAC) or (iii) bacteria in the cytosol (CYT). We aimed to explore how these stages differentially activate the transcriptional host immune response at the levels of small bulk cell populations (20 cells) and of single cells. To distinguish these stages within a heterogeneous layer of cells challenged with Shigella, we set up a workflow combining fluorescence-based cell sorting with multiplex qPCR analysis (Fig. 1a; see Fig. S1 in the supplemental material). The workflow uses dsRed-fluorescent Shigella in conjunction with the CCF4/β-lactamase FRET assay, previously developed by our laboratory (8). This assay is able to detect infection by a single bacterium; monitors bacterial vacuolar rupture at high temporal resolution, resulting in a fluorescence switch from 535 nm (green) to 450 nm (blue); and is compatible with FACS-based sorting (Fig. 1a; see Fig. S1 in the supplemental material) (22). We adjusted the MOI to yield 40 to 60% infected cells at 3 h postinfection (p.i.), thus obtaining robust transcriptional host responses and also providing enough bystander cells (Fig. 1a and data not shown). We decided against the use of gentamicin, an antibiotic used to kill bacteria in the medium, to avoid the possibility of host response signaling to inactivated bacteria. Consequently, the infection was not synchronized, explaining the proportion of cells at the membrane-bound and vacuolar stages. To measure host responses, we assembled a set of expression assays for genes characterizing the onset, regulation, and execution of inflammatory signaling, apoptosis, survival, and cell cycle signaling, as well as of stress response, repair, and lipid metabolism (see Table S1 and Fig. S2 in the supplemental material). Briefly, our set contained subsets of inducible PRMs and components of signaling hubs, as well as of their downstream molecules, including a subset of cytokines and chemokines, which indicate activation of different PRM signaling pathways. The selected pathways include signaling via NF-κB, MAPK, IRF3, Gadd45a, ATF3, Xbp1, and CDKN1A (see Fig. S2 in the supplemental material). In addition, we chose two distinct reference genes (see Fig. S3a and b and further details in the supplemental material).
FIG 1.
Identification of localization-dependent transcriptional signatures during Shigella infection, combining fluorescence reporters and microfluidics. (a) Workflow for treatment, cell sorting, and subsequent transcriptional analysis of uninfected (CO) cells or cells at distinct stages of dsRed-expressing Shigella (+S.f. dsRed) infection (BY, VAC, and CYT). The conditions were distinguished using the CCF4/β-lactamase FRET approach and dsRed-fluorescent bacteria and were sorted by FACS according to their specific fluorescent properties, as indicated. The sorted samples were further processed for multiplex qPCR analysis. (b) PCA plot showing transcriptional signatures of quadruplicates of bulk cell samples (dots) at the indicated stages of WT Shigella infection (BY, VAC, and CYT) or of the uninfected control (CO). PC1 and PC2 and their percentages of captured variance are shown. The lines between samples indicate the two nearest neighbors. Representative data from one out of three independent experiments are shown (see Fig. S4a and b and Movies S1 to S3 in the supplemental material). (c) Heat map showing median ΔCT values of bulk cell samples transformed into a row Z-score of the indicated genes under the indicated conditions of Shigella WT infection (BY, VAC, and CYT) or of an uninfected control (CO). All genes with at least one significant gene expression change under one of the conditions are shown. The color key indicates the row Z-scores ranging from +2 (high expression; yellow) to −2 (low expression; blue). Data from two independent experiments at a P value cutoff of 0.05 are shown (see Table S2 in the supplemental material).
To obtain high-content transcriptional information, we used microfluidic devices for multiplex qPCR and subjected the collected data to a custom computational data analysis. To this end, we established a protocol to obtain robust transcriptional responses at the levels of bulk and single cells (see Fig. S3 and Results in the supplemental material for a detailed quality assessment). Each bulk sample contained 20 cells in the same condition (one of the three stages of infection or uninfected [CO] control cells), which were collected in quadruplicate. To produce statistically strong data sets for single-cell analysis, we collected 20 individual cells per condition.
We analyzed transcriptional profiles of bulk cells using PCA, a mathematical procedure that transforms high-dimensional data into three principal components, PC1 to PC3, with minimal loss of information (see Fig. S4a in the supplemental material). Each data point represents the transcriptional information from 96 genes of one bulk cell sample. Each succeeding PC shows the largest possible uncorrelated variance of all data at a given significance cutoff. This allows the identification of patterns contained in the data set, such as sample similarities or differences. After applying stringent significance cutoffs (P < 0.005 and q < 0.05), the data points separated into four independent clusters, each corresponding to the four previously sorted conditions of Shigella-treated cells and the uninfected control (Fig. 1b; see Movie S1 in the supplemental material). This illustrates the fact that distinct and significantly relevant transcriptional signatures correspond to uninfected cells and to the individual stages of WT Shigella infection, i.e., the bystander, the vacuolar, and even the cytosolic stages. Furthermore, we identified 42 out of 96 measured host genes that were significantly (P < 0.05) differentially regulated during at least one of the previously determined individual stages of Shigella infection (Fig. 1c; see Table S2 and Fig. S4b in the supplemental material). Taken together, these data suggest that Shigella WT infection activates distinct gene expression signatures depending on the stage of bacterial infection.
We also applied our workflow to basolateral Shigella infection of polarized intestinal epithelial (Caco-2) cells as a more physiological cell culture infection model (see Fig. S4c to f in the supplemental material). In these experiments, we included genes specific for polarized epithelial cells, such as regulators and components of tight junctions and additional mediators of the host response (see Table S1 in the supplemental material). Basolateral infection via porous membranes of transwell plates resulted in only 4 to 7% infection rates, explaining a rather heterogeneous, less reliable bystander population (data not shown). Nonetheless, infection of polarized Caco-2 cells uncovered distinct localization-dependent transcriptional signatures of infected cells (see Fig. S4e and f and Table S3 in the supplemental material). Many signatures of genes examined in both cell types were largely similar (e.g., CYP1B1, JunB, Xbp1, and CCL2). In some cases, however, we observed differences of differential expression of some genes at specific locations between cell types. In general, this argues for HeLa cells being a robust, representative, and simple cell culture model for the tracking of transcriptional immune response analysis of our chosen genes upon Shigella infection.
Activation of a gene expression signature in bystander cells requires host cell invasion.
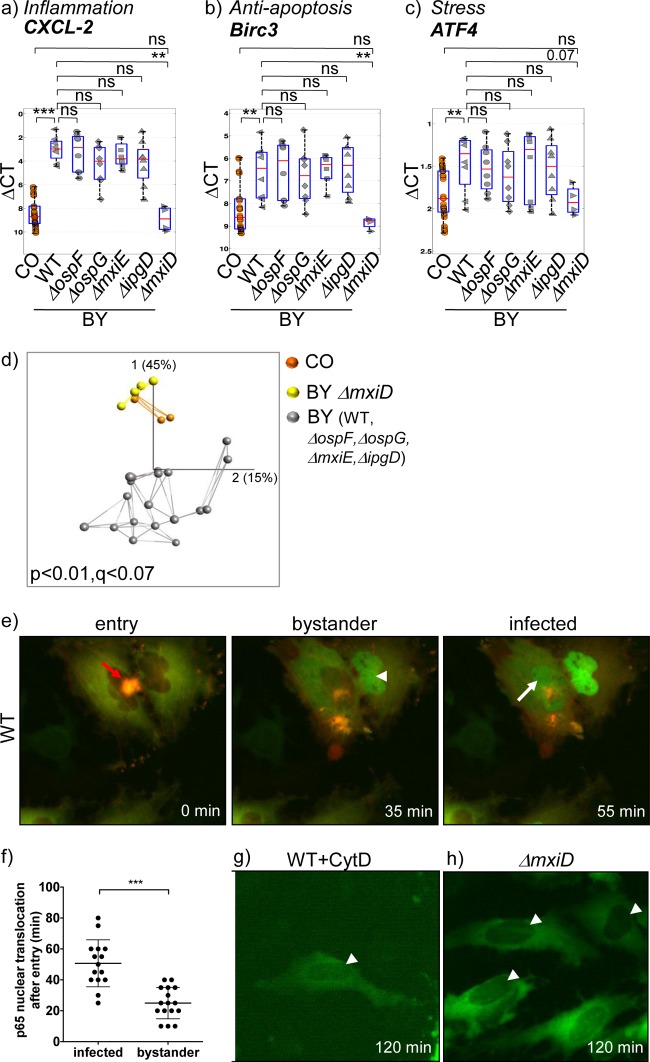
One striking feature of the localization-dependent gene expression signatures we found in bulk cell samples was the detected bystander response of noninfected neighboring cells (Fig. 1b and c). Until now, bystander responses upon microbial infection have been investigated only in the context of inflammatory signaling (15, 16). Here, we explored the bystander response of bulk cell samples on a broader scale of 96 genes from different host response pathways. Our analysis identified 20 genes significantly altered in bystander cells in comparison to uninfected controls (compare columns CO and BY WT in Table S4 in the supplemental material). They were all upregulated and comprised three main gene families (Fig. 2a to c): (i) proinflammatory cytokines and chemokines (e.g., CXCL-2 and IL-8), as expected, including components of the implicated signaling pathways (e.g., cJun); (ii) predominantly antiapoptosis genes (e.g., Birc3); and (iii) genes of the cellular stress response, including major transcription factors (e.g., ATF3).
FIG 2.
Bystander cells exhibit an internalization-dependent and effector-independent signature. (a to c) Box plot analysis of expression of representative proinflammatory (a), antiapoptosis (b), and stress response (c) genes in uninfected (CO) and bystander (BY) cells of samples treated with WT S. flexneri or the indicated mutants. The box plots show ΔCT values of quadruplicates of bulk cell samples from two independent experiments, the median expression values (red lines), and the 25% and 75% quartiles (boxes). ***, P < 0.001; **, P < 0.005; ns, not significant; Mann-Whitney U test. (d) PCA plot of bulk cell transcriptional signatures (dots) of bystander cells (BY) from samples treated with the indicated Shigella strains and of uninfected control cells (CO). Connections of the two nearest neighbors (lines) and percentages of the captured variability for each PC are shown. Representative data from one out of three independent experiments are shown. (e) Time-lapse analysis of p65 nuclear translocation upon Shigella infection. HeLa cells expressing p65-YFP (green) and actin-mOrange (red) were challenged with WT Shigella at an MOI of 20, and images were taken every 5 min. The images show merged channels and are representative of the first occurrence of entry (focus formation; 0 min) and of p65 nuclear translocation in bystander (35 min; indicated by the arrowhead) and in infected (55 min; indicated by the arrow) cells. The time scale was normalized to the onset of bacterial entry (see Movie S4, top, in the supplemental data). (f) Quantification of the time after entry of WT Shigella at which p65 nuclear translocation occurred. The data show mean values from multiple sites of two independent experiments run in duplicate. ***, P < 0.001 by Student's t test. (g and h) Analysis of p65 localization in p65-YFP-expressing HeLa cells 120 min p.i. upon cytochalasin D treatment and WT Shigella infection (g) or ΔmxiD strain infection (h) at an MOI of 20 (see Movie S4, middle and bottom, respectively, in the supplemental data). The representative images show absence of p65 nuclear translocation (indicated by the arrowheads).
We reasoned that the injected T3SS effectors may play a role in the activation and outcome of bystander responses. Surprisingly, box plot analysis of all identified bystander signatures showed strong and highly similar inductions, with P values of ≥0.05 for the different injected effector mutants compared to WT bacteria (Fig. 2a to c; see Table S4 in the supplemental material). In contrast, infection with the secretion-deficient, noninvasive ΔmxiD mutant did not induce bystander responses. Accordingly, PCA of transcriptional signatures in bystander cells challenged with the different pathogen strains resulted in two major clusters (Fig. 2d). The first cluster grouped signatures of untreated control cells together with those of cells challenged with the noninvasive ΔmxiD strain, demonstrating that the presence of extracellular bacteria did not significantly affect the overall epithelial cell gene expression profile. The second cluster, which was clearly separated along the PC1 axis, comprised the transcriptional responses of cells challenged with the WT, ΔospF, ΔospG, ΔmxiE, or ΔipgD strain, indicating strong activation of a distinct transcriptional signature that shows minimal differences between the transcriptional signatures activated by these bacteria. Together, these results suggest that the internalization of the pathogen is required to activate a bystander response and that the amplitude of bystander activation, at least for the tested genes, is independent of the injected bacterial effectors in infected cells.
To investigate if internalization of bacteria causes the bystander response, we performed single-cell live imaging using a reporter plasmid for the onset of inflammation signaling through the nuclear translocation of the NF-κB subunit p65 (Fig. 2e to h; see Movie S4 in the supplemental material). Upon entry of WT Shigella, p65 nuclear translocation first occurred in bystander cells at 25 (±10) min p.i. before its onset in infected cells at 50 (±15) min p.i. (Fig. 2e and f; see Movie S4, upper sequence, in the supplemental material). In contrast, inhibition of bacterial entry by cytochalasin D treatment (Fig. 2g; see Movie S4, middle, in the supplemental material) or challenge with ΔmxiD (Fig. 2h; see Movie S4, bottom, in the supplemental material) did not lead to p65 nuclear translocation in any cells, corroborating our hypothesis that bacterial internalization causes the bystander effect, although by an unknown mechanism.
Immune response pathways are differentially regulated depending on the stage of Shigella infection.
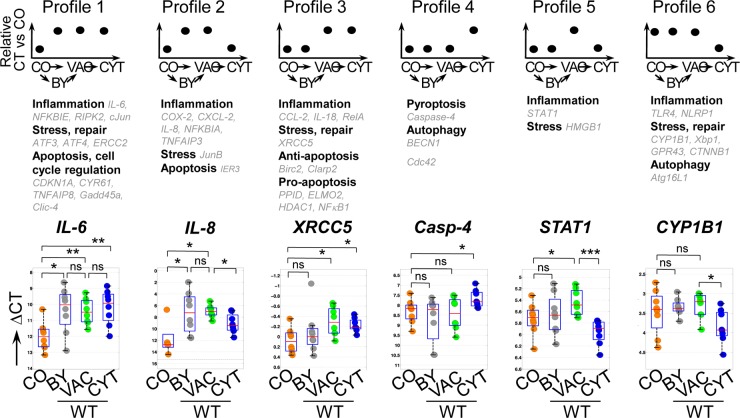
Next, we aimed to identify patterns of regulation of the distinct host transcriptional signatures of bulk cell samples, which might be determined by the bacterial localization, bacterial effector activities, or both. To this end, we developed a procedural-profiling model (see Fig. S5a in the supplemental material) by which each gene expression signature (Fig. 1c) is assigned to one profile, depending on the significance of transcriptional changes during the possible successive stages of WT Shigella infection (i.e., CO→VAC→CYT or CO→BY→VAC→CYT, as depicted in Fig. 3, top). This procedure yields a theoretical maximum of 34 distinct profiles. Surprisingly, the 42 signatures shown in Fig. 1c were assigned to only six profiles (profiles 1 to 6) among the 34 possibilities (Fig. 3). Profiles 1 and 2 contained all genes with increased expression in bystander cells, which were also induced in infected cells during the vacuolar stage, with a subset of genes with decreased expression during cytosolic infection (profile 2). Interestingly, no profile of genes activated only in bystander cells could be identified for our chosen gene set. The other four profiles displayed genes uniquely regulated in infected cells, which included an increase of gene expression either at the vacuolar stage (profiles 3 and 5) or at the cytosolic stage (profile 4). Profiles 5 and 6 display genes that showed decreased expression upon the switch to cytosolic localization. Of note, 50 signatures did not change significantly at any of the successive stages and hence were assigned to profile 0 (see Fig. S5b in the supplemental material). Furthermore, for a few proinflammatory cytokine genes (CXCL-1, CCL20, TNF-α, and IL-1α) no expression was detected in control cells, and hence, neither a statistical test nor profiling could be performed (data not shown). Together, these profiles suggest that the host immune response to Shigella infection is tailored to a few expression patterns, possibly reflecting signaling pathways, whose regulation is finely tuned depending on the bacterial localization. Furthermore, the observation of (i) genes regulated in either infected cells, bystander cells, or both or (ii) those altered only upon direct infection led us to define these sets of genes as (i) external-danger regulated (profiles 1 and 2) and (ii) internal-danger regulated (profiles 3 to 6).
FIG 3.
Identification of six distinct localization-dependent gene expression patterns during Shigella infection in bulk cells. The identified transcriptional signatures obtained from bulk cells (Fig. 1c) were classified into profiles using the computational procedure summarized in Fig. S5 in the supplemental material. In this method, each transcriptional difference of all analyzed genes upon the transitions CO→BY, BY→VAC, and VAC→CYT was given a score, as follows: −1 for a significant decrease in expression, +1 for a significant increase in expression, and 0 for no significant change at a P value of <0.05 (Mann-Whitney U test). (Top) Subsequently, the genes were classified into the corresponding profiles according to the decision tree derived from our computational procedure (see Fig. S5 in the supplemental material). (Middle) The genes were organized by their main annotation. (Bottom) Box plots showing the ΔCT values of representative genes, i.e., IL-6, IL-8, XRCC5, Caspase-4, STAT1, and CYP1B1. Quadruplicates of bulk cell samples from two independent experiments, with the median expression values (red lines), as well as the 25% and 75% quartiles (boxes). *, P < 0.05; **, P < 0.005; ***, P < 0.001; ns, not significant; Mann-Whitney U test.
We next assessed the compositions of genes assigned to each profile and their known or presumed functions (Fig. 3, middle). We observed that expression of genes from multiple published Shigella-induced inflammatory pathways was increased in bystander cells and early after bacterial invasion in infected cells, during the vacuolar stage. Expression of some inflammatory genes remained increased at the cytosolic stage (e.g., IL-6, profile 1). However, expression of a subset of these genes was shut down at the cytosolic stage of infection (e.g., IL-8 and TLR4, profiles 2 and 6), possibly via bacterial effector activities or host cytosolic mechanisms targeting the corresponding signaling pathways. This suggests that in both bystander and infected cells a multilayered inflammatory response is activated through external danger and that the signaling pathways involved are differentially regulated during the cytosolic stage of infection through internal danger. Expression of type I IFNs, well known to be activated upon recognition of nucleic acids in the cytosol, did not significantly alter throughout WT Shigella infection (see Fig. S5b, profile 0, in the supplemental material). In contrast, expression of STAT1, a transcription factor downstream of IFN signaling, was decreased at the cytosolic stage, indicating targeting of the IRF3 pathway in WT infection (profile 5).
Analogously to inflammatory genes, stress genes were found in multiple profiles with similar patterns of differential induction or downregulation at distinct infection stages (e.g., ATF3, XRCC5, or CYP1B1, profiles 1, 3, and 6, respectively). Thus, although many factors of the stress response were differentially regulated exclusively at the cytosolic stage (profile 6), as expected (1, 23), a number of major stress-related transcription factors and their downstream targets were also induced at the bystander and vacuolar stages. This suggests that both stress and inflammatory genes are regulated via overlapping or even identical signaling hubs at all infection stages. Generally, stress responses during bacterial infections are initiated by diverse stimuli, most notably upon amino acid starvation in the cytosol or induction of stress at the endoplasmic reticulum (ER) (23, 24). We observed that the transcriptional response to cytosolic, but not to vacuolar, WT Shigella infection resembles the amino acid starvation-induced stress response and involves regulation by the stress receptor GCN2 (data not shown). This response overlapped with a PERK-dependent ER stress response at the vacuolar stage (data not shown).
Moreover, while mainly antiapoptosis genes were induced in bystander cells and at the vacuolar stage (e.g., CYR61 and Birc2, profiles 1 and 3), the response switched toward an expression program promoting cell death upon cytosolic infection (e.g., Caspase-4 and NF-κB1, profile 4). These results provide evidence that external danger during Shigella infection mainly sustains antiapoptosis, while prolonged internal danger leads to a shift toward an active proapoptosis program within the host cell. Finally, bacterial translocation to the cytoplasm also resulted in regulation of autophagy genes (e.g., Atg16L, profile 6), as expected (23, 25).
Together, the dissection of the dynamics of host gene expression signatures revealed a complex and finely tuned adaptation of distinct immune response pathways. It allows the distinction between external- and internal-danger-regulated signaling pathways.
OspF subverts specific intracellular signatures from different gene families and host pathways.
To decrypt the roles of the injected T3SS effectors in the establishment of the dynamic gene expression profiles, we compared their induced stage-dependent transcriptional signatures in bulk cell samples. PCA plots of ΔospF strain-induced vacuolar and cytosolic signatures no longer clearly separated as independent clusters and shared nearest-neighbor connections (compare Fig. S6a in the supplemental material to Fig. 1b). In contrast, responses to ΔipgD, ΔospG, or ΔmxiE strain infection did not result in a loss of localization-dependent signatures for infected cells (compare Fig. S6b to d in the supplemental material to Fig. 1b). Furthermore, jointly plotting the WT and mutant strains demonstrated distances between the WT- and ΔospF strain-induced signatures larger than those between WT- and ΔipgD, ΔospG, or ΔmxiE mutant-infected cells (compare Fig. S6e to Fig. S6f to h and Movie S5 to Movies S6 to S8 in the supplemental material). Therefore, we conclude that, of the tested effectors, OspF had the broadest impact on the Shigella-induced host immune responses at both the vacuolar and the cytosolic stages.
We also examined the broad scale of the effector-induced changes in localization-dependent intracellular responses using subtractive heat maps. These maps indicate if gene expression is higher (yellow) or lower (blue) upon infection with the respective mutants compared to WT infection (Fig. 4a to d). Infection with the ΔospF strain led to modulation of more genes (25 genes in total) than infection with the ΔipgD (17 genes), ΔospG (9 genes), or ΔmxiE (15 genes) strain, respectively (Fig. 4e). Moreover, upon ΔospF strain infection, expression of the majority of genes was significantly increased compared to WT infection during both vacuolar (14 genes) and cytosolic (17 genes) localization, portraying OspF as a master regulator dampening host gene expression independent of the stage of infection. Of note, in addition to inflammatory genes, this included genes for the cellular integrated stress and repair responses, cell cycle regulation, antiapoptosis, and lipid metabolism, suggesting that OspF impacts gene expression for a broad spectrum of host responses. In particular, OspF strongly altered the expression of genes assigned to profiles 1 and 2 that were also regulated by external danger (Fig. 3). These genes are regulated via the MAPK signaling pathways, most likely downstream of Nod1 activation, in agreement with the known impact of OspF on MAPK activity (13).
Upon ΔipgD infection, fewer genes were dampened at the cytosolic stage than at the vacuolar stage (10 versus 2 genes) (Fig. 4b and e) compared to WT infection. Hence IpgD, in contrast to OspF, predominantly dampens host transcriptional signatures only at the cytosolic stage of infection. Interestingly, this included a subset of genes that were assigned to profile 0 upon WT infection (see Fig. S5b in the supplemental material), i.e., type I IFN genes, CXCL10, and HBD3 (Fig. 4b). This highlights a previously unrecognized role of IpgD in the host cytosolic type I IFN signaling pathways. Our data on ΔospG strain-infected cells showed that genes involved in antiapoptosis and (oxidative) stress were significantly less expressed than during WT infection, predominantly at the vacuolar stage of infection (Fig. 4c and e), implying a role of OspG in positively regulating survival. Finally, compared to WT infection, the ΔmxiE-activated transcriptional response displays both decreased expression of apoptosis genes at the vacuolar stage (as observed for ΔospG strain infection) and enhanced expression of inflammatory genes at the cytosolic stage (as observed for ΔipgD strain infection), suggesting that MxiE reflects the effects of multiple secondary effectors, in agreement with its role as a transcriptional activator for secondary bacterial effectors (Fig. 4d and e).
Single-cell analysis revealed that Shigella disrupts coordinated gene expression via the effector OspF.
We turned to single-cell analysis to explore the impact of bacterial effectors on the coordination of localization-dependently expressed genes. We performed adequate controls and preprocessing of the single cell data obtained (as detailed in Materials and Methods and Results and Fig. S7 in the supplemental material). We aimed at distinguishing detection biases resulting from nonspecific CT measurements of weakly expressed genes and valid CT measurements of genes showing low or absent expression in the respective individual cells. To do this, we first assessed the concordance of single-cell and bulk cell results by comparing the average CTbc (obtained from bulk cell replicates containing 20 cells per bulk cell sample) to the cumulative CT of single cells (cCTsc), a computed CT value corresponding to the sum of expression values from 20 single cells (see Fig. S7a in the supplemental material). Then, we determined a CT cutoff (CTmax) above which all CTs were regarded as nonspecific, and thus nonexpressed, in order to improve the global concordance of single- and bulk cell measurements (see Fig. S7b in the supplemental material). To determine a suitable CTmax, we evaluated two global measures of concordance, the absolute mean difference between the CTbc and the cCTsc (see Fig. S7b, red line, in the supplemental material) and the average root mean square deviation (RMSD; see Fig. S7b, blue line, in the supplemental material) for a range of CT cutoffs. This identified the CTmax as 23, the application of which removed nonspecific measurements and retained over 90% of all single-cell expression data, thus providing single-cell expression data that were highly concordant with the collected bulk cell data (Fig. 5a; see Fig. S7b, green line, and Materials and Methods in the supplemental material). Next, we performed box plot and statistical analyses of single-cell measurements using the log LRT, which simultaneously tests for differences in mean expression and the proportion of cells expressing a given gene (26) (see the supplemental material). This demonstrated that the differential, localization-dependent gene expression signatures largely mirrored the results obtained from bulk cells (Fig. 5b; see Fig. S7c and d in the supplemental material). However, we observed exceptions for some inflammatory genes under certain conditions, e.g., IL-8 expression in bystander cells, for which the cCTsc was strongly overestimated (see Fig. S7c and d in the supplemental material). This highlights the importance of taking single-cell measurements for transcriptional analysis of heterogeneous populations. In order to analyze the distribution of expression measurements over individual single cells and to obtain information about whether a detected differential gene expression results from differences between distinct subpopulations, we employed violin plot analysis. The CTsc values of some genes, e.g., NFKBIA and CXCL2, showed a multimodal distribution (Fig. 5b, arrows), suggesting the existence of subpopulations expressing these genes in different quantities. In contrast, the CTsc values of B2M and JunB were log-normally distributed around the mean (Fig. 5b, arrowheads), which points to the expected stochastic distribution of constant gene expression in single cells.
These findings raised the question of coordination of gene expression patterns upon bacterial infection within a single cell. To assess whether genes of entire pathways were regulated concomitantly within a cell, we carried out an analysis of inflammatory, apoptosis, and stress gene pairs to check for correlation within single cells (Fig. 5c to e). To do this, we computed both the Spearman's rank correlation coefficients and empirical P values of the absolute measured correlation for all possible pairs of genes for inflammation, apoptosis, or stress (Fig. 5c to e; see the supplemental material for further details of how correlation measurements were set up). Heat maps give information about the degree of increased (orange), decreased (blue), or unchanged (gray) correlation between all pairs of genes of a given pathway during individual infection stages (e.g., CO→BY or CO→VAC). This analysis revealed a strong increase of gene pair correlations of proinflammatory genes, but not of apoptosis or stress genes, at the bystander stage (CO→BY) upon infection with either Shigella WT or mutant strains (Fig. 5c). This indicates concomitant regulation of the respective gene pairs in bystander cells of only the inflammatory responses, which were independent of bacterial effectors. Furthermore, no concomitant coordination of inflammatory, apoptosis, or stress genes was measured in cells infected with WT Shigella at the vacuolar (CO→VAC) or cytosolic (CO→CYT and VAC→CYT) stages (Fig. 5c to e). In contrast, ΔospF infection led to a strong increase in correlations of proinflammatory gene pairs at both the vacuolar and cytosolic stages (Fig. 5c). Moreover, correlations also increased for gene pairs of apoptosis at the cytosolic stage and of stress response genes at the vacuolar stage during ΔospF strain infection (Fig. 5d and e). Together, this suggests that the bacterial effector OspF is capable of disrupting the potentially highly synchronized gene expression of proinflammatory, apoptosis, and stress genes in infected cells. Further, upon infection with ΔospG and ΔmxiE strains, correlations of apoptosis and stress genes strongly decreased at the cytosolic stage (Fig. 5d and e), indicating that the bacterial effectors OspG and MxiE maintain the concomitant expression of apoptosis and stress genes.
The bacterial effector OspF and infection of the cytosolic compartment reduce concomitant host gene expression.
We further investigated the impact of OspF on gene expression correlations of individual gene pairs of the proinflammatory host response (see Fig. S7e in the supplemental material). Heat maps showed the degree of correlation (green) or anticorrelation (purple) of expression of indicated gene pairs that showed concomitant expression for at least at one of the indicated conditions (CO, BY, VAC, or CYT). This revealed that upon WT infection, fewer gene pairs (7 pairs) were concomitantly expressed, and correlations were largely weakened or even lost at the cytosolic stage (see Fig. S7e, left, in the supplemental material). In contrast, more gene pairs (14 pairs) with concomitant expression were observed during ΔospF infection (see Fig. S7e, right, in the supplemental material). However, analogously to WT infection, we also observed a decrease or loss of significant correlation of most gene pairs at the cytosolic stage during ΔospF strain infection. This suggests that coordination of concomitant gene expression is weakened both by the bacterial effector OspF and upon cytosolic localization. Of note, the correlative expression pattern within a cluster of genes consisting of IL-8, CXCL-2, NFKBIA, IER3, CXCL-1, and cJun was considerably stronger upon ΔospF infection, underlining the targeted action of OspF in the disruption of this specific cluster of genes.
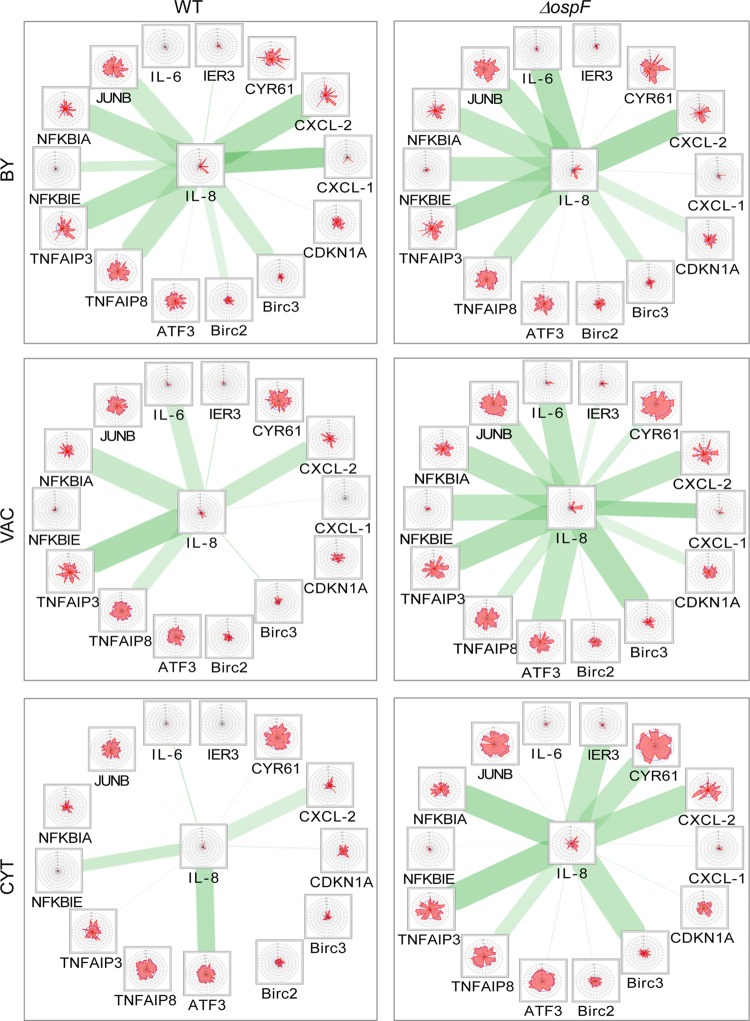
Since OspF has been localized to the IL-8 promoter (13), we performed network analysis of all gene pairs from our data set that show correlations with IL-8 (Fig. 6). Cytoscape analysis depicted expression data for the indicated genes in 40 single cells (shown as radar plots) and their degrees of correlation with IL-8 (indicated by the thickness of the green edges) at the indicated stages of WT or ΔospF strain infection. Surprisingly, this revealed strong correlations of IL-8 expression, not only with other inflammatory genes, but also with genes related to apoptosis or the stress response. This was evident within bystander cells upon both WT and ΔospF strain infection. In WT-infected cells, most of these correlations were strongly affected at the vacuolar stage and were mostly lost at the cytosolic stage (e.g., IL-8 and CXCL-2). In contrast, in ΔospF strain-infected cells, more and stronger gene pair correlations with IL-8 were observed at the vacuolar stage. This was followed by the loss of some correlations at the cytosolic stage (IL-8 and IL-6), while other correlations remained strong (e.g., IL-8 and TNFAIP3). This illustrates how both the bacterial effector OspF and the infection of the host cytosol have a disruptive role in the coordination of gene expression within the IL-8 network.
FIG 6.
The bacterial effector OspF and the cytosolic infection stage disrupt concomitant gene expression within the IL-8 network. The cytoscape networks show all the genes that have at least one significant correlation with IL-8 for at least one of the indicated stages of infection (BY, VAC, and CYT) with WT (left) or ΔospF (right) Shigella. The radar plots indicate expression threshold (ET) values (CTmax − CTsc; red spikes) and median expression (blue lines) corresponding to the magnitude of expression of the indicated genes within 40 individual cells from two independent experiments. Each radar plot corresponds to the expression of the respective gene in a single cell. The green edges represent the existence and strength of concomitant expression between the genes within the same cell connected by the edge.
Taking the data together, single-cell analysis revealed that Shigella targets the coordination of host gene expression within specific pathways during host cell invasion. Importantly, both the bacterial compartmental switch to the host cytosol and the bacterial effector OspF led to the disruption of coordinated gene expression patterns from proinflammation, apoptosis, and stress pathways.
DISCUSSION
In this study, we hypothesized that individual stages of bacterial infection lead to activation of distinct transcriptional signatures. We established a robust and comprehensive workflow to correlate different stages of S. flexneri infection with the activated host immune response signatures of two different epithelial cell lines. FACS-based single-cell sorting, multiplex qPCR analysis, and rigorous computational analysis at the bulk and single-cell levels enabled us to delineate how specific factors differentially act on host epithelial cells during the infectious process, i.e., (i) the bacterial invasion itself, (ii) pathogen localization, and (iii) the injected T3SS effectors.
We opted for medium-throughput multiplex qPCR, the gold standard for gene expression analysis. This proved to be highly favorable for reliable, sensitive, and reproducible comparison of representative “reporter” genes for specific gene families in intact single cells. Currently, powerful methodologies for transcriptional analysis from hundreds of genes (microarrays) or by whole transcriptomes (RNA-seq) in bulk cells exist. However, although single-cell RNA-seq is becoming more refined (27), it still remains challenging to get unbiased results for weakly expressed genes. We decided to measure transcriptional responses at 3 h p.i., as they gave robust gene expression values for most sorted bulk and single cells compared to earlier time points (data not shown). By that time, cells at different stages of infection, including the short-lived vacuolar phase, could be isolated due to the nonsynchrony of the invasion process. Using our previously developed FRET approach combined with fluorescent bacteria, we were able to distinguish these stages and could measure host responses at distinct stages of bacterial infection.
We deciphered distinct host immune signatures that correspond to individual stages of bacterial infection, i.e., the bystander, the vacuolar, and the cytosolic stages, thus confirming our hypothesis. These localization-dependent signatures were similar for both HeLa and polarized Caco-2 cells (Fig. 1; see Fig. S4 in the supplemental material), a trend that also appeared to be true for bystander responses, although we could not always isolate a reliable bystander population from Caco-2 cells.
The bystander response consisted of activated inflammatory, apoptosis, and stress response pathways and thus appeared more complex than anticipated (15, 16) (Fig. 2). Moreover, all genes induced in bystander cells were also markers of early intracellular infection. However, in contrast to host responses of infected cells, the onset and amplitude of the bystander response were independent of a subset of injected bacterial effectors. We propose that the bystander response is activated very early upon infection to mount a comprehensive host immune response that escapes the bacterial immune offensive strategies. None of the tested Shigella mutants, including the ΔmxiE strain, which does not express about half of the injected bacterial effectors, displayed an altered bystander response. Furthermore, bystander responses were shown previously to be induced by other bacteria, including Listeria monocytogenes (16). Therefore, it is very likely that bacterial invasion into cells is the event inducing a bystander response in neighboring, uninfected cells. This defines invasion as a pattern of pathogenesis, possibly discriminating pathogenic from nonpathogenic threats (Fig. 7). Internalization of pathogens as an inducer for bystander activation would allow epithelial cells to avoid premature and undue inflammation in response to nonpathogenic bacteria, such as commensals. It remains to be investigated if noninvading pathogens, such as enterohemorrhagic Escherichia coli (EHEC), induce such bystander responses. Moreover, according to our hypothesis of an early-onset bystander response, inflammatory activation occurred much earlier, and in a stronger fashion, in bystander than in infected cells. Thus, we propose, in line with previous reports (15, 16), that a signal upstream of the signaling pathways hijacked by bacterial effectors (e.g., MAPK and NF-κB) is propagated to noninfected cells to cause bystander activation. This signal might be either a bacterium-derived, intracellularly processed PAMP or a host-derived molecular mechanism generated upon bacterial internalization, such as cytoskeletal reorganization. Interestingly, during L. monocytogenes infection, which produces fewer actin rearrangements during invasion, less bystander activation was observed (16, 28). However, in contrast to previous studies that proposed a gap junction-mediated signal transfer to bystander cells, we observed bystander activation in cells without gap junctions (15, 16). This puts forward the existence of alternative signaling routes for the potential mediator. One candidate could be the rapid release and paracrine action of eATP, which is known to induce both inflammation and apoptosis responses (10). Together, our findings on the bystander population emphasize its role as a potent and integral part of the host epithelial cell immune response, and it can thus be considered an “immunological compartment” that differentially senses and exhibits distinct immune responses (Fig. 7). This bystander compartment may be a front-line host strategy to escape or to compensate for the effector-mediated attack in infected cells.
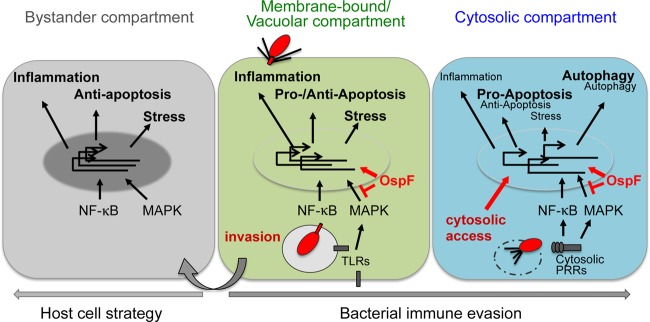
FIG 7.
Compartmentalized host epithelial immune response to S. flexneri infection. Epithelial cells infected with Shigella sense different patterns of pathogenesis (indicated in red) during internal danger, e.g., invasion, perturbation of cellular processes by bacterial effectors, or cytosolic access. This results in activation and differential modulation of multilayered host response gene expression (boldface, increased expression; smaller lightface, decreased expression) depending on the stage of bacterial infection, i.e., the vacuolar (green) and the cytoplasmic (blue) stages. The bacterial effector OspF impacts host signaling pathways and disrupts the coordination of concomitant gene expression as an important bacterial immune evasion strategy, most notably at the cytosolic stage of Shigella infection. Noninfected cells in proximity to infected cells (gray) sense bacterial invasion in the infected cells as a pattern of pathogenesis leading to concomitant expression of external-danger-regulated inflammatory, stress and antiapoptosis genes. Thus, bystander cells can be regarded as an immunological compartment (see the text for a detailed description).
The vacuolar and cytosolic stages were distinguished from the bystander stage through the modulation or additional activation of proinflammatory, pro- and antiapoptosis, stress, and repair genes (Fig. 7). Progress of the bacteria to the cytosolic compartment caused a distinct change in the transcriptional profiles of all tested responses (Fig. 1 and 3). Of note, as vacuolar rupture occurs within 5 to 15 min upon invasion, observed differences in gene expression are unlikely to result from differences in the duration of infection (8). This late stage of regulation included autophagy, stress, repair, and apoptosis genes. The cytosolic access of bacteria includes vacuolar-membrane damage, leakage of bacterial products, bacterial intracellular spread, and replication in the cytosol. Exploitation of the host cytoskeleton and nutrient starvation as a result of cytosolic access cause signaling changes due to host receptors, which detect bacterial MAMPs and DAMPs. These stimuli individually and differentially actuate the cytosolic recognition machinery, rather than membrane bound, leading to a number of host responses, including stress, autophagy, inflammation, and apoptosis, thus explaining our results, and as hypothesized (22, 23, 25, 29).
Systematic classification of gene expression profiles reflected functional pathways, which are differentially regulated depending on the stage of infection, on bacterial effectors, or on both (Fig. 3). In conjunction with the analysis of the bacterial-mutant-induced transcriptional signatures (Fig. 4), this sheds light on how the injected effectors impact the activated transcriptional signatures, depending on the bacterial localization. With respect to inflammatory genes, activation of a multilayered immune response was identified, which was differently affected by the cytosolic bacterial localization and by the bacterial effectors OspF and/or IpgD. First, the strong and the partial inhibition of inflammatory genes of profile 2 (e.g., IL-8) and of profile 1 (e.g., IL-6), respectively, by the bacterial effector OspF in infected cells identified their regulation via MAPK, and most likely NF-κB, signaling pathways (Fig. 7). Second, expression of a subset of these genes appeared to depend on the bacterial effector IpgD, suggesting their regulation via multiple signaling pathways. Third, infection with the ΔipgD strain revealed a further, previously underestimated inflammatory immune response mediated by the IRF3-IFN pathway, which was undermined during cytosolic WT Shigella infection. The molecular mechanisms of both the IpgD-dependent activation of inflammatory genes and the suppression of type I IFNs during cytosolic infection require further investigation. Our analysis on the localization-dependent inflammatory signaling during Shigella infection revealed the general shutdown of multiple signaling pathways during cytosolic infection. Moreover, this led us to the classification of externally and internally danger-regulated signaling pathways, which discriminate between general danger originating from Shigella infection in the environment or acute danger of direct infection. The former is represented by, e.g., MAPK and NF-κB signaling, which was activated in bystander and infected cells; the latter includes IRF3 signaling or apoptosis- and autophagy-regulating pathways activated only in infected cells. A large diversity of OspF-modulated genes was observed in this and other studies, suggesting direct and indirect effects on MAPKs and other substrates targeted by this effector (30). The identification of such novel non-MAPK-related OspF substrates is, however, beyond the scope of this study.
Our correlation analysis of single cells demonstrated that each of the bacterium-induced host transcriptional programs is highly coordinated within a cell (Fig. 5). We discovered that Shigella is able to disrupt this via the action of a single T3SS effector, OspF (Fig. 5 and 6). This occurred only within the boundaries of the infected cell and not in bystander cells (Fig. 7). Thus, our multiplex single-cell analysis brings novel insight into the relevance of compartmentalization to coordination of immune responses: (i) it highlights once more the important role of bystander cells as an immunological compartment that establishes and maintains a coordinated immune response to pathogenic bacteria, (ii) it suggests how bacterial effectors target the coordination of specific host gene expression programs, and (iii) it underscores the disruption of concomitant gene expression as a bacterial strategy to subvert host immune responses. In this context, OspF has been reported to alter chromatin recruitment of NF-κB to the IL-8 promoter (13). Nevertheless, our combined single- and bulk cell analysis suggests a larger impact of OspF on the modulations of transcriptional host responses. This is also supported by a study measuring its broad impact on the host protein phosphorylation status (30). Single-cell analysis has been crucial for the discovery of new subpopulations during early developmental stages (31) and of novel myeloid cell lineages (32), as well as for transcriptional correlation analysis during the cell cycle (33). Our work is the first report on transcriptional networks of bacterium-infected single cells highlighting how both distinct infection stages and bacterial effectors shape the compartmentalized, coordinated host immune response.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Centre Immunologie Humain (CIH) at the Institut Pasteur, particularly Milena Hasan and Valentina Libri. We thank all members of the DIHP group for helpful discussions, in particular Jennifer Fredlund for critical reading of the manuscript.
This work was supported by an ERC starting grant to Jost Enninga (Rupteffects, no. 261166), by the CARNOT Pasteur-MIE (program “ExpressGene”), and by the ANR (BioEmergence “GenExpress”). Juliane Lippmann was funded by an EMBO long-term postdoctoral fellowship and by a “Bourse Roux” fellowship from Institut Pasteur. Jost Enninga and Benno Schwikowski are members of the LabEx consortium IBEID, which funded part of Frederik Gwinner's work.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00574-15.
REFERENCES
- 1.Vance RE, Isberg RR, Portnoy DA. 2009. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Randow F, MacMicking JD, James LC. 2013. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science 340:701–706. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Blander JM, Sander LE. 2012. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol 12:215–225. doi: 10.1038/nri3167. [DOI] [PubMed] [Google Scholar]
- 5.Buchrieser C, Glaser P, Rusniok C, Nedjari H, de Hauteville H, Kunst F, Sansonetti P, Parsot C. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol 38:760–771. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- 6.Le Gall T, Mavris M, Martino MC, Bernardini ML, Denamur E, Parsot C. 2005. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology 151:951–962. doi: 10.1099/mic.0.27639-0. [DOI] [PubMed] [Google Scholar]
- 7.Ehsani S, Santos JC, Rodrigues CD, Henriques R, Audry L, Zimmer C, Sansonetti P, Van Nhieu GT, Enninga J. 2012. Hierarchies of host factor dynamics at the entry site of Shigella flexneri during host cell invasion. Infect Immun 80:2548–2557. doi: 10.1128/IAI.06391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ray K, Bobard A, Danckaert A, Paz-Haftel I, Clair C, Ehsani S, Tang C, Sansonetti P, Van Nhieu GT, Enninga J. 2010. Tracking the dynamic interplay between bacterial and host factors during pathogen-induced vacuole rupture in real time. Cell Microbiol 12:545–556. doi: 10.1111/j.1462-5822.2010.01428.x. [DOI] [PubMed] [Google Scholar]
- 9.Pendaries C, Tronchère H, Arbibe L, Mounier J, Gozani O, Cantley L, Fry MJ, Gaits-Iacovoni F, Sansonetti PJ, Payrastre B. 2006. PtdIns5P activates the host cell PI3-kinase/Akt pathway during Shigella flexneri infection. EMBO J 25:1024–1034. doi: 10.1038/sj.emboj.7601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puhar A, Tronchère H, Payrastre B, Van Nhieu GT, Sansonetti PJ. 2013. A Shigella effector dampens inflammation by regulating epithelial release of danger signal ATP through production of the lipid mediator PtdIns5P. Immunity 39:1121–1131. doi: 10.1016/j.immuni.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 11.Pruneda JN, Smith FD, Daurie A, Swaney DL, Villén J, Scott JD, Stadnyk AW, Le Trong I, Stenkamp RE, Klevit RE, Rohde JR, Brzovic PS. 2014. E2∼Ub conjugates regulate the kinase activity of Shigella effector OspG during pathogenesis. EMBO J 33:437–449. doi: 10.1002/embj.201386386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Dong N, Hu L, Shao F. 2013. The Shigella type three secretion system effector OspG directly and specifically binds to host ubiquitin for activation. PLoS One 8:e57558. doi: 10.1371/journal.pone.0057558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, Muchardt C, Parsot C, Sansonetti PJ. 2007. An injected bacterial effector targets chromatin access for transcription factor NF-κB to alter transcription of host genes involved in immune responses. Nat Immunol 8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou JM, Shao F. 2007. The phosphothreonine lyase activity of a bacterial type III effector family. Science 315:1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- 15.Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. 2013. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasper CA, Sorg I, Schmutz C, Tschon T, Wischnewski H, Kim ML, Arrieumerlou C. 2010. Cell-cell propagation of NF-κB transcription factor and MAP kinase activation amplifies innate immunity against bacterial infection. Immunity 33:804–816. doi: 10.1016/j.immuni.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 17.Kim DW, Lenzen G, Page A-L, Legrain P, Sansonetti PJ, Parsot C. 2005. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A 102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mavris M, Page A-L, Tournebize R, Demers B, Sansonetti P, Parsot C. 2002. Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol Microbiol 43:1543–1553. doi: 10.1046/j.1365-2958.2002.02836.x. [DOI] [PubMed] [Google Scholar]
- 19.Allaoui A, Ménard R, Sansonetti PJ, Parsot C. 1993. Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect Immun 61:1707–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allaoui A, Sansonetti PJ, Parsot C. 1993. MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri lpa invasins. Mol Microbiol 7:59–68. doi: 10.1111/j.1365-2958.1993.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 21.Sorensen M, Lippuner C, Kaiser T, Misslitz A, Aebischer T, Bumann D. 2003. Rapidly maturing red fluorescent protein variants with strongly enhanced brightness in bacteria. FEBS Lett 552:110–114. doi: 10.1016/S0014-5793(03)00856-1. [DOI] [PubMed] [Google Scholar]
- 22.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, Enninga J. 2012. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog 8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, Carneiro LAM, Yang C, Emili A, Philpott DJ, Girardin SE. 2012. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11:563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 24.Pillich H, Loose M, Zimmer KP, Chakraborty T. 2012. Activation of the unfolded protein response by Listeria monocytogenes. Cell Microbiol 14:949–964. doi: 10.1111/j.1462-5822.2012.01769.x. [DOI] [PubMed] [Google Scholar]
- 25.Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T, Cossart P. 2011. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem 286:26987–26995. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, Gottardo R. 2013. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, Schwartz S, Fowler B, Weaver S, Wang J, Wang X, Ding R, Raychowdhury R, Friedman N, Hacohen N, Park H, May AP, Regev A. 2014. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pizarro-Cerda J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Casson CN, Copenhaver AM, Zwack EE, Nguyen HT, Strowig T, Javdan B, Bradley WP, Fung TC, Flavell RA, Brodsky IE, Shin S. 2013. Caspase-11 activation in response to bacterial secretion systems that access the host cytosol. PLoS Pathog 9:e1003400. doi: 10.1371/journal.ppat.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmutz C, Ahrne E, Kasper CA, Tschon T, Sorg I, Dreier RF, Schmidt A, Arrieumerlou C. 2013. Systems-level overview of host protein phosphorylation during Shigella flexneri infection revealed by phosphoproteomics. Mol Cell Proteomics 12:2952–2968. doi: 10.1074/mcp.M113.029918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. 2010. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Dong S, Maiella S, Xhaard A, Pang Y, Wenandy L, Larghero J, Becavin C, Benecke A, Bianchi E, Socié G, Rogge L. 1 July 2013. Multi-parameter single-cell profiling of human CD4+FOXP3+ regulatory T cell populations in homeostatic conditions and during graft-versus-host disease. Blood doi: 10.1182/blood-2013-02-482539. [DOI] [PubMed] [Google Scholar]
- 33.Zopf CJ, Quinn K, Zeidman J, Maheshri N. 2013. Cell-cycle dependence of transcription dominates noise in gene expression. PLoS Comput Biol 9:e1003161. doi: 10.1371/journal.pcbi.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.