Abstract
Human monoclonal antibody CH58 isolated from an RV144 vaccinee binds at Lys169 of the HIV-1 Env gp120 V2 region, a site of vaccine-induced immune pressure. CH58 neutralizes HIV-1 CRF_01 AE strain 92TH023 and mediates ADCC against CD4 + T cell targets infected with CRF_01 AE tier 2 virus. CH58 and other antibodies that bind to a gp120 V2 epitope have a second light chain complementarity determining region (LCDR2) bearing a glutamic acid, aspartic acid (ED) motif involved in forming salt bridges with polar, basic side amino acid side chains in V2. In an effort to learn how V2 responses develop, we determined the crystal structures of the CH58-UA antibody unliganded and bound to V2 peptide. The structures showed an LCDR2 structurally pre-conformed from germline to interact with V2 residue Lys169. LCDR3 was subject to conformational selection through the affinity maturation process. Kinetic analyses demonstrate that only a few contacts were responsible for a 2000-fold increase in KD through maturation, and this effect was predominantly due to an improvement in off-rate. This study shows that preconformation and preconfiguration can work in concert to produce antibodies with desired immunogenic properties.
Keywords: HIV-1, Gp120, V2, RV-144, Antibody maturation, Germline, Unmutated ancestor
Graphical abstract
Highlights
-
•
With only 2-3% mutation from germline, the HIV-1 antibody CH58 developed neutralizing and ADCC capabilities.
-
•
The LCDR2 Glu–Asp motif of the RV144 antibody CH58 is pre-conformed from germline to interact with the gp120 V2 loop.
-
•
Affinity and neutralization gains resulted from tuning local interactions rather than gross sequence or structure changes.
-
•
Structural analyses show the second light chain complementarity determining region Glu–Asp motif of the CH58 antibody isolated from an RV144 vaccinee is optimally pre-conformed from germline to interact with the gp120 V2 loop. The increased binding affinity and neutralization capacity of the mature antibody compared to its germline precursor were achieved with only 2–3% mutation from germline, and the fact that these gains appeared to be a result of the tuning of local interactions rather than gross sequential or conformational changes provides hope that a rational immunogen design for HIV-1 treatment may become a reality.
1. Introduction
The RV144 human immunodefiency virus (HIV) vaccine efficacy trial utilized a regimen combining recombinant canarypox vector ALVAC-HIV (i.e. expressing HIV type 1 (HIV-1) Gag, Pro and membrane-linked gp120 (vCP1521)) with HIV clades B and E gp120 envelope (Env) glycoprotein (Rerks-Ngarm et al., 2009), and showed an estimated vaccine efficacy of 31.2% (Rerks-Ngarm et al., 2009). Gp120 V1V2 antibodies were correlates of decreased transmission risk for RV144 (Haynes et al., 2012), primarily due to V2 antibodies (Karasavvas et al., 2012), and high levels of antibody dependent cellular cytotoxicity (ADCC) in the absence of high Env IgA levels were inversely correlated with infection risk (Haynes et al., 2012). A molecular sieve analysis demonstrated Lys169 within the V1V2 region to be a site of immune pressure with an estimated 48% vaccine efficacy when the challenging HIV strain matched the vaccine at this position (Rolland et al., 2012).
As a result, the gp120 V1V2 region has been of intense interest owing to its immunogenic promise; however, V1V2 has proven difficult to characterize structurally. V1V2 has been observed in crystal structures in a variety of conformations. Bound to antibodies CH58 and CH59 isolated from RV144 vaccinees, V2 peptides demonstrated helix and loop-helix conformations respectively (Liao et al., 2013). When presented in the context of a scaffold, the antibody-bound V1V2 has adopted stranded conformations, notable not only because of the differing conformation but also because some of these antibodies were broadly neutralizing (McLellan et al., 2011, Pancera et al., 2013). The stranded V2 conformation was recapitulated in pre-fusion Env trimer structures (Julien et al., 2013, Pancera et al., 2014); however, the predominant responses to it as an antigen have been with glycan V3 antibodies (Pancera et al., 2014). Thus interest remains in directing an immune response against V1V2.
The V2 antibody CH58 has been biochemically and immunologically characterized (Liao et al., 2013). CH58 mediates ADCC against tier 2 virus-infected CD4+ T cell targets (Haynes et al., 2012), has a footprint at a site of immune pressure (gp120 Lys169) (Rolland et al., 2012), and like PG9 and CH01 (Liao et al., 2013, McLellan et al., 2011) binds the gp120 V2 region involving Lys169. CH58 cross-blocks binding of PG9 and CH01 V1V2 broadly neutralizing antibodies (bNAbs) to Env, but CH58 does not have bnAb activity and only neutralized the tier 1 HIV-1 CRF_01 AE strain 92TH023 that was included in ALVAC as a prime vaccine immunogen (Liao et al., 2013, McLellan et al., 2011, Rerks-Ngarm et al., 2009, Walker et al., 2009). Importantly, the germline precursor to CH58 has been inferred and shows a relatively low number of mutations leading to its functions: only 11 mutations occurred from germline to mature antibody in both light and heavy (Fab component) chains (Liao et al., 2013). Furthermore, CH58 has been found to bear a key Glu–Asp (ED) pair in its second light chain complementarity determining region (LCDR2) (Liao et al., 2013) that is conserved among humans and rhesus macaques (Wiehe et al., in press). This ED motif is predominantly present in V2 antibodies where the negatively charged Glu and Asp side chains may form salt bridges with the positively charged Lys and Arg side chains prevalent in variants of the V2 sequence when presented as a linear epitope such as CH59, HG107, and HG120 (Liao et al., 2013, Wiehe et al., in press). Moreover, the presence of this ED motif within the preconformed LCDR2 suggests a pathway in which germline antibodies bearing it can recognize basic residues within V2 (North et al., 2011, Wiehe et al., in press).
The relatively low number of mutations between the inferred, unmutated ancestor of CH58 (CH58-UA) and the mature CH58 suggested this relationship as ripe for study in the area of antibody lineages. In terms of structure, it has been established that germline precursors tend to display greater flexibility in their idiotopes than mature antibodies, the rationale being that evolution tends to favor greater diversity in immune responses so that it can react to new threats (Foote and Milstein, 1994). Even through the maturation process, mature antibodies can exist in an equilibrium of two or more structural isomers, only one of which is the binding conformer; stated differently, mutations that accumulate during antibody maturation can work cooperatively to narrow conformational ensembles (Foote and Milstein, 1994, Manivel et al., 2000, Zimmermann et al., 2006). In support of the notion of promiscuous germline antibodies, it has been specifically found that germline antibodies have HCDR3 sequences capable of adopting a wide range of conformations (Babor and Kortemme, 2009). However, this cannot be taken to mean that mutations impacting immunogenicity must necessarily occur within CDRs. In fact mutations that impact functionality have been shown to occur in any of the CDRs as well as in the IgG framework regions (Fera et al., 2014, Klein et al., 2013). Recently, it has been demonstrated that antibody paratopes with CDRs preconfigured (rigidified) through the maturation process are likely to experience large affinity gains through maturation (Schmidt et al., 2013). The mechanism for that particular study (the CH65-67 lineage of influenza HA antibodies) was rigidification of the antibody paratope through key somatic mutations at the HCDR3 anchorpoints (Schmidt et al., 2013).
Examining the detailed kinetics found within antibody affinity maturation, seminal work in the hapten system showed that the association rate rather than affinity was more important in affinity maturation (Foote and Milstein, 1994). In agreement with this, a study of various antibodies from early primary and secondary responses suggested that antibody maturation was a process driven to improve the association but not dissociation step of binding (Manivel et al., 2000). Recently, the study of the CH65-67 lineage of influenza HA antibodies demonstrated a 30–40-fold increase in association rate with maturation and a decrease in dissociation rate of significantly lesser magnitude (Schmidt et al., 2013, Xu et al., 2014). There are also cases where improved dissociation rates are the primary factor observed in affinity gains associated with antibody maturation. This scenario has the benefit of being weighted toward antibodies with flexible domains and HIV antibodies in particular. Study of the evolution of the antifluorescein antibody 4-4-20 showed that the paratope was localized from a heterogeneous ensemble of conformations to a single conformation by introducing mutations that acted cooperatively and over significant distances to rigidify the protein (Zimmermann et al., 2006). Kinetic analysis showed a modestly higher improvement in the dissociation rate of this Ab–Ag complex compared to concomitant improvement in association rate. Also, VH mutations were found that introduced H-bonding between Ab and Ag that could account for this anchoring effect. Second, kinetic analysis using a group of anti-(4-hydroxy-3-nitrophenyl)acetyl monoclonal antibodies from within a single lineage revealed that whereas association rates changed at most by a factor of two through affinity maturation, dissociation rates decreased by over an order of magnitude (Sagawa et al., 2003). Recent work on antibodies against IFNβ (a potential side effect of treatment with IFNβ for multiple sclerosis) showed that sera containing sustained neutralizing antibodies (NAbs) demonstrated significantly higher binding responses and slower dissociation rates than sera containing transient NAbs and that the anti-IFNβ antibody maturation response acted over time to decrease dissociation rates thus increasing relative antibody binding affinity (Gibbs et al., 2014, Gibbs and Oger, 2008). Importantly, kinetic analysis of IgG framework region reversion mutations in HIV-1 bNAbs VRC01, NIH45-46, 12A21, and 3BNC117 (Scheid et al., 2011, Wu et al., 2011, Zhou et al., 2010, Zhou et al., 2013) showed that the loss of affinity appeared to be primarily due to an increased dissociation rate (Klein et al., 2013). Lastly, the same effect was shown for the unmutated ancestor of the anti-gp41 MPER antibody 2F5 (Alam et al., 2011).
Given the fact that CH58 is only modestly mutated from its inferred ancestor CH58-UA, the pair presented itself as an ideal study in antibody lineages to explore these issues of structural rigidity, preconfiguration, preconformation, and binding affinity improvements. In addition, interest in CH58 and similar antibodies against linear V2 epitopes remains high because even though their neutralization capacities are limited, they are easy to induce (Haynes et al., 2012, Liao et al., 2013, Rerks-Ngarm et al., 2009, Wiehe et al., in press); whereas broadly neutralizing antibodies are not induced by current HIV-1 vaccine candidates (Mascola and Haynes, 2013). For this study, we have determined the crystal structures of CH58-UA in both its unliganded state and in complex with a gp120 peptide representing the nominal V2 epitope. Comparative analysis of these structures in concert with likewise unliganded and liganded mature CH58 structures showed that of the 11 total mutations from unmutated ancestor, nine mutations occurred in CDRs and only a subset of those show any direct impact on contacts with antigen. Importantly, our kinetic analyses those few contacts were responsible for a 2000-fold increase in KD for the mature CH58 over CH58-UA, and this effect was predominantly due to a largely decreased off-rate that could be related back to specific contacts gained by virtue of just a few functionally important mutations.
2. Materials & methods
2.1. Protein production
The Fab fragment of CH58-UA was produced recombinantly as previously described (Nicely et al., 2010). In short, Fab chains were generated by PCR using light and heavy chain genes as templates with appropriate primer pairs, and cloned into pcDNA3.1/hygro (Liao et al., 2006). Recombinant Fabs were produced in 293F cells by transient transfection then purified using methods described previously (Nicely et al., 2010). For structural and SPR studies, after affinity capture using LambdaCapture (BAC), the Fab was further purified via gel filtration chromatography using a HiLoad 26/60 Superdex 200 pg 26/60 column at 2 mL/min with a buffer of 10 mM Hepes pH 7.2, 50 mM NaCl, 0.02% NaN3. Fab peak elution fractions were pooled and exchanged into ddH2O via five dilute-concentrate cycles and brought to final concentrations of 15.0 mg/mL.
Gp120165–186 (LRDKKQKVHALFYKLDIVPIED) wild-type and Ala-substituted peptides were originally synthesized for ELISA by CPC Scientific, and a similar peptide albeit with a biotin tag (gp120166–186; “171”) was synthesized for SPR experiments. Gp120165–182 (LRDKKQKVHALFYKLDIV), gp120165–182 Leu179Ala (LRDKKQKVHALFYKADIV) and gp120165–182 Leu179Ala,Ile181Ala (LRDKKQKVHALFYKADAV) were synthesized with C-terminal linkers (− GGGK–) and biotin tags for BLI experiments, and the best binding of the three synthesized with an acetylated N-terminus and amidated C-terminus for crystallography to be used to form a complex with CH58-UA where it was dissolved in neat DMSO to 100 mg/ml then added to Fab in solution at a 1:3 Fab:peptide molar ratio prior to setting of crystallization experiments.
2.2. Surface plasmon resonance (SPR)
Epitope mapping experiments in SPR were performed on a BIAcore 4000 (BIAcore Inc, Piscattaway, NJ) instrument at 25 °C. Using a Series S CM5 chip, wild-type and Ala-substituted peptides were amine-coupled directly on the chip surface. Data analyses were performed using the Biacore 4000 evaluation and BIAevaluation 4.1 software (BIAcore) as previously described (Alam et al., 2007, Alam et al., 2011). Monoclonal antibodies were used in these experiments. Binding responses of the irrelevant respiratory syncytial virus (RSV) antibody Synagis were used to subtract out responses due to non-specific interactions as standard good practice.
2.3. BioLayer interferometry (BLI)
BLI measurements were made using a ForteBio OctetRed 96 instrument and streptavidin sensors at 25 °C. Data analyses were performed using ForteBio data analysis 7 software. The gp120165–182 peptide sensors were prepared by dipping streptavidin sensors into wells containing biotinylated gp120165–182 peptides (5 μg/ml) for 300 s. The peptide-loaded sensors were washed in PBS buffer (pH 7.4) for 60 s before obtaining baseline. In the screening experiments, binding of the CH58-UA and mature CH58 Fabs (at 10 μg/ml) to gp120165–182 peptide (wild-type and Ala mutants) sensors were monitored for 300 s and the dissociation was followed for 300 s. The influenza hemagglutinin-specific antibody CH65 was used as negative control to subtract out binding due to non-specific interactions with sensors as standard good practice. The binding signal during the last 20 s of the dissociation phase was averaged and used to calculate normalized binding. The affinity measurements of CH58-UA and mature CH58 Fabs to the wild-type and Leu179Ala,Ile181Ala mutant peptides were carried out by performing binding titrations (Fab concentrations ranged from 1 to 20 μg/ml). The gp41 MPER specific 13H11 Fab (1–20 μg/ml) binding to the wild-type and Leu179Ala,Ile181Ala mutant peptides sensors was used in parallel to subtract out binding due to non-specific interactions with the sensors. The subtracted binding curves were fitted globally to a 1:1 binding model to obtain association (ka), dissociation (kd) rate constants and the apparent dissociation constant (KD).
2.4. Circular dichroism spectroscopy
The circular dichroism (CD) spectra of the WT and mutant gp120165–182 peptides were measured on an Aviv model 202 spectropolarimeter using a 1 mm path length quartz cuvette. The peptides were solubilized in PBSF buffer (19 mM phosphates pH 7.4, 100 mM NaF) at 0.1 mg/ml concentration and CD spectra were recorded at 25 °C. Three scans of the CD spectra of each peptide were averaged and the CD signal from PBSF buffer was subtracted out.
2.5. Crystallography
Unliganded CH58-UA Fab was crystallized in a low ionic strength screen (Harris et al., 1995) in a drop composed of 0.3 μl 15 mg/ml protein plus 0.3 μl 25 mM MES pH 6.0, 1.5% PEG 3350 over a reservoir of 50 μl 24% PEG 3350 at 20 °C. These crystals were cryoprotected using a solution of 30% PEG 3350, 30% ethylene glycol. Crystals of CH58-UA Fab–gp120165–182 Leu179Ala,Ile181Ala were observed over a reservoir of 0.2 M calcium acetate hydrate, 20% PEG 3350 in a drop composed of 0.2 μl 12 mg/ml protein plus 0.4 μl reservoir. These crystals were cryoprotected using a solution of 25% PEG 3350, 30% ethylene glycol. Diffraction data were collected at SER-CAT with an incident beam of 1 Å in wavelength. Datasets were reduced in HKL-2000 (Otwinowski and Minor, 1997). Matthews analysis suggested two Fabs in the asymmetric unit of the unliganded CH58-UA structure and only one in that of the liganded CH58-UA structure (Matthews, 1968). The structures were phased by molecular replacement in PHENIX (Terwilliger et al., 2008) using as source models the mature CH58 Fab structure (Liao et al., 2013). Rebuilding and real-space refinements were done in Coot (Emsley et al., 2010) with reciprocal space refinements in PHENIX (Adams et al., 2010) and validations in MolProbity (Lovell et al., 2003).
2.6. Accession codes
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 4RIR (CH58-UA), and 4RIS (CH58-UA complex).
3. Results
3.1. Increased KD with maturation was primarily due to off-rate improvement
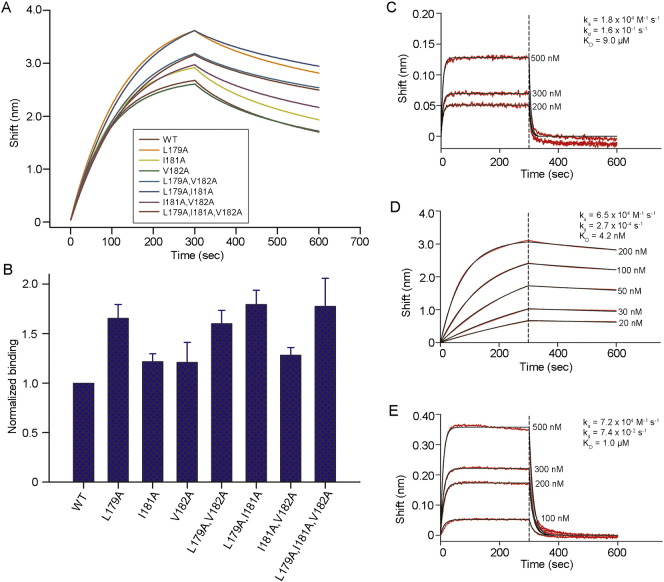
The nominal epitope for the CH58-UA antibody was mapped in surface plasmon resonance (SPR) using AE.CM244V2-171 peptide (gp120165–186 = LRDKKQKVHALFYKLDIVPIED) and its sequentially Ala-substituted derivatives (Fig. 1A). The results agreed with those seen for the mature CH58 (Fig. 1B) except that some Ala substitutions produced notably higher binding responses to CH58-UA (Liao et al., 2013). The three V2 residues that conferred the strongest binding to CH58-UA when mutated to Ala were Leu179 (a ~ 6.1-fold increase in the SPR binding response compared to wild-type peptide), Ile181 (a ~ 3.9-fold increase in the same), and Val182 (a ~ 3.4-fold increase in the same).
Fig. 1.
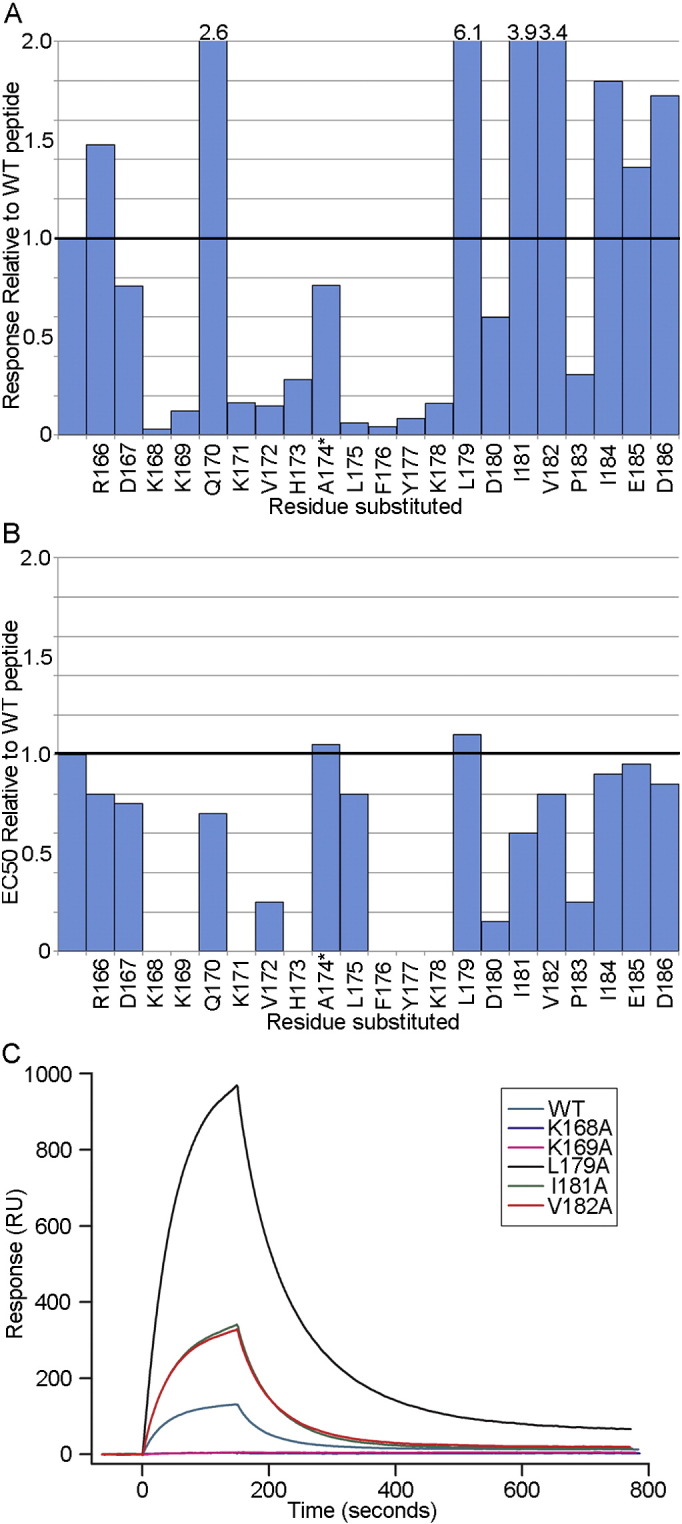
Epitope mapping of CH58-UA and CH58 mAbs. A) The epitope for the CH58-UA monoclonal antibody was mapped in SPR using sequentially Ala-substituted AE.CM244V2-171 peptides (gp120166–186). The response from each experiment was normalized to that of wild-type peptide for comparative purposes. The graph is truncated at a two-fold response ratio to emphasize losses of binding. The substitutions that gave responses above two-fold are specified over their columns. The asterisk next to Ala174 indicates that it was substituted with Glu to distinguish that peptide from wild-type and to impart a contrasting charge character to the predominantly positively charged V2 peptide N-terminal region. B) The epitope for the mature CH58 mAb was previously mapped in ELISA (Liao et al., 2013) and is included here for comparative purposes. C) Example SPR sensograms are shown for some of the Ala-substituted peptides in the CH58-UA study. Wild-type peptide binding levels were taken into account to normalize data for presentation in this single plot.
The SPR epitope mapping data served as a basis for a second round of targeted Ala mutations. The Leu179Ala, Ile181Ala, and Val182Ala mutations were screened singly and in combinations using biolayer interferometry (BLI) with a modestly shorter gp120165–182 peptide to identify improved potential binding partners for a liganded CH58-UA structure (Fig. 2, Tables S1–S3). SPR was performed with monoclonal antibodies whereas BLI was performed with Fabs. Fabs are known to display weaker binding to antigens compared to mAbs owing to avidity effects. Thus it was important to include experiments of mature CH58 (mAb or Fab as appropriate) binding wild-type peptide to serve as a point of reference. In our BLI experiments, we observed an average KD value of 4.6 nM for wild-type gp120 peptide binding mature CH58 compared to a 11.0 μM KD for the same peptide binding to the CH58-UA (Table S1). This represented an approximate 2000-fold gain in binding with maturation. In detail, the association rate (ka) for wild-type gp120 peptide in binding mature CH58 had an average value of 6.6 × 104 M− 1 s− 1 compared to one of 1.6 × 104 M− 1 s− 1 for CH58-UA with the same peptide, representing a modest 4-fold gain of on-rate with maturation (Table S2). Compare that to the dissociation rate (kd) for wild-type peptide with mature CH58 of 3.0 × 10− 4 s− 1 versus one of 1.7 × 10− 1 s− 1 for the same peptide with CH58-UA, a ~ 600-fold decrease in off-rate. Thus the improved KD values from CH58-UA to mature antibody were predominantly a result of decreasing off-rates.
Fig. 2.
Binding of CH58-UA and CH58 to wild-type and Ala-substituted gp120165–182 peptides. A) Results from BLI showing binding between CH58-UA Fab fragment and gp120165–182 V2 peptide variants. B) The shift in wavelength during the last 20 s of the dissociation phase of CH58-UA binding to the peptides shown in panel A was averaged and normalized to that of wild type peptide for comparative purposes. The error bars denote the standard deviation in the values determined from triplicate measurements. C) Representative BLI sensograms are shown for detailed kinetic experiments with wild-type gp120165–182 V2 peptide and CH58-UA Fab. Likewise, D) mature CH58 Fab with wild-type peptide and E) CH58-UA Fab with gp120165–182 Leu179Ala,Ile181Ala. In these experiments, the gp41 MPER-specific 13H11 Fab was used as negative control to subtract non-specific binding to the peptide as standard good practice. The ka, kd and derived KD values were obtained by global fitting of specific binding sensograms to a 1:1 binding model. Example measurements are reported in these panels. Average KD values are reported in the text for duplicate (CH58-UA Fab) and triplicate (CH58 Fab) measurements.
In terms of finding a peptide suitable for structural work with CH58-UA, the double Ala mutant Leu179Ala,Ile181Ala conferred the largest binding enhancement to CH58-UA (Fig. 2B). The average association rate for this mutant peptide with CH58-UA was 9.5 × 105 M− 1 s− 1 compared to one of 1.6 × 104 M− 1 s− 1 for wild type peptide, representing a 60-fold increase in on-rate (Table S3). In contrast, the average dissociation rates were 7.2 × 10− 2 s− 1 for CH58-UA with the mutant peptide and 1.7 × 10− 1 s− 1 for wild-type peptide, indicating a mere 2-fold decrease in off-rate. However, the enhanced on-rate resulting from these two mutations manifested in an average KD of 0.9 μM compared to one of 11.0 μM for the wild-type peptide. This approximate ~ 10-fold increase in binding was sufficient to generate a complex suitable for crystallography. To examine whether the enhanced affinity of the Leu179Ala,Ile181Ala double mutant might be due to any change toward preferred structure of the peptide itself, we recorded the circular dichroism (CD) spectra of wild-type gp120165–182, gp120165–182 Leu179Ala and gp120165–182 Leu179Ala,Ile181Ala peptides (Fig. 3). All three peptides exhibited similar CD profiles. Therefore, the selected mutations (single and double) did not appear to affect peptide secondary structure in solution. Lastly, an alignment of 4907 Env sequences from the LANL HIV sequence database (www.hiv.lanl.gov) showed that alanines at the 179, 181 and 182 positions in the V2 region are rare or non-existent with frequencies of 0.77% (38/4907), 0% (0/4907) and 1.61% (79/4907), respectively.
Fig. 3.
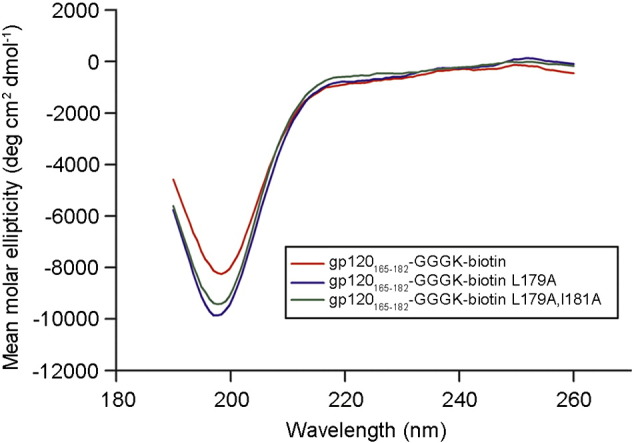
CD data for gp120165–182 peptide and two of its Ala-substituted variants. CD spectra for the gp120165–182 peptide and its Ala-substituted variants Leu179Ala and Leu179Ala, Ile181Ala.
3.2. The ED motif is part of a pre-configured LCDR2
The crystal structures of unliganded CH58-UA Fab and CH58-UA Fab in complex with gp120165–182 Leu179Ala,Ile181Ala peptide were solved by molecular replacement and refined to resolutions of 2.5 and 2.3 Å, respectively (Table 1). The statistics for model refinements were typical of other crystal structures at these resolutions (Urzhumtseva et al., 2009). Notably, HCDR3 was ordered in the liganded CH58 structure and disordered in the unliganded CH58 structure (Liao et al., 2013); in comparison, HCDR3 was ordered in the liganded CH58-UA structure though in a different conformation than that seen with the mature CH58. HCDR3 was also ordered in the unliganded CH58-UA structure although it appeared that crystal contacts played a role in helping to stabilize the observed conformation. The HCDR3 conformational difference was reflected in the RMSD values for pairwise structural comparisons: the RMSD between the Fv fragments of CH58-UA and mature CH58 was 0.284 Å, whereas the RMSD between the unliganded and liganded CH58-UA structures was 0.963 Å.
Table 1.
Data collection and refinement statistics for the structures.
| Structure | CH58-UA | CH58-UA–gp120165–182 Leu179Ala,Ile181Ala |
|---|---|---|
| Data collection | ||
| Space group: | P21a | P21 |
| Cell dimensions | ||
| a, b, c (Å): | 53.7, 103.7, 101.2 | 55.7, 53.9, 73.6 |
| α, β, γ (°): | 90.0, 100.5, 90.0 | 90.0, 108.9, 90.0 |
| Resolution (Å): | 50–2.5 (2.54–2.50) | 50–2.3 (2.34–.30) |
| Rmerge (%): | 13.1 (42.6)b | 11.4 (36.6) |
| < I/σ > | 7.9 (2.0) | 19.0 (3.4) |
| Completeness (%): | 97.4 (93.9) | 99.9 (99.5) |
| Redundancy: | 3.3 (3.0) | 6.1 (5.2) |
| Refinement | ||
| Total # reflections: | 36,924 | 18,762 |
| Unique # reflections: | 33,979 | 18,258 |
| Rwork/Rfree (%): | 22.0/28.9 (30.8/43.0) | 16.1/24.6 (19.3/31.4) |
| Average B factor (Å2): | 26.35 | 30.25 |
| Nonhydrogen atomsc: | 6578 | 3461 |
| Water molecules: | 112 | 239 |
| R.M.Scuu deviations | ||
| Bond lengths (Å): | 0.005 | 0.007 |
| Bond angles (°): | 0.943 | 1.136 |
| ψ, φ favored (%): | 96.6 | 96.2 |
| ψ, φ allowed (%): | 3.0 | 3.4 |
| zψ, φ outlying (%): | 0.4 | 0.5 |
This crystal had two Fabs in the asymmetric unit.
Values in parentheses are for the highest resolution shells.
Not including water molecules.
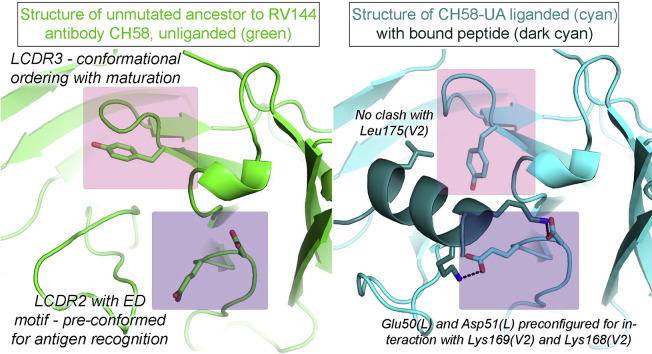
The conserved ED motif is present in LCDR2 of both the CH58-UA and CH58 antibodies at residue positions Glu50 and Asp51 (Figure S1). LCDR2 adopted the same overall conformation in the unliganded and liganded forms of both CH58-UA and CH58 (Fig. 4). While LCDR2 is the least structurally variable of the six CDRs (North et al., 2011), the ED motif was found to occur in only two human germline light chain sequences, both Vλ (3–10 and 6–57), and both bearing the ED motif at one amino acid position into the LCDR2 region (xEDxxxxx). This positions both side chains to orient outward from the core IgG fold and into the antibody paratope (Fig. 4A). In terms of specific interactions, the CH58-UA liganded structure showed a salt bridge of 2.8 Å between Lys169(V2) and Glu50(L) (Fig. 4B). The amine functional group on the Lys169 side chain additionally found a favorably electronegative environment for binding courtesy of Tyr32 in LCDR1 and Asp100B, Ser100C, Ser100D, and the backbone carbonyl of Gly100E in HCDR3. Asp51(L) formed a salt bridge with Lys168(V2) also 2.8 Å in length (Fig. 4B), and likewise the amine functional group of the Lys168(V2) side chain found favorable electrostatic interactions with the carbonyls of Ala29(L) and Asn31(L) in LCDR1. None of these LCDR1 and LCDR2 interactions changed appreciably in the mature CH58 complex (Fig. 4B) (Liao et al., 2013).
Fig. 4.
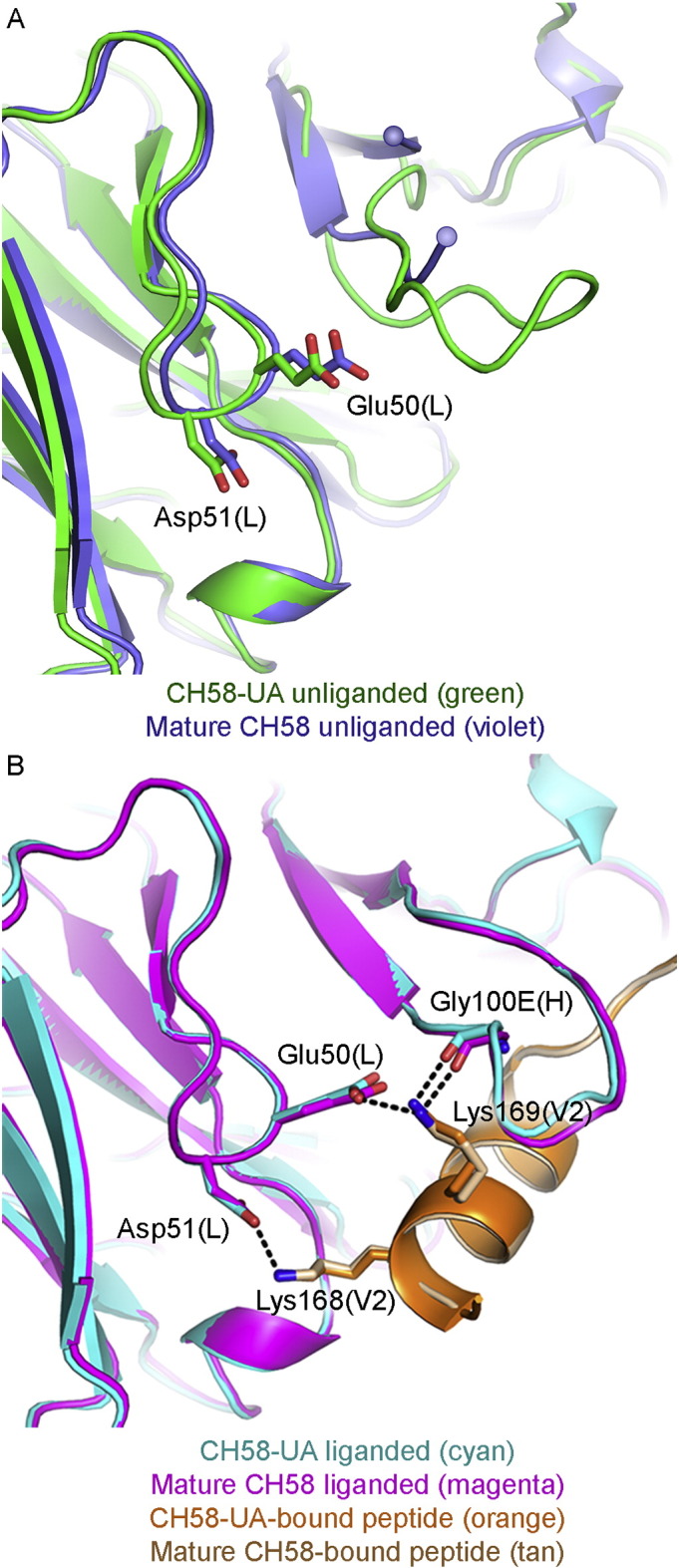
LCDR2 and ED motif interactions. A) The unliganded structures of CH58-UA and mature CH58 (with superimposed V2 peptide) showed that the Lys169 salt bridge with Glu50 in LCDR2 was pre-configured. In contrast, the disordered HCDR3 (indicated by spheres) offered no stable contacts. B) The liganded CH58-UA and mature CH58 structures showed that the hydrogen bond between Gly100E(H) and Lys169(V2) required ordering of HCDR3, whereas the ability to form the salt bridge between Glu50(L) in LCDR2 and Lys169(V2) was pre-conformed in CH58-UA.
3.3. Specific antibody-antigen interactions were gained through maturation
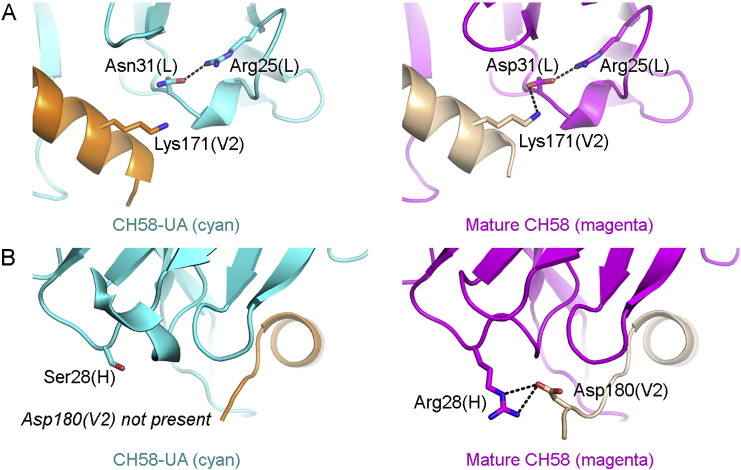
In total, there were five key salt bridges between antibody and peptide in the liganded CH58 structure (Liao et al., 2013). Three of these salt bridges – Lys168(V2) to Asp51(L), Lys169(V2) to Glu50(L), and Lys178(V2) to Asp54(H) – were present in the liganded CH58-UA complex. The two additional salt bridges, Lys171(V2) to Asp31(L) and Asp180(V2) to Arg28(H), were formed during affinity maturation (Fig. 5). The light chain residue Asp31(L) was an Asn in CH58-UA. The Asn residue formed a preferential H-bond with the nearby light chain residue Arg25(L), and this intra-chain association was uninterrupted upon introduction of ligand (Fig. 5A). However, the mature antibody had an Asp residue in place of the germline Asn, and the Asp side chain accommodated a bifurcated H-bond to both Arg25(L) and V2 peptide residue Lys171(V2). The second noted salt bridge, Asp180(V2) to Arg28(H), represented a different scenario. In CH58-UA, Ser28(H) lacked both the reach and the electrostatic character to establish a salt bridge with Asp180(V2) (Fig. 5B). In the mature CH58, the Arg side chain rectified both these problems and established an H-bond with Asp180(V2). It is likely that this interaction anchored the C-terminal portion of the V2 peptide in the mature complex, and conversely failed to anchor it in the CH58-UA complex thus suggesting why the peptide C-terminus is disordered in that structure. Lastly, this salt bridge with Asp180 is potentially significant in that this residue was part of the putative α4β7 binding interface with gp120 (Arthos et al., 2008). The tripeptide motif LDV was identified as part of the α4β7 binding interface, and this motif appears in gp120 V2 and the liganded CH58 structure as LDI (gp120179–181) (Liao et al., 2013, Zeller et al., 2001). The salt bridge at Asp180 is indeed likely to be responsible for CH58 blocking α4β7, and we have previously reported that CH58 can block α4β7 binding to V2 (Liao et al., 2013).
Fig. 5.
Specific polar interactions between antibody and epitope gained through maturation. A) Asp31(L) in mature CH58 forms a salt bridge with Lys171(V2) and is additionally stabilized by a salt bridge with Arg25(L). In CH58-UA the salt bridge with Arg25(L) is present as are the proximity and geometry necessary to interact with V2, except that Asn31(L) clearly lacks the functional group necessary. B) Arg28(H) in mature CH58 forms a salt bridge with Asp180(V2), helping to anchor the C-terminus of the V2 peptide. However in CH58-UA, Ser28(H) is unable to make any such interaction with V2 peptide. In fact the C-terminus of the peptide in the CH58-UA complex structure is disordered without this interaction, and Asp180(V2) is not present in the structure as a result.
3.4. Conformational selection of LCDR3
The paratope of liganded CH58-UA displayed a pocket in which the hydrophobic side chain of Leu175(V2) bound (Fig. 6A). This pocket was present in both the unliganded and liganded mature structures as well, though LCDR3 was in a slightly different conformation (Fig. 6B). However, the Leu175(V2) hydrophobic pocket was not present in the paratope of the unliganded CH58-UA because LCDR3 adopted a conformation there which put the side chain of Tyr91(L) in a position that eliminated the character of the pocket in terms of both its shape and its hydrophobicity; in essence, the Tyr91(L) side chain in the unliganded CH58-UA structure caused a steric clash with Leu175(V2) of overlaid peptide (Fig. 6A). This may suggest that the maturation process selected an optimal conformation for LCDR3 that would not require an energetic penalty associated with conformational change on binding. It is noted that movement in the Tyr91(L) side chain as it appeared in the unliganded CH58-UA structure required concomitant movement of the side chain of Tyr100G in HCDR3. Tyr100G exhibited a different rotamer in the mature CH58 structure that was likely influenced by the proximity of Phe100I(H) which in turn was limited by Gly35(H). Heavy chain residue 35 is the site of a Gly → Val mutation (Figure S1). Thus it is apparent that structural rearrangements in LCDR3 may well be linked to those in HCDR3 through the anchorpoints of each CDR and with both affected in some way by mutations occurring with maturation; however as HCDR3 was disordered in the liganded mature CH58 structure (Liao et al., 2013), it is impossible to draw definitive conclusions about HCDR3 and its role in ligand binding. However, it seems clear that this was a case of conformational limiting for LCDR3 (Boehr et al., 2009, Foote and Milstein, 1994, Jimenez et al., 2003, Manivel et al., 2000, Zimmermann et al., 2006).
Fig. 6.
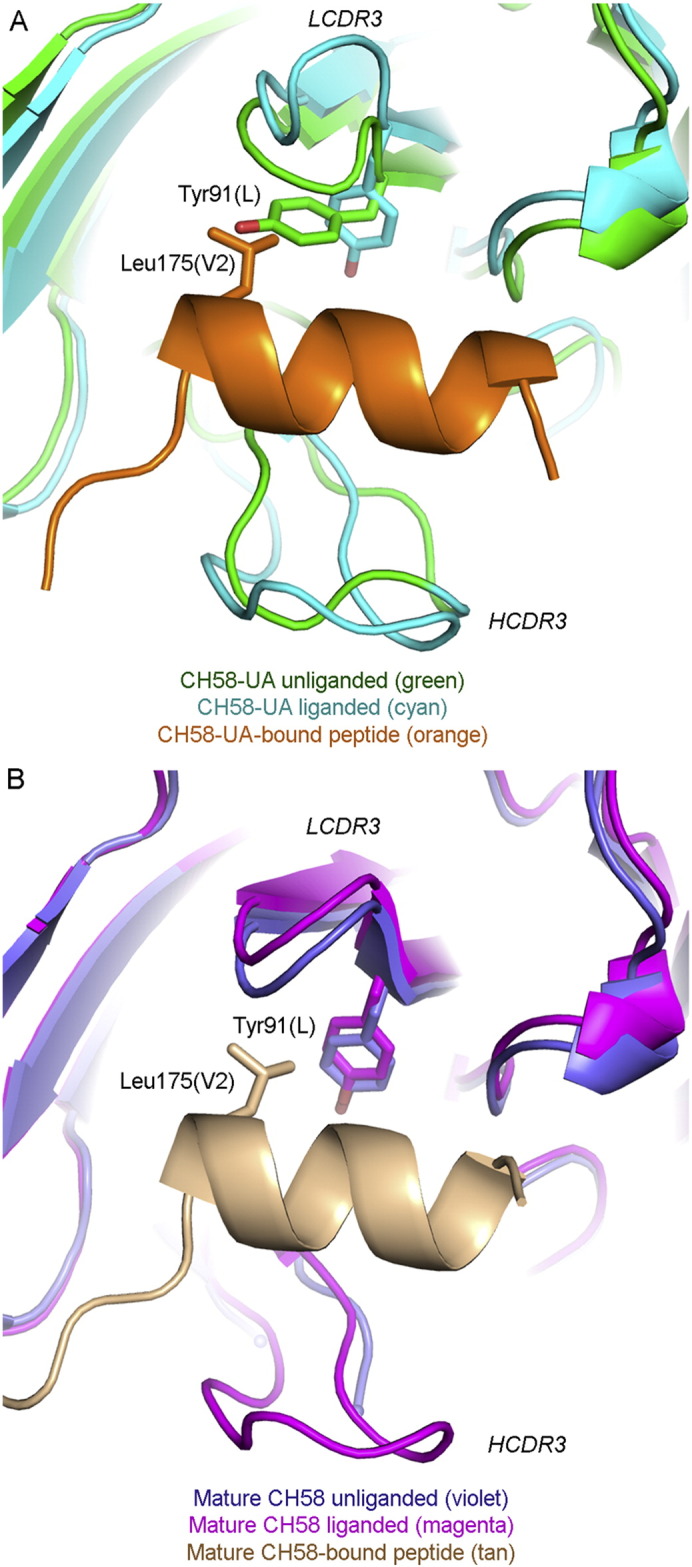
LCDR3 conformational ordering through maturation. A) In CH58-UA, LCDR3 exhibited different conformations between its unliganded and liganded states. In a superposition between the unliganded (green) and liganded (cyan and orange as labeled) structures, Tyr91(L) in the unliganded conformation clashed with overlaid Leu175(V2) from the complex structure. HCDR3 is additionally indicated for reference. B) In the mature CH58, LCDR3 exhibited the same conformation in both unliganded and liganded states, and this conformation differed from either of those observed in the CH58-UA structures. HCDR3 is also labeled here, though it was disordered in the unliganded mature CH58 structure (Liao et al., 2013).
4. Discussion
The overall purpose of the study was to scrutinize the development of the CH58 antibody from its germline precursor in the context of understanding V2 immonogenic responses. The increased affinity of the mature CH58 antibody over its inferred unmutated ancestor CH58-UA appeared to depend in part on tuning of local interactions. The introduction of new salt bridges as well as mutations from affinity maturation resulted in subtle structural changes that propagated throughout the paratope, including modest but effective conformational selections and rotamer adjustments. The conformation of LCDR2 containing the ED motif was nearly identical in all CH58 lineage structures — CH58-UA and mature CH58, unliganded and liganded. The ED motif also appeared in the CH59 antibody lineage, where Asp51(L) formed a salt bridge with Lys169(V2) (Liao et al., 2013). Similar to the CH58 lineage structures, the amine functional group of Lys169 was met favorably in the V2 antibody CH59 paratope by electronegativity conveyed by Asp51(L) in LCDR2 and Ser66(L), as well as the backbone carbonyls of Leu28(L) and Pro29(L). The low level of conformational diversity in LCDR2 (North et al., 2011) and the invariance of the positioning of the ED motif within the LCDR2 germline alleles within which it appears represent two levels of conservation (structure and sequence) for this key motif. Moreover, preconfiguration of an antibody paratope can mitigate the energetic penalty paid upon association between the mature antibody and its antigen (Schmidt et al., 2013). Conceptually, the same penalty is avoided when an element of the paratope is preconformed from germline, as is the case with LCDR2 in CH58-UA, and when the large gain in affinity for antigen (KD) through maturation is predominantly attributable to a decreased off-rate (kd). Preconformed motifs in germline alleles tend to be preferred in anti-HIV antibodies as they mimic HIV epitopes or otherwise present favorable structural paratope features. CH58 and antibodies like it are unique from CD4 binding site or gp41 MPER antibodies in that restricted pairing of light and heavy chain alleles occurs at the light chain V gene level (Wiehe et al., in press). For example, with VRC-01, there is a steric requirement that the LCDR3 be short, but that is independent of the light chain V gene used since its particular LCDR3 depends on rearrangement, not just the V gene used; whereas, in instances where LCDR2 bears the ED motif (CH58 and CH59), the motif encoded strictly by the V gene.
Mutations in both the CDRs and the framework region played important roles in conformational selection of LCDR3 in the CH58 lineage, reflecting previous research showing that the influenza antibody CH65 lineage rigidified its flexible HCDR3 through maturation (Schmidt et al., 2013). The importance of somatic mutations may be in their ability to foster a certain degree of flexibility necessary to promote optimal antigen binding (Klein et al., 2013). For instance, a recent report of a framework mutation in the anti-HIV antibody lineage producing CH103 that culminated in a shift between the L and H domains (Fera et al., 2014). With CH58, a network of structural changes involving such mutations acted to configure the conformation of LCDR3 from one that would only tolerate antigen binding to one that was optimal for it.
The lineage producing CH58 showed an affinity gain primarily being due to improvements in kd. The dissociation rate improved in going from CH58-UA to CH58 almost 600-fold compared to an association rate improvement of only about 4-fold, the net result being an affinity gain from 11.0 μM to 4.6 nm. The anti-HIV antibodies VRC01, NIH45-46, 12A21, and 3BNC117 displayed similar patterns. They did not have values reported for on- and off-rates; however, it was qualitatively evident from published SPR raw data that affinity gains with maturation in all four cases were primarily due to improvements in off-rate (Klein et al., 2013). CH59 showed a dramatically improved off-rate compared to its inferred unmutated ancestor (1.5 × 10− 4 s− 1 for the mature CH59 compared to 7.4 × 10− 2 s− 1 for CH59-UA) that was entirely responsible for the affinity gain observed in the mature antibody (Wiehe et al., in press). The unmutated ancestor of the gp41 MPER antibody 2F5 was inferred to stem from one of two germline variants, either of which was equally likely owing to the high degree of mutation observed in 2F5 along with uncertainty inherent to the inference method (Alam et al., 2011). 2F5-UA variant 1 bore residue D54(H) in its HCDR2 whereas variant 2 showed N54(H) (Alam et al., 2011). The mature 2F5 had D54(H) like variant 1. The association rates of variant 1 and mature were similar (1.8 × 104 M− 1 s− 1 vs 2.7 × 104 M− 1 s− 1 respectively). Variant 2 had an association rate an order of magnitude lower (3.9 × 103 M− 1 s− 1). However, both UA variants had dissociation rates comparable (1.1 × 10− 3 s− 1 for variant 1, 2.6 × 10− 3 s− 1 for variant 2), whereas the mature 2F5 antibody had a dissociation rate two orders of magnitude lower (3.1 × 10− 5 s− 1). It was this significant decrease in the off-rate that was found to drive the higher affinity that the mature had for antigen. Of note, the uncertainty between the two 2F5-UA variants illustrates either of two important concepts in the present study: either the higher association rate of UA variant 1 belied the benefit of preconformed paratope features because 2F5 relies on D54(H) along with D56(H) to establish key salt bridges with its epitope in the gp41 MPER; or UA variant 2 illustrates how salt bridges and other specific contacts gained in the maturation process help to dramatically improve off-rate between Ab–Ag thus increasing affinity.
In looking at non-HIV antibody lineage affinity gains, fitting the same category of predominant off-rate improvements is a series of anti-(4-hydroxy-3-nitrophenyl)acetyl antibodies where the most mature member of the set had a 5-fold improved off-rate over the least mature member, with a worse on-rate for the same pairwise comparison (Sagawa et al., 2003). Though less exemplary, the antifluorescein antibody 4-4-20 showed a 6-fold improved off-rate and a 2-fold on-rate over its germline (Zimmermann et al., 2006). Also, anti-IGNβ antibodies in the sera of patients with sustained IGNβ treatment were found on average to have improved off-rates compared to the sera from patients transiently treated (Gibbs et al., 2014). In contrast, the flu antibodies CH65 and CH67 showed affinity gains over their shared unmutated ancestor that were predominantly due to on-rate improvements (Schmidt et al., 2013). More detailed kinetic analyses are needed before the field can draw significant conclusions on the topic, but it may be that antibodies with elements of preconformation and key Ab–Ag contacts gained through maturation in their idiotopes tend to be driven by improvements in off-rate, whereas antibodies with paratopes that require structural ordering (preconfiguration) in addition to the previous factors see comparably higher gains in on-rate.
HIV broadly neutralizing antibodies tend to show high levels of mutation from germline and are rare or difficult to induce (Kwong and Mascola, 2012, Mascola and Haynes, 2013). In contrast, modestly mutated antibodies from the RV144 vaccine efficacy trial produced the strongest ADCC responses (Bonsignori et al., 2012, Haynes et al., 2012). CH58 is not a broadly neutralizing antibody, however it is a facile, dominant-response antibody induced by the RV144 vaccine (Haynes et al., 2012, Karasavvas et al., 2012, Liao et al., 2013). In neutralizing isolated strains and mediating ADCC, it may confer some protective benefit. Moreover, its ability to block interaction between gp120 and α4β7 suggests that although many HIV-1 strains do not appear to bind α4β7 (Perez et al., 2014), CH58 and antibodies like it may restore the natural proliferation of B cells that is subverted by any α4β7 interaction (Jelicic et al., 2013, Tassaneetrithep et al., 2014). Thus it is hoped that vaccine regimens can be developed to include CH58-like responses. In this regard, a recent publication has shown that the combination of CH58 with 7B2, a gp41 immunodominant loop antibody, can increase the capture of infectious virions (Liu et al., 2014).
In summary, structural and kinetic analyses of CH58-UA and mature CH58 showed that a key feature of the paratope, LCDR2, was preconformed for antigen recognition at a site of immune pressure (V2 Lys169) and presented a conserved motif optimal for interacting with a reciprocal motif in gp120 V2. Another factor in maturation was conformational selection of LCDR3, resulting in an optimal binding site for antigen. A third factor was the gain of salt bridges between antibody and antigen specific to mutations in the antibody that occurred with maturation. The primary goal of follow-up studies to RV144 is learning how to improve on it. One such improvement would be to increase the breadth of V2 responses. To learn how to manipulate V2 responses, it is key to know how they develop. In the case of the lineage producing CH58, it is clear that specific residues within the V2 region are critical contacts with both the germline ancestor and the mature antibody.
Author contributions
The CH58 antibody was isolated by M.B. and K-K.H. and characterized by M.B., H-X.L., K-K.H., S.M.A., N.I.N., K.W., and T.B.K. in collaboration with S.R-N., S.N., P.P., J.K., M.L.R., R.J.O., N.L.M. and J.H.K. B.F.H. conceived the study. T.B.K. inferred the UA protein sequence. H-X.L. designed the protein-expressing constructs. N.I.N and H-X.L. performed transfections and protein purifications. N.I.N., S.M.D., S.M.A. and B.F.H. designed experiments. F.H.J. and S.M.D. performed SPR and BLI experiments and analyzed results with S.M.A, N.I.N. and K.W. N.I.N. performed all aspects of crystallography and structure determination. N.I.N. and K.W. analyzed the structural models in the context of other data. N.I.N. and B.F.H. wrote the manuscript with input from K.W., and all authors reviewed it.
Acknowledgements
This work was supported by a Collaboration for AIDS Vaccine Discovery grant to B.F.H. from the Bill and Melinda Gates Foundation (number OPP1033098). Crystallography was performed in the Duke University X-ray Crystallography Shared Resource. The use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. We express our gratitude to Stephen C. Harrison from Boston Children's Hospital for manuscript review.
Footnotes
The opinions herein are those of the authors and should not be construed as representing the official views of the Departments of Defense or the Army.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.06.016.
Contributor Information
Nathan I. Nicely, Email: nathan.nicely@duke.edu.
Barton F. Haynes, Email: hayne002@mc.duke.edu.
Appendix A. Supplementary data
Supplementary material.
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.M., McAdams M., Boren D., Rak M., Scearce R.M., Gao F., Camacho Z.T., Gewirth D., Kelsoe G., Chen P. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.M., Liao H.X., Dennison S.M., Jaeger F., Parks R., Anasti K., Foulger A., Donathan M., Lucas J., Verkoczy L. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. J. Virol. 2011;85:11725–11731. doi: 10.1128/JVI.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthos J., Cicala C., Martinelli E., Macleod K., Van Ryk D., Wei D., Xiao Z., Veenstra T.D., Conrad T.P., Lempicki R.A. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat. Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- Babor M., Kortemme T. Multi-constraint computational design suggests that native sequences of germline antibody H3 loops are nearly optimal for conformational flexibility. Proteins. 2009;75:846–858. doi: 10.1002/prot.22293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr D.D., Nussinov R., Wright P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M., Pollara J., Moody M.A., Alpert M.D., Chen X., Hwang K.K., Gilbert P.B., Huang Y., Gurley T.C., Kozink D.M. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J. Virol. 2012;86:11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fera D., Schmidt A.G., Haynes B.F., Gao F., Liao H.X., Kepler T.B., Harrison S.C. Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains. Proc. Natl. Acad. Sci. U. S. A. 2014;111:10275–10280. doi: 10.1073/pnas.1409954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote J., Milstein C. Conformational isomerism and the diversity of antibodies. Proc. Natl. Acad. Sci. U. S. A. 1994;91:10370–10374. doi: 10.1073/pnas.91.22.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs E., Oger J. A biosensor-based characterization of the affinity maturation of the immune response against interferon-beta and correlations with neutralizing antibodies in treated multiple sclerosis patients. J. Interf. Cytokine Res. 2008;28:713–723. doi: 10.1089/jir.2008.0144. [DOI] [PubMed] [Google Scholar]
- Gibbs E., Karim M.E., Oger J., the Steering Committee of the B.s. Antibody dissociation rates are predictive of neutralizing antibody (NAb) course: a comparison of interferon beta-1b-treated Multiple Sclerosis (MS) patients with transient versus sustained NAbs. Clin. Immunol. 2014;157:91–101. doi: 10.1016/j.clim.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Harris L.J., Skaletsky E., McPherson A. Crystallization of intact monoclonal antibodies. Proteins. 1995;23:285–289. doi: 10.1002/prot.340230218. [DOI] [PubMed] [Google Scholar]
- Haynes B.F., Gilbert P.B., McElrath M.J., Zolla-Pazner S., Tomaras G.D., Alam S.M., Evans D.T., Montefiori D.C., Karnasuta C., Sutthent R. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelicic K., Cimbro R., Nawaz F., Huang da W., Zheng X., Yang J., Lempicki R.A., Pascuccio M., Van Ryk D., Schwing C. The HIV-1 envelope protein gp120 impairs B cell proliferation by inducing TGF-beta1 production and FcRL4 expression. Nat. Immunol. 2013;14:1256–1265. doi: 10.1038/ni.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez R., Salazar G., Baldridge K.K., Romesberg F.E. Flexibility and molecular recognition in the immune system. Proc. Natl. Acad. Sci. U. S. A. 2003;100:92–97. doi: 10.1073/pnas.262411399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J.P., Cupo A., Sok D., Stanfield R.L., Lyumkis D., Deller M.C., Klasse P.J., Burton D.R., Sanders R.W., Moore J.P. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasavvas N., Billings E., Rao M., Williams C., Zolla-Pazner S., Bailer R.T., Koup R.A., Madnote S., Arworn D., Shen X. The Thai Phase III HIV Type 1 Vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res. Hum. Retrovir. 2012;28:1444–1457. doi: 10.1089/aid.2012.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Diskin R., Scheid J.F., Gaebler C., Mouquet H., Georgiev I.S., Pancera M., Zhou T., Incesu R.B., Fu B.Z. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Mascola J.R. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Sutherland L.L., Xia S.M., Brock M.E., Scearce R.M., Vanleeuwen S., Alam S.M., McAdams M., Weaver E.A., Camacho Z. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Bonsignori M., Alam S.M., McLellan J.S., Tomaras G.D., Moody M.A., Kozink D.M., Hwang K.K., Chen X., Tsao C.Y. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Williams L.D., Shen X., Bonsignori M., Overman R.G., Moody M.A., Liao H.X., Stieh D.J., McCotter K.L., French A.L. Capacity for infectious HIV-1 virion capture differs by envelope antibody specificity. J. Virol. 2014;88:5165–5170. doi: 10.1128/JVI.03765-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell S.C., Davis I.W., Arendall W.B., 3rd, de Bakker P.I., Word J.M., Prisant M.G., Richardson J.S., Richardson D.C. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Manivel V., Sahoo N.C., Salunke D.M., Rao K.V. Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity. 2000;13:611–620. doi: 10.1016/s1074-7613(00)00061-3. [DOI] [PubMed] [Google Scholar]
- Mascola J.R., Haynes B.F. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol. Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B.W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- McLellan J.S., Pancera M., Carrico C., Gorman J., Julien J.P., Khayat R., Louder R., Pejchal R., Sastry M., Dai K. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicely N.I., Dennison S.M., Spicer L., Scearce R.M., Kelsoe G., Ueda Y., Chen H., Liao H.X., Alam S.M., Haynes B.F. Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region. Nat. Struct. Mol. Biol. 2010;17:1492–1494. doi: 10.1038/nsmb.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B., Lehmann A., Dunbrack R.L., Jr. A new clustering of antibody CDR loop conformations. J. Mol. Biol. 2011;406:228–256. doi: 10.1016/j.jmb.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski A., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in enzymology 276. Macromol. Crystallogr. A. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pancera M., Shahzad-Ul-Hussan S., Doria-Rose N.A., McLellan J.S., Bailer R.T., Dai K., Loesgen S., Louder M.K., Staupe R.P., Yang Y. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1–V2-directed antibody PG16. Nat. Struct. Mol. Biol. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M., Zhou T., Druz A., Georgiev I.S., Soto C., Gorman J., Huang J., Acharya P., Chuang G.Y., Ofek G. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez L.G., Chen H., Liao H.X., Montefiori D.C. Envelope glycoprotein binding to the integrin alpha4beta7 is not a general property of most HIV-1 strains. J. Virol. 2014;88:10767–10777. doi: 10.1128/JVI.03296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., Premsri N., Namwat C., de Souza M., Adams E. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Rolland M., Edlefsen P.T., Larsen B.B., Tovanabutra S., Sanders-Buell E., Hertz T., deCamp A.C., Carrico C., Menis S., Magaret C.A. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagawa T., Oda M., Ishimura M., Furukawa K., Azuma T. Thermodynamic and kinetic aspects of antibody evolution during the immune response to hapten. Mol. Immunol. 2003;39:801–808. doi: 10.1016/s0161-5890(02)00282-1. [DOI] [PubMed] [Google Scholar]
- Scheid J.F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T.Y., Pietzsch J., Fenyo D., Abadir A., Velinzon K. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A.G., Xu H., Khan A.R., O'Donnell T., Khurana S., King L.R., Manischewitz J., Golding H., Suphaphiphat P., Carfi A. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc. Natl. Acad. Sci. U. S. A. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassaneetrithep B., Tivon D., Swetnam J., Karasavvas N., Michael N.L., Kim J.H., Marovich M., Cardozo T. Cryptic determinant of alpha4beta7 binding in the V2 loop of HIV-1 gp120. PLoS ONE. 2014;9:e108446. doi: 10.1371/journal.pone.0108446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P.V., Moriarty N.W., Zwart P.H., Hung L.W., Read R.J., Adams P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urzhumtseva L., Afonine P.V., Adams P.D., Urzhumtsev A. Crystallographic model quality at a glance. Acta Crystallogr. D Biol. Crystallogr. 2009;65:297–300. doi: 10.1107/S0907444908044296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Phogat S.K., Chan-Hui P.Y., Wagner D., Phung P., Goss J.L., Wrin T., Simek M.D., Fling S., Mitcham J.L. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehe K., Easterhoff D., Luo K., Nicely N.I., Bradley T., Jaeger F.H., Dennison S.M., Zhang R., Lloyd K.E., Stolarchuk C. 2014. Phylogenetic Conservation of Antibody Light Chain-restricted Recognition of the Site of Immune Pressure in an HIV-1 Vaccine Trial Immunity. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N.S., Louder M., McKee K. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Schmidt A.G., O'Donnell T., Therkelsen M.D., Kepler T.B., Moody M.A., Haynes B.F., Liao H., Harrison S.C., Shaw D.E. Key mutations stabilize antigen-binding conformation during affinity maturation of a broadly neutralizing influenza antibody lineage. Proteins. 2014;83:771–780. doi: 10.1002/prot.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller Y., Mechtersheimer S., Altevogt P. Critical amino acid residues of the alpha4 subunit for alpha4beta7 integrin function. J. Cell. Biochem. 2001;83:304–319. doi: 10.1002/jcb.1197. [DOI] [PubMed] [Google Scholar]
- Zhou T., Georgiev I., Wu X., Yang Z.Y., Dai K., Finzi A., Kwon Y.D., Scheid J.F., Shi W., Xu L. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Zhu J., Wu X., Moquin S., Zhang B., Acharya P., Georgiev I.S., Altae-Tran H.R., Chuang G.Y., Joyce M.G. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann J., Oakman E.L., Thorpe I.F., Shi X., Abbyad P., Brooks C.L., 3rd, Boxer S.G., Romesberg F.E. Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13722–13727. doi: 10.1073/pnas.0603282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.