Abstract
The purpose of this study was to determine if mitochondrial dysfunction plays a role in diabetic nephropathy (DN), a kidney disease which affects > 100 million people worldwide and is a leading cause of renal failure despite therapy. A cross-sectional study comparing DN with diabetes patients without kidney disease (DC) and healthy controls (HCs); and renal mesangial cells (HMCs) grown in normal and high glucose, was carried out. Patients with diabetes (DC) had increased circulating mitochondrial DNA (MtDNA), and HMCs increased their MtDNA within 24 h of hyperglycaemia. The increased MtDNA content in DCs and HMCs was not functional as transcription was unaltered/down-regulated, and MtDNA damage was present. MtDNA was increased in DC compared to HC, conversely, patients with DN had lower MtDNA than DC. Hyperglycaemic HMCs had fragmented mitochondria and TLR9 pathway activation, and in diabetic patients, mitophagy was reduced. Despite MtDNA content and integrity changing within 4 days, hyperglycaemic HMCs had a normal bio-energetic profile until 8 days, after which mitochondrial metabolism was progressively impaired. Peripheral blood mononuclear cells (PBMCs) from DN patients had reduced reserve capacity and maximal respiration, loss of metabolic flexibility and reduced Bioenergetic Health Index (BHI) compared to DC. Our data show that MtDNA changes precede bioenergetic dysfunction and that patients with DN have impaired mitochondrial metabolism compared to DC, leading us to propose that systemic mitochondrial dysfunction initiated by glucose induced MtDNA damage may be involved in the development of DN. Longitudinal studies are needed to define a potential cause–effect relationship between changes in MtDNA and bioenergetics in DN.
Keywords: Diabetic nephropathy, Mitochondrial DNA, Hyperglycemia, Mitochondrial dysfunction, Bioenergetic deficit, Bioenergetic health index (BHI)
Highlights
-
•
Diabetic nephropathy may be a disease of acquired MtDNA damage and bioenergetic deficit.
-
•
MtDNA content is increased in blood cells of diabetes patients and hyperglycaemic renal cells.
-
•
Hyperglycaemia leads to renal cell MtDNA damage and subsequent bioenergetic dysfunction.
-
•
Diabetic nephropathy patients have reduced circulating MtDNA , BHI and metabolic flexibility bioenergetic dysfunction and reduced metabolic flexibility and BHI.
1. Introduction
Diabetic nephropathy (DN) affects ~ one-third of patients with diabetes and develops over a long period of clinical silence (Ritz and Orth, 1999). With the epidemic rise in the incidence of diabetes currently affecting more 350 million people, > 100 million people are at risk of DN and of these 30% are likely to progress to end stage renal failure despite therapy (IDF, 2013, Wild et al., 2004). Therefore there is an urgent need to understand the underlying mechanisms of damage in the diabetic kidney in order to design novel therapeutics.
Clinical randomised studies such as the Diabetes Control and Complications Trial and the Epidemiology of Diabetes Interventions and Complication have shown that early intensive glycemic control reduces risk, whereas prolonged hyperglycaemia can increase the long term risk of diabetic complications (Kilpatrick et al., 2009). Hyperglycaemia is therefore accepted as a major mediator of renal damage and activates several complex and overlapping biochemical pathways resulting in abnormal signalling in cells (Aronson, 2008, Brownlee, 2001). Diabetes results in increased risk of numerous other complications which affect major organs, including eyes (retinopathy), heart (diabetic cardiomyopathy), blood vessel (peripheral vascular disease) and brain (dementia) (Stratton et al., 2000). The multi-organ impact of diabetes complications resembles mitochondrial genetic disease (Moraes et al., 1991, Wallace, 1999) and suggests a systemic dysfunction in the body.
Mitochondria, cellular organelles in the cytosol of eukaryotic cells, harbour their own circular DNA genome which is located outside the nuclear genome, and require both mitochondrial and nuclear genome encoded proteins to function. Mitochondria produce energy in the form of ATP via oxidative phosphorylation (OXPHOS) (Wojtczak and Zabłocki, 2008), therefore the number of mitochondria in a cell is related to the energy requirements of the cell and can vary depending on many factors such as the environment and redox balance of the cell (Michel et al., 2012). Mitochondrial dysfunction can affect key cellular functions, result in a variety of diseases (Wallace, 1999), and altered mitochondrial DNA (MtDNA) levels have been reported in a wide range of human disease (Malik and Czajka, 2013).
We and others have reported changes in circulating MtDNA in patients with DN (Lee et al., 2009, Malik et al., 2009). Reduced renal mitochondrial function was postulated in a urinary metobolomics study showing that mitochondrial metabolites were reduced in the urine of DN patients (Sharma et al., 2013), however, it was suggested that the basis for reduced mitochondrial content/function would be difficult to address in clinical samples. Therefore, despite some evidence that mitochondrial dysfunction may be involved in DN, there is no direct evidence in clinical samples and it is unclear whether measuring mitochondrial function in patients may have any translational potential. Furthermore the underlying molecular mechanisms are not understood.
If systemic mitochondrial dysfunction is associated with DN, then we would predict systemic changes in the body which may be the basis of renal microvascular complications in diabetes. Peripheral blood mononuclear cells (PBMCs) from patients have been used widely as surrogates and various parameters associated with DN can be detected in these cells (Hofmann et al., 1999, Ihm et al., 1997, Sourris et al., 2010, Yi et al., 2014); our assumption is that PBMC mitochondria will resemble systemic changes in the body and therefore will act as surrogate cells for kidney tissue. Recently, methodology for measuring bioenergetics in live fractionated blood cells have been developed and it has been suggested that these may be utilised to measure oxygen consumption rates (OCR) and extra cellular flux analysis (ECAR) to indicate potential mitochondrial dysfunction (Chacko et al., 2013) It has been proposed that ECAR and OCR values can be combined using a formula as a single value that defines an individual's bioenergetic health index (BHI) which could be used as a prognostic/diagnostic indicator of metabolic stress.
In the current paper our major aim was to determine if mitochondrial dysfunction can be detected in patients with DN and to elucidate the potential mechanisms by which hyperglycaemia may affect renal mitochondria. To expand our in-vivo observations we also utilised an experimental in-vitro cell model system to examine the effects of hyperglycaemia on renal mitochondria. We used primary human mesangial cells (HMCs) as transformation can affect cellular bioenergetics. Importantly, glomerular mesangial cells are a target of both metabolic and haemodynamic perturbations in diabetes (Clarkson et al., 2002, Murphy et al., 1999) and have been widely utilised as models of DN. We therefore used PBMCs to examine systemic changes in diabetes and HMCs to examine effect of diabetes on renal mitochondria.
2. Materials and Methods
2.1. Human Subjects
Patients were recruited between 2008 and 2014 with written informed consent from Guy's and St Thomas' hospital clinics under ethical approval from the regional Research Ethics Committee (REC; ref number 07/H0806/120). The cross-sectional study adhered to the Ethical Principles for Medical Research Involving Human Subjects, World Medical Association Declaration of Helsinki. A random blood glucose level of ≥ 11.1 or a fasting blood glucose level of ≥ 7 mmol/l was considered to be indicative of diabetes. Type 1 diabetes (T1D) and type 2 Diabetes (T2D) were defined as follows: T1D: onset before age 35, insulin therapy within 6 months of diagnosis and no breaks in insulin therapy > 6 months; T2D: onset after age 35, controlled by diet or established oral hypoglycaemic treatment and/or insulin. Albumin/creatinine ratio (ACR) was used to assess the level of albuminuria, ACR < 2.5 mg/mmol for men and ACR < 3.5 mg/mmol for women, was defined as cut off for normo to microalbuminuria. Glomerular filtration rate (GFR) was assessed using the Modification of Diet in Renal Disease (MDRD) formula (Stoves et al., 2002). For controls without nephropathy, we used patients with type 1 diabetes and type 2 diabetes with ≥ 20 or ≥ 10 years of diabetes duration, respectively, without a history of albuminuria, with normal renal function and normal blood pressure (≤ 130/80 mm Hg) and taking no antihypertensive agents. HC (n = 39) with no history of the disease or current medication were recruited with informed consent, and were age and sex matched with the patient study group. Sample size was calculated from a pilot experiment using Cohens d, which showed that n = 39 for each group was adequate to power the MtDNA study. Table 1 shows the baseline characteristics of subjects used in this study.
Table 1.
Baseline characteristics of healthy controls and diabetic patients with and without nephropathy with MtDNA content.
| Variable | Healthy controls (HCs) (n = 39) |
Diabetic controls (DCs) (n = 45) |
Diabetic nephropathy(DN) (n = 83) |
|---|---|---|---|
| Type of diabetes (T1D:T2D) | NR | 27:18 | 31:52 |
| Age (years) | 52 ± 25 | 49 ± 14 | 61 ± 14⁎⁎ |
| Gender (female:male) | 20:19 | 28:17 | 37:46 |
| Diabetes duration (years) | NR | 22 ± 10 | 22 ± 13 |
| BMI (kg/m2) | 23 ± 4 | 27 ± 4# | 30 ± 7⁎⁎## |
| HbA1c (%) | ND | 10 ± 9 | 8 ± 1 |
| ACR (mg/mmol) | ND | 0.8 ± 0.6 | 18 ± 50⁎⁎ |
| eGFR (ml min− 1 1.73 m− 2) | ND | 103 ± 21 | 66 ± 34⁎⁎ |
| Systolic BP (mm Hg) | ND | 122 ± 12 | 133 ± 17⁎⁎ |
| Diastolic BP (mm Hg) | ND | 72 ± 9 | 72 ± 9 |
| Cholesterol (mmol/l) | ND | 4.4 ± 0.8 | 3.9 ± 0.9⁎ |
| MtDNA content | 34 ± 9 | 64 ± 75# | 43 ± 52⁎⁎ |
| MtDNA (Median) |
32 (35) | 38 (469)# | 26 (298)⁎⁎⁎ |
Data are means ± SD, except where otherwise indicated.
P < 0.05 compared with DC.
P < 0.01 compared with DC.
P < 0.001 compared with DC.
P < 0.05 compared with HC.
P < 0.01 compared with HC.
2.2. Primary Human Glomerular Mesangial Cells
Human kidneys unsuitable for transplantation or normal portion from renal carcinoma were used as a source of primary human mesangial cells (HMCs) which were cultured and characterised as previously described (Gruden et al., 1997, Thomas et al., 2000) and grown in Dulbecco's Modified Eagle Medium (DMEM, Sigma Aldrich) supplemented with 10% FBS, ITS and antibiotics. Cells were seeded at an equal density (1 × 105/well) in 6-well plates in DMEM containing either 5 mM (normal, NG) or 25 mM (high, HG) glucose for varying time points. 20 mM mannitol in 5 mM glucose DMEM was used as an osmotic control in all experiments.
2.3. Real Time Quantitative PCR
Genomic DNA or RNA templates converted to cDNA were used for real time qPCR using SYBR green (Qiagen) as previously described (Shahni et al., 2013). All primer sequences used in the study are listed (Table S3). Absolute quantification was carried out in the presence of dilution standards for each gene. The mRNA copy numbers were calculated by dividing the mean values of target gene relative to the mean values of β-actin (the most stably expressed gene), MtDNA copy numbers were assessed as mitochondrial to nuclear gene ratio using Beta-2 microglobulin (B2M) as the nuclear control (Malik et al., 2011).
2.4. Extracellular Flux Analysis
Extracellular flux analysis is defined as the measurement of cellular bioenergetics (respiratory activity), based on the measurement of two major energy pathways OXPHOS and glycolysis (Chacko et al., 2013, Ferrick et al., 2008).
The metabolic profiles of cultured HMCs and freshly isolated human PBMCs s were assessed using the XFe96 Seahorse analyser and XF cell mito stress test kit (Seahorse Biosciences). Oligomycin (ATP synthase blocker) was used to measure ATP turnover and to determine proton leak, the mitochondrial uncoupler — FCCP (carbonyl cyanide 4-[trifluoromethoxy] phenylhydrazone) was used to measure maximum respiratory function (maximal OCR). Reserve capacity was calculated as maximal OCR minus the basal respiration. Rotenone (inhibitor of complex I) and Antimycin A (a blocker of complex III), were used to measure non-mitochondrial respiration (Brand and Nicholls, 2011, Dranka et al., 2011). The data generated for OCR and ECAR in PBMCs were used to calculate the Bioenergetic Health Index as described recently by Darley-Usmar group (Chacko et al., 2014) using the BHI = log (reserve capacity) × (ATP-linked) / (non-mitochondrial) × (proton leak).
2.5. Mitochondrial DNA Damage
DNA damage was quantified using the elongase method (Furda et al., 2014) by comparing the relative amplification of an 8.9 kb region relative to a 127 bp region in the mitochondrial genome using specific primers (Table S3). For the Surveyor Nuclease method (Bannwarth et al., 2006), amplicon A was amplified (Table S3) and digested with Surveyor nuclease. Gel electrophoresis was used in both methods to detect MtDNA damage.
2.6. Measurement of ROS Production, Cell Viability and Apoptosis in Human Mesangial Cells
Intracellular ROS in was measured using cell-permeant 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) oxidation, cell viability was measured using CellTiter-Glo luminescence assay (Promega, UK) in a microplate reader according to the manufacturer's instructions.
2.7. Assessment of Mitochondrial Network
Cells were grown in 96-well plates with clear bottoms, fixed with paraformaldehyde and visualized by staining with Mitotracker Red (Invitrogen) according to the manufacturer's instructions. Cells were viewed under Nikon Eclipse Ti-E Inverted Microscope, using CFI S Plan Fluor ELWD 20 ×/0.45NA objective. The stained mitochondrial network was assessed using ImageJ software (version 1.47q: National Institutes of Health; www.rsb.info.nih.gov/ij) using an image-processing algorithm (Koopman et al., 2005). Two different parameters, formfactor F as a measure of degree of branching (calculated as a perimeter2 / 4Π ∗ area) aspect ratio AR (measure of mitochondria length) were used to quantify mitochondrial morphology.
2.8. Statistics
Analysis was performed using GraphPad (GraphPad Software, Inc.) and IBM SPSS for Windows software. The distribution of the data was tested using the Kolmogorov–Smirnov test (graph pad) and histograms in SPSS and parametric tests were used on raw or log transformed data. For parametric analysis, groups were compared using t-test (2 groups) or one way ANOVA with post-hoc Tukey's multiple comparison test (> 2 groups). For non-parametric analysis, groups were compared using Mann–Whitney (2 groups) or Kruskal Wallis with Dunn's post-hoc test with Bonferroni correction (> 2 groups). Data are presented as mean ± Standard error of the mean and as median for non-normally distributed data.
2.9. Author Contributions
AC and SA generated the cell and human data respectively and are joint first authors, the study was conceived and designed by AM with the help of LG and PJ, KP undertook MtDNA damage experiments, the data analysis was undertaken by AC/SA/AM and evaluated by all authors, the statistical analysis was supported by FR, all authors were involved in the drafting and revision of the manuscript.
2.10. Funding
This work was partly funded by an EFSD/Janssen Programme for the Study of the Role of the Kidney in Diabetes grant; AC and SA are supported by KCL PhD scholarships.
3. Results
3.1. Detection of Changes in Mitochondrial DNA and mRNAs in Blood Samples From DN Patients
We first set out to establish if we could confirm and extend ours and another study showing altered circulating MtDNA in DN patients (Lee et al., 2009, Malik et al., 2009). Using small volumes of peripheral blood, our assays comprised of measurement of MtDNA content, and mRNA content of various mitochondrial-encoded and nuclear-encoded mRNAs including genes encoding various mitochondrial subunits, involved in biogenesis, fusion, fission and mitophagy. In addition we assessed the integrity of MtDNA in these samples.
Using a cross-sectional study design, we compared 3 groups of subjects: healthy controls (HCs, n = 39) were volunteers with no history of any disease, diabetes controls (DCs, n = 45) comprised of patients with ≥ 20 years diabetes duration, normal renal function and no history of albuminuria, and diabetic nephropathy patients (DN, n = 83) with a history of or current albuminuria (Table 1). The HC group were age and sex matched with the diabetes patients (Table S1) and had a lower BMI (P < 0.05). The DN patients were older, had higher BMI, albumin/creatinine ratio (ACR), and systolic blood pressure and lower eGFR than DC. We used both T1D and T2D patients in this study and analysed the groups combined as hyperglycaemia plays a role in progression of renal disease independently of the type of diabetes. Analysis of T1D and T2D patients separately showed the same trends (Table S5).
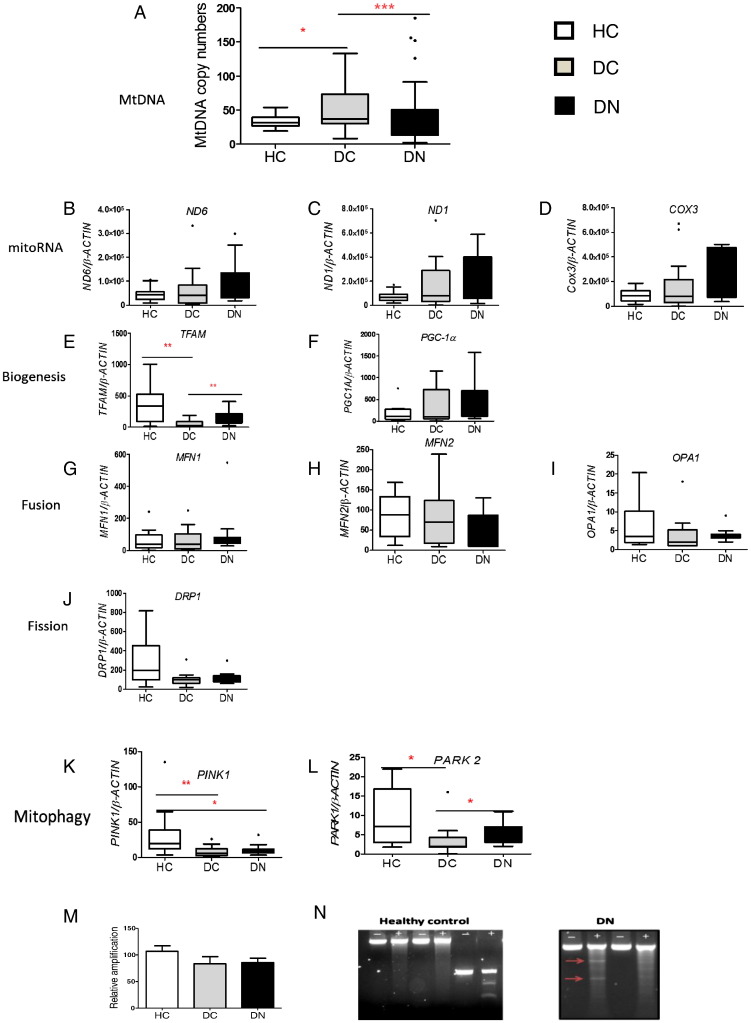
MtDNA content was quantified as the ratio of mitochondrial genome to nuclear genome (Malik et al., 2011). DC had significantly higher MtDNA content compared to HC (P < 0.05) whereas the DN patients had reduced MtDNA compared to DC (P < 0.001, Fig. 1A). We examined if the decreased MtDNA in DN patients was a consequence of other parameters, as age, BMI, systolic blood pressure, eGFR and cholesterol, were significantly different between the DC and DN groups (Table 1). After adjusting for these variables, stepwise binary regression analysis showed that eGFR, systolic blood pressure and MtDNA levels remained independently associated with DN (P = 0.009) however, age, BMI, and cholesterol were no longer associated with DN. As reduced eGFR and elevated blood pressure are well known risk markers for DN (Alaveras et al., 1997), our data strongly suggest that MtDNA levels represent a new risk marker for DN and the associated increase in cardiovascular morbidity and mortality of these patients.
Fig. 1.
Diabetes associated changes in mitochondrial DNA content and mitochondrial life cycle in circulating cells. DNA and RNA were isolated from 0.1 ml and 1 ml respectively of peripheral blood from HC (n = 39), DC (n = 45) and DN (n = 83) groups. Data are presented as box plots (except for M) with whiskers showing median, quartiles and range which were analysed using non-parametric Kruskal–Wallis test with Dunn's multiple comparison test. (A) MtDNA content. Real time qPCR was carried out to determine MtDNA content as mitochondrial to nuclear (B2M) ratio. *P < 0.05, ***P < 0.001. (B–L) Quantitative analysis of mitochondrial (mito mRNA) and nuclear encoded mRNAs. RNA was converted to cDNA and used as template and real time qPCR was used to quantify mRNA copy numbers relative to 1000 copies of reference gene β-actin.*P < 0.05, **P < 0.01. (M) MtDNA damage assessed using the elongase method in HC (n = 10), DC (n = 14) and DN (n = 16), data are presented as mean ± SEM. (N) Representative agarose gels showing amplicon A (−) and after surveyor nuclease digestion (+). Digested bands in DN patients are illustrated with red arrows.
To investigate if the changes in circulating MtDNA were accompanied by altered transcription of mitochondrial mRNAs, we quantified mitochondrial genome coded OXPHOS subunits; NADH oxidase subunit VI (ND6), NADH oxidase subunit I (ND1) and cytochrome c oxidase subunit 3 (COX3); and nuclear encoded mitochondrial mRNAs involved in mitochondrial biogenesis (mitochondrial transcription factor A — TFAM, Peroxisome proliferator-activated receptor gamma coactivator 1 alpha — PGC1A), fission (Dynamin-1-like protein — DRP1), fusion (Mitofusin 1 and 2-MFN1 and MFN2, Optic atrophy 1 — OPA1) and mitophagy (PTEN-induced putative kinase 1 — PINK1, parkin RBR E3 ubiquitin protein ligase — PARK2, Fig. 1B–L).
As the amount of MtDNA is known to be directly proportional to the amount of mRNA (Hock and Kralli, 2009, Williams, 1986) we expected to see changes in mitochondrial encoded mRNAs. However, despite altered MtDNA levels in DC and DN, we did not see any significant alterations in mRNAs levels of mitochondrial encoded subunits (Fig. 1B–D) or in most nuclear encoded mitochondrial mRNAs (Fig. 1F–J), suggesting that the altered MtDNA levels are not functional. This idea is further supported by the reduced TFAM levels (Fig. 1E), and a significant reduction in mitophagy mRNAs in diabetes patients (Fig. 1K and L), suggesting that removal of dysfunctional mitochondria is hampered. Despite having higher MtDNA, DC patients have reduced TFAM compared to HC, and despite having lower MtDNA, DN patients have higher TFAM than DC, supporting the idea that the MtDNA may comprise of a mixture of functional and non-functional molecules. One possibility is that MtDNA in these samples comprises of both intact and damaged/mutated MtDNA i.e. there could be a level of heteroplasmy. The integrity of the MtDNA was determined in a subset of patients, using two methods: a PCR based “elongase method” (Furda et al., 2014) and the surveyor nuclease method (Bannwarth et al., 2006). Using the elongase method, we found that relative amplification was lower in DC (84 ± 50) and DN (85 ± 33) compared to HC (107 ± 37), but this difference was not significant (P > 0.05 Fig. 1M). Using surveyor nuclease, we detected mismatches in 5 out of 10 DN patients, but none in 10 HC patients (Fig. 1N). We were unable to confirm presence of mutations by Sanger sequencing. As surveyor nuclease can detect > 3% of heteroplasmy whereas with Sanger sequencing detection levels are > 20% (Bannwarth et al., 2006), our data suggest that the DN patients have damaged MtDNA genomes present at between 3% and 20% of the total MtDNA content.
3.2. Reduced Mitochondrial Metabolism in PBMCs From DN Patients
The data above showed that DN patients have reduced circulating MtDNA compared to DC patients and an increased level of heteroplasmy. It is possible that this could affect cellular energy production. Therefore we examined whether there is any difference in mitochondrial metabolism by determining the bioenergetic profile of HC, DC, and DN groups. The baseline characteristics of this subset of patients (Table S2) show similar trends to the whole study group (Table 1). We used freshly purified PBMCs which were used for the assays within 2 h of collection from patients. Basal, ATP-linked, maximal oxygen consumption rate (OCR), reserve capacity and extracellular acidification rate (ECAR) were measured using a Seahorse XFe96 analyser. OCR, an indicator of mitochondrial respiration, and ECAR, representative of glycolysis, were normalised to the cell number of PBMCs from HC (n = 10), DC (n = 14) and DN patients (n = 16).
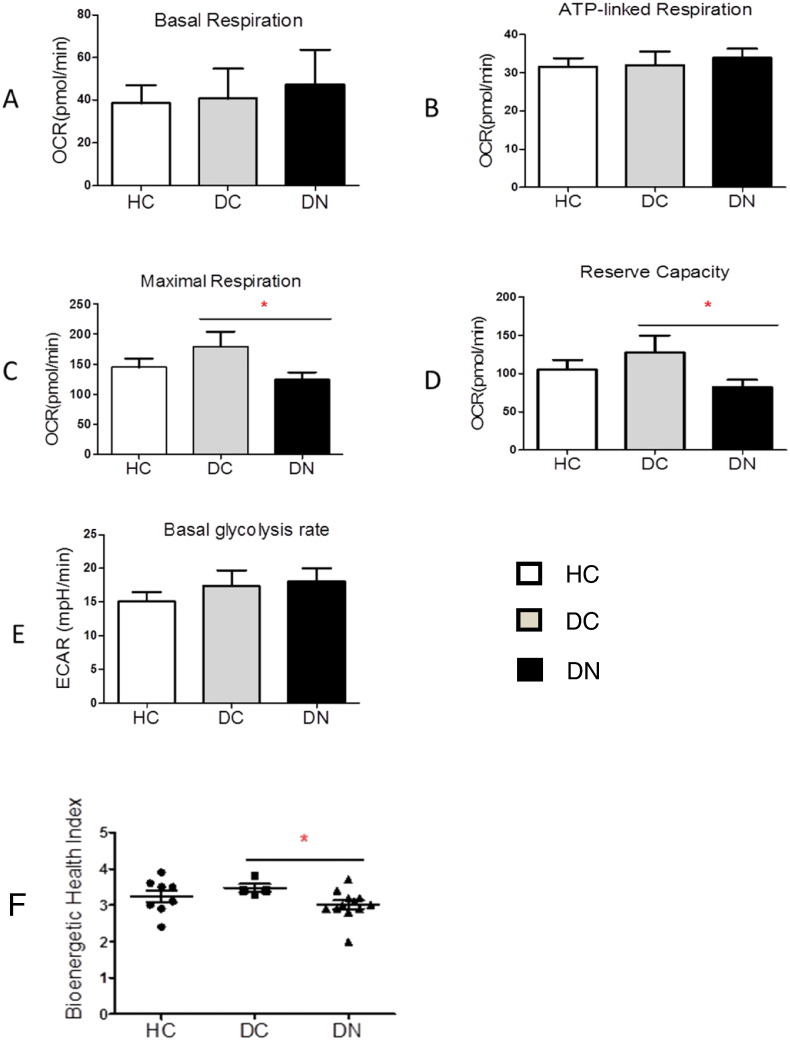
Basal respiration (Fig. 2A), ATP-linked respiration (Fig. 2B), basal glycolytic rate (Fig. 2E), proton leak and non-mitochondrial respiration (data not shown) were similar in the 3 groups. However, both maximal respiration and reserve capacity were significantly reduced by ~ 40% (P < 0.05) in DN patients (Fig. 2C and D). These data suggest that whilst energy production capability under normal physiological conditions is likely to be similar in the 3 groups, reduced reserve capacity and reduced maximal respiration in DN patients are suggestive of a compromised response to stress.
Fig. 2.
Dysfunctional metabolic response in live peripheral blood mononuclear cells (PBMCs) from patients with diabetic nephropathy. PBMCs from HC (n = 10), DC (n = 14) and DN (n = 16) were isolated, seeded at 3 × 105 cells/well, and the Seahorse XFe96 extracellular flux analyser was used to measure oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). Data presented as mean ± SEM and analysed by one-way ANOVA with Tukey's test. (A) Basal respiration rate. (B) ATP-linked respiration after ATP synthase blocker (oligomycin) injection. (C) Maximal (uncoupled) respiration rate measured after the FCCP injection *P < 0.05. (D) Reserve capacity, measured as a difference between maximal and basal respiration, *P < 0.05. (E) Basal ECAR (glycolysis). (F) BHI calculated for HC (n = 8), DC (n = 5) and DN (n = 12) groups, *P < 0.05.
3.3. Loss of Metabolic Flexibility in Diabetic Nephropathy
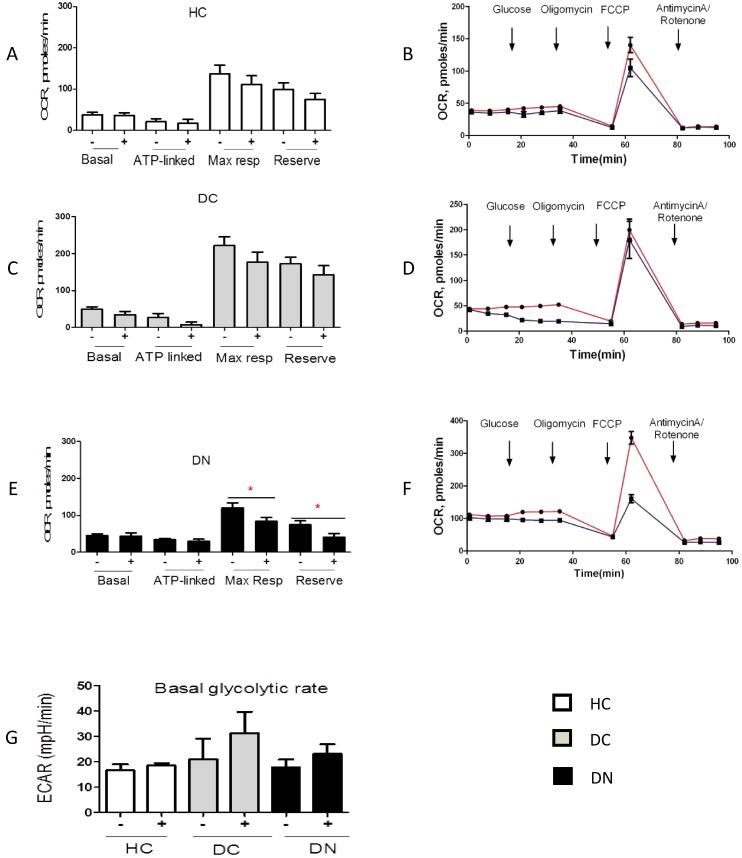
To investigate whether DN patients have a reduced response to stress, we determined the effect of acute applied stress (Fig. 3A–G) in PBMCs. Cell chambers were injected with high glucose (20 mM) and the change in mitochondrial respiration was measured immediately. Acute glucose loading had no impact on PBMCs from HC and DC, with a trend for slight but non-significant reduction in basal, ATP-linked, maximal respiration and reserve capacity, showing that these cells are able to handle acute stress (Fig. 3A–D). In contrast, PBMCs from DN patients were sensitive to acute glucose loading (Fig. 3E and F), as although there was no significant change in basal and ATP-linked respiration, both maximal respiration and reserve capacity were significantly decreased (P < 0.05). Maximal respiration (119.8 ± 39.7) decreased 40% (83.7 ± 29.1, P = 0.05) and reserve capacity (74.4 ± 30) decreased ~ 50% (40.2 ± 0.7, P = 0.04) after acute load in DN patients (n = 8). There was no significant difference in basal ECAR between the three groups in high glucose and normal glucose (Fig. 3G).
Fig. 3.
The loss of metabolic flexibility in diabetic nephropathy patients. Live PBMCs from HC (n = 4), DC (n = 4) and DN (n = 8) were isolated, seeded at 3 × 105 cells/well and analysed using the Mito Stress Test (Seahorse) in the Seahorse XFe96 analyser. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were determined without (−, red) and with (+, blue) acute glucose load. Data are expressed as mean ± SEM, n = 12–20 assay replicates per sample (A, C, D). Independent Student's t-test. (A) Mean OCR values in HC samples. (B) Representative example from a single HC sample. (C) Mean OCR values DC samples. (D) Representative example from a single DC patient. (E) Mean OCR in DN patients. (F) Representative example from a single DN patient. (G) Mean ECAR from HC, DC and DN.
As this data shows that DN patients have a compromised metabolic response, with a reduced maximal respiration and reserve capacity compared to HC and DC, we sought to determine if there is any translational potential in identifying DN patients using the bioenergetics response of PBMCs. Therefore, the bioenergetics data (Fig. 2 A–D) were used to calculate the Bioenergetic Health Index (BHI) as described recently by the Darley-Usmar group (Chacko et al., 2014) (Fig. 2F). The mean (+/− SD) BHI value for DN patients (3.0 ± 0.4, n = 12, P = 0.01) was significantly lower than DC (3.4 ± 0.2, n = 5) (Fig. 2F, Table S4).
3.4. Hyperglycaemia-induced Changes in Mitochondrial DNA and mRNAs in Human Renal Glomerular Mesangial Cells
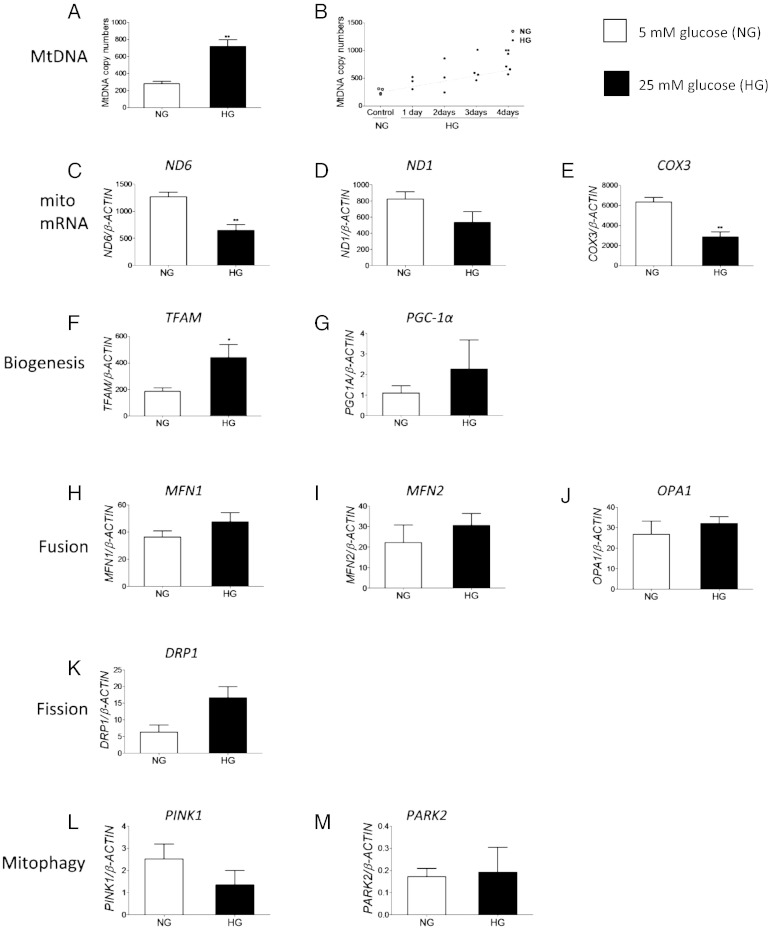
The data presented above suggested that in PBMCs from DN patients there may be a dis-connect between MtDNA levels and mitochondrial mRNA levels. As PBMCs may be reflective of systemic changes in the body (Rudkowska et al., 2011), we wanted to determine if diabetes could affect renal mitochondria. To investigate this we used primary glomerular mesangial cells (HMCs) and examined the effect of hyperglycaemia on their mitochondria. Growth of HMCs in 25 mM glucose (HG) for 4 days resulted in a ~ 2.5-fold increase in cellular MtDNA content (717 ± 157) compared to cells cultured in 5 mM glucose (NG) (280 ± 53, n = 4, P < 0.01, Fig. 4A). A time course experiment showed that MtDNA content began to increase within 24 h of incubation in HG, and this increase was statistically significant (P = 0.02) after 72 h (Fig. 4B).
Fig. 4.
Glucose-induced changes in mitochondrial DNA content and mitochondrial gene transcription in kidney cells. HMCs were cultured in 5 mM glucose (NG) and 25 mM glucose (HG) for 4 days and used to prepare DNA and RNA. Real-time qPCR was carried out to determine MtDNA content using the DNA as template, and expressed as the mitochondrial to the nuclear genome ratio. For mRNA quantification the RNA was converted to cDNA and used as template, mRNA copy numbers for each gene were determined relative to 1000 copies of reference gene β-actin. Data are presented as mean ± SEM, n > 3 independent experiments. Independent Student's t-test. (A) Quantitative measurement of the MtDNA content in HMCs after 4 days, **P < 0.01. (B) MtDNA content in HMCs measured every 24 h, *P < 0.05, **P < 0.01. (C–M) Quantitative analysis of mitochondrial (mito mRNA) and nuclear encoded mRNAs in HMCs, *P < 0.05, **P < 0.01.
To examine whether increased MtDNA content after 4 days of growth in HG was accompanied by increased transcription of mitochondrial mRNAs, we quantified mitochondrial genome encoded OXPHOS subunits, and nuclear encoded mitochondrial mRNAs involved in mitochondrial biogenesis, fission, fusion and mitophagy.
Surprisingly, there was a more than 50% reduction in ND6 and COX3 mRNA in HG (Fig. 4C and E, P < 0.01), and no significant change was observed in ND1 mRNA (Fig. 4D, P > 0.05), suggesting that increased MtDNA did not result in increased mitochondrial genome transcription. TFAM mRNA was significantly increased by ~ 140% (Fig. 4F, P < 0.05), but the levels of the remaining mRNAs did not change in HG (P > 0.05, Fig. 4G–M). No changes were observed at the protein level when mitochondrial respiratory complexes I–V were measured via Western blot (data not shown).
3.5. Functional Consequences of Hyperglycaemia in Kidney Cells
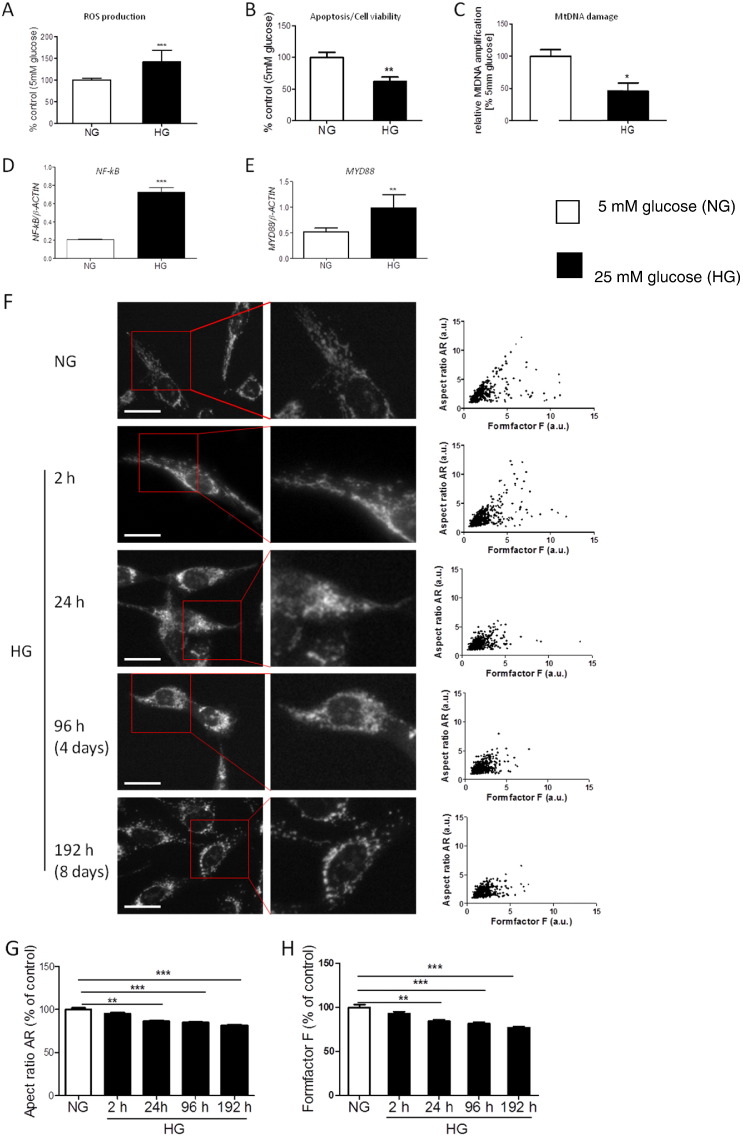
Since nuclear encoded mRNAs involved in the mitochondrial life cycle did not respond to hyperglycaemia in the time frame in which the MtDNA increase was seen; we examined intracellular reactive oxygen species (ROS) (measured using DCF fluorescence which is an assay of generalised oxidative stress and not mitochondrial specific, Kalyanaraman et al., 2012), cell viability, MtDNA damage, and mitochondrial morphology in order to elucidate the underlying mechanisms.
There was a significant increase in ROS in HMCs grown in HG (Fig. 5A, P < 0.001). In parallel to the increased ROS, cell viability was significantly reduced in HMCs grown in HG (Fig. 5B). As the increase in ROS was paralleled with the increase in MtDNA (Fig. 4A) and a reduction in MtDNA transcription (Fig. 4C–E), we speculated that ROS might damage the MtDNA, and that the damaged MtDNA may then activate inflammation via the TLR9 pathway (Oka et al., 2012). MtDNA damage was significantly increased in cells grown in HG at 4 days (Fig. 5C, P < 0.05), and at the same time both NF-κB and MYD88 mRNAs were significantly increased (Fig. 5D and E, P < 0.001, P < 0.01), suggesting activation of the TLR9 pathway.
Fig. 5.
Functional consequences of hyperglycaemia in kidney cells. HMCs were cultured in 5 mM glucose (NG) and 25 mM glucose (HG) for the times shown. (A–B) ROS production and cell viability shown as a % of control. Data are presented as mean ± SEM, n = 17–21 observations. Non-parametric Mann Whitney test, **P < 0.01, ***P < 0.001. (C) Quantitative analysis of MtDNA damage assessed using the elongase method. Data are presented as mean ± SEM, n = 2, 5–6 observations/experiment. Mann–Whitney test, *P < 0.05. (D, E) mRNA expression analysis of NF-kB, MYD88 in HMCs. RNA was converted to cDNA and used as template, mRNA copy numbers for each gene were determined relative to 1000 copies of reference gene β-actin. Data are presented as mean ± SEM, n = 3. Independent Student's t-test, **P < 0.01, ***P < 0.001. (F) Mitochondrial morphology in HMCs exposed to high glucose. Mitochondria were labelled with MitoTracker Red CMXRos and images were captured at magnification × 20. Representative images of each condition shown in left panel. Mitochondrial length (aspect ratio — AR) was plotted against mitochondrial degree of branching (formfactor F) and shown on the right panel as scatter plots. Scale bar, 50 μm. (G–H) Quantitative analysis of aspect ratio AR and formfactor F shown as a % of the control. Data are presented as mean ± SEM, n = 2, > 14 cells/experiment. Non-parametric Kruskal–Wallis test with Dunn's multiple comparison test, **P < 0.01, ***P < 0.001.
MitoTracker Red staining was used to examine the effect of HG on the mitochondrial network in HMCs. Images were acquired at different time points (Fig. 5F). In cells grown in NG, the mitochondrial network was seen as a mixture of elongated and connected mitochondria (Fig. 5F top panel), and no significant changes were observed during the duration of the experiment (data not shown). When cells were grown in HG, their mitochondria became more rounded and fragmented (Fig. 5F). Plotting the values for both mitochondrial length/width (aspect ratio AR) versus degree of branching (formfactor F) showed a progressive reduction in both parameters with increased time of HG exposure (Fig. 5F right panel). The aspect ratio was significantly down regulated in cells exposed to HG, and appeared to change within 24 h (Fig. 5G, P < 0.01). This change became highly significant after a longer time of exposure, with the appearance of more mitochondria of a rounded shape (Fig. 5G, P < 0.001). Formfactor F values were also significantly decreased, following 24 h of exposure to HG with the lowest values at day 8 (Fig. 5H, P < 0.001). These results suggest that incubation in HG leads to increased oxidative stress, MtDNA damage, reduced cellular viability, mitochondrial fragmentation and loss of mitochondrial network connection indicating mitochondrial dysfunction.
3.6. Reduced Metabolism in Kidney Cells Grown in High Glucose
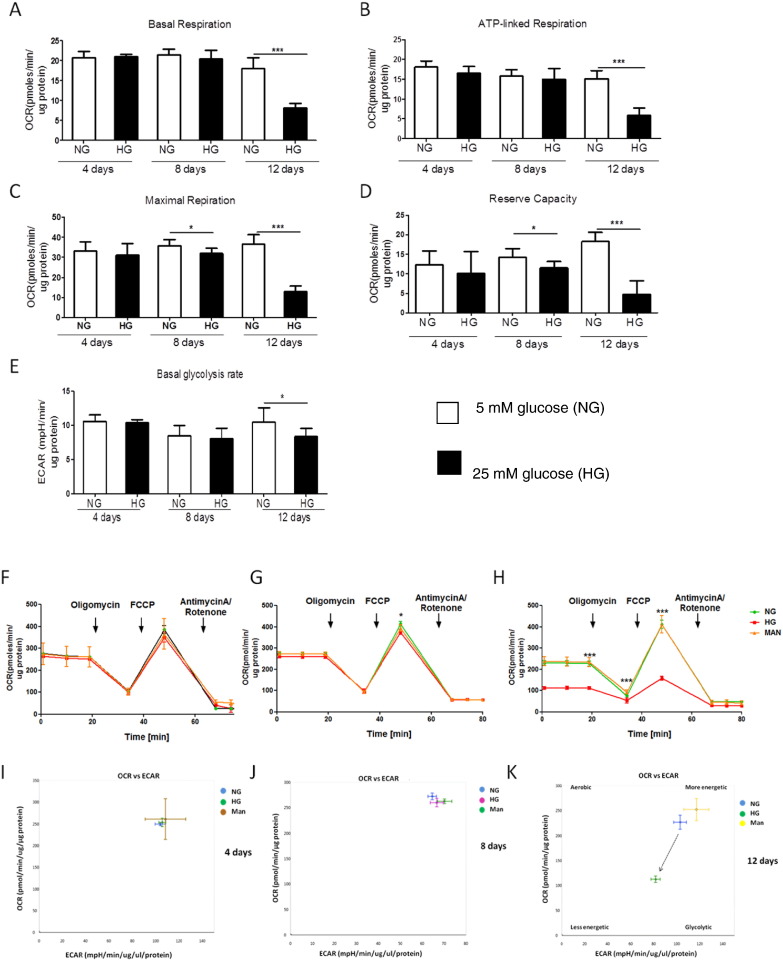
To assess the effect of continued hyperglycaemia on mitochondrial bioenergetics in renal cells, HMCs were incubated in NG and HG for 4, 8 and 12 days, and basal, ATP-linked, maximal OCR, reserve capacity and ECAR were measured using a Seahorse XFe96 analyser (Fig. 6A–H). OCR and ECAR were both normalised to the protein content as HMCs are growing cells, and presented as pmolesO2/min/μg protein.
Fig. 6.
Reduced metabolism in kidney cells grown in high glucose. HMCs were grown in 5 mM glucose (NG, open bars) and 25 mM glucose (HG, solid bars) for 4, 8 and 12 days, and then plated at a density of 3.5 × 104 cells/well one day prior to the experiment. The Seahorse XFe96 extracellular flux analyser was used to measure oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), all data were normalised to the protein content and shown as pmoles/min/ug protein. (A) Quantitative measurement of the basal OCR in HMCs, ***P < 0.001. (B) ATP-linked OCR after oligomycin injection. (C) Maximal (uncoupled) OCR after FCCP injection, *P < 0.05, ***P < 0.001. (D) Quantitative measurement of the reserve capacity, measured as a difference between maximal and basal respiration rates, *P < 0.05, ***P < 0.001. (E) Basal ECAR (glycolysis), *P < 0.05. (F–H) Representative mito stress test runs showing mitochondrial respiration in HMCs cultured in NG, HG and osmotic control (Man) at 4, 8 and 12 days respectively. (I–K) Representative analysis of the basal OCR plotted against basal ECAR in HMCs cultured in NG, HG and osmotic control (Man) at 4, 8 and 12 days respectively. HMCs grown in HG for 12 days had lower OCR/ECAR ratio, metabolic shift is indicated by an arrow. Data are presented as mean ± SEM, n = 6–8 replicates for 2 independent experiments. Independent Student's t-test.
We could detect no differences in the bioenergetic profile of cells incubated in HG for 4 days, as basal, ATP-linked, maximal respiration and basal glycolytic rate were similar between cells grown in NG and HG (Fig. 6A–F). Cells exposed to HG for 8 days had similar basal and ATP-linked respiration and the basal glycolytic rate, but had a reduced maximal respiration (P < 0.05, Fig. 6A–E and G). After 12 days of incubation in HG, cells showed significantly reduced basal, ATP-linked, maximal respiration and basal glycolytic rate (P < 0.05, Fig. 6A–E and H). No changes were observed in the proton leak and non-mitochondrial respiration for any of the time points (data not shown). The reserve capacity was unchanged at 4 days, but was reduced in cells grown in HG after 8 and 12 days (P < 0.05, Fig. 6D and F–H). Mitochondrial respiration in cells incubated in HG was independent of osmolarity or culture time (data not shown). Comparison of basal respiration rate with basal glycolytic rate showed no difference at 4 and 8 days in HG (Fig. 6I and J). However, after 12 days, the cells grown in HG had reduced OCR and ECAR (P < 0.05) which shows that they have shifted to a less metabolically active state (Fig. 6K).
4. Discussion
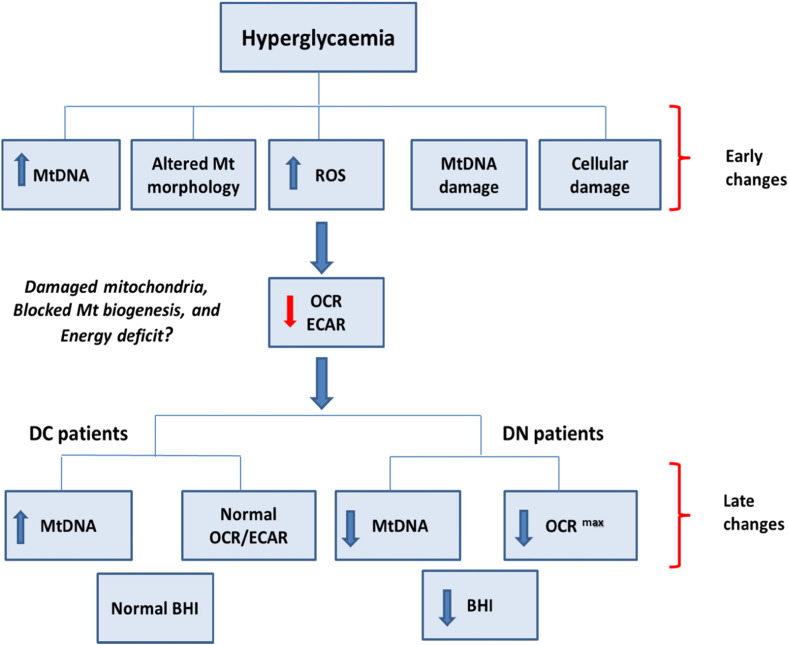
Diabetes increases the risk of multi-organ complications affecting various cell types and is a major cause of blindness, renal failure, and cardiovascular disease (Gordin et al., 2012, He and King, 2004). Our study suggests that systemic mitochondrial dysfunction and glucose induced changes in MtDNA parallel diabetic kidney disease. The role of MtDNA mutations in mitochondrial genetic disease (Hudson et al., 2014, Niaudet, 1998, Tuppen et al., 2010, Wallace and Chalkia, 2013) and of MtDNA as an inflammatory molecule (Collins et al., 2004, Zhang et al., 2010) has been demonstrated. Specific MtDNA mutations can lead to diabetes and kidney disease in patients with mitochondrial genetic disease (D'Aco et al., 2013, Mazzaccara et al., 2012, Seidowsky et al., 2013), suggesting that acquired MtDNA damage may play a role in DN. We present novel evidence showing that glucose induced increase in MtDNA in renal cells precede mitochondrial dysfunction, suggesting a novel cascade of events contributing to mitochondrial dysfunction in patients with DN (Fig. 7).
Fig. 7.
Early glucose induced changes in mitochondria may lead to diabetic nephropathy: a schematic diagram with each box referring to key findings of this paper given in brackets. The early changes refer to data obtained from cultured renal cells. The late changes refer to data obtained from PBMCs from HC, DC and DN patients. The evidence supporting this schematic in the paper: hyperglycaemia leads to increased MtDNA content (Fig. 4A), altered mitochondrial morphology (Fig. 5, F–H), increased intracellular reactive oxygen species (ROS Fig. 5A), mitochondrial DNA damage (Fig. 5C), cellular damage measured as apoptosis/viability (Fig. 5B), and inflammation (Fig. 5, D and E) as early changes in cells. These changes precede and are followed by reduced OCR (basal, maximal, ATP-linked), reserve capacity and ECAR (Fig. 6, A–E). These changes may result in damaged mitochondria, blocked mitochondrial biogenesis and an energy deficit. The long term consequences of such changes could be as shown in the bottom panel. In DC patients these changes are seen as an increase in MtDNA (Fig. 1A) and normal metabolism (Fig. 2, A–E) however DN patients have decreased MtDNA (Fig. 1A) accompanied with a dysfunctional metabolic response (Fig. 2A–E) resulting in reduced BHI (Fig. 2F). There is evidence of reduced mitophagy in both DC and DN patients (Fig. 1, K and L).
We were interested in comparing DC patients, who had a long duration of diabetes but did not develop kidney disease, with the DN group, who had developed kidney disease, in order to determine if susceptibility for the development of diabetic kidney disease may involve mitochondrial dysfunction. Our data shows that it is possible using PCR based assays to detect differences in MtDNA and mitochondrial mRNAs between diabetes patients who develop kidney disease and those that don't. We used extracellular flux analysis, as previously described (Ferrick et al., 2008), to measure cellular respiration, and showed, using live PBMCs, that DN patients have a diminished bioenergetic response. We also converted the bioenergetics data to the single value “Bioenergetic Health Index (BHI)” which has recently been proposed as a new biomarker for assessing patient health with both prognostic and diagnostic value (Chacko et al., 2014), and we found that DN patients had significantly reduced BHI. To our knowledge this is the first report demonstrating altered BHI in patients, supporting the hypothesis that mitochondrial dysfunction is involved in DN. In fact, there have been very few studies reporting metabolic function in live PBMCs and none specifically in diabetic complications (Hartman et al., 2014, Maynard et al., 2013). We have detected a clear difference in metabolic function between the DC and DN group and our data suggests that inability to cope with energy demand due to MtDNA damage could be a key factor in the progression of DN.
To establish whether hyperglycaemia can affect renal mitochondria and contribute to mitochondrial dysfunction, we used primary glomerular mesangial cells, widely used as an experimental model of DN (Clarkson et al., 2002, Thomas et al., 2000). The data derived from the HMC experiments supports the hypothesis that glucose can lead to alterations in MtDNA and mitochondrial function in renal cells and may be considered as representative of an early stage in diabetes. Exposure of HMCs to hyperglycaemia led to changes in mitochondrial morphology and increased MtDNA levels with 24 h, after 4 days there was reduced transcription of the mitochondrial genome, increased ROS and MtDNA damage, and activation of the NF-κB pathway, and after 8–12 day HMCs displayed reduced mitochondrial metabolism. These results suggest that the observed similar defect seen in PBMCs occur at a systemic level and could be the cause of the changes we see in mitochondrial parameters in PBMCs from DN patients. Circulating MtDNA levels have been previously reported to be increased in diabetes but there have been contradictory reports in DN (Lee et al., 2009, Malik et al., 2009), possibly due to the fact that earlier studies used mitochondrial primers which may amplify nuclear pseudogenes with high homology to MtDNA (Malik and Czajka, 2013, Malik et al., 2011). In the current study, using unique primers and a protocol designed for accurate MtDNA quantification from blood (Ajaz et al., 2014) we found that DC patients had significantly higher levels of circulating MtDNA compared to HC. Several mitochondrial- and nuclear-encoded mitochondrial mRNAs remained unchanged, apart from reduced TFAM, PINK1 and PARK2, suggesting that the increased MtDNA in DC patients may not be fully functional, and that mitophagy may be impaired. The inhibition of mitophagy and the presence of damaged MtDNA are very likely to lead to a decrease in mitochondrial quality. The DN patients had significantly reduced MtDNA compared to DC. Binary regression analysis showed that eGFR, A/C ratio, and MtDNA levels were independently associated with DN. As eGFR and A/C ratio are well established risk markers of DN, our data suggests that the MtDNA changes could be a risk marker for DN; prospective studies will have to answer this question in the future.
A key finding of our study is the demonstration that exposure to HG leads to a rapid increase in mesangial cell MtDNA, accompanied by unchanging/decreased transcription of mitochondrial encoded mRNAs and little change in nuclear encoded mitochondrial mRNAs in renal cells. The amount of MtDNA is known to be directly proportional to the amount of mitochondrial encoded mRNAs (Hock and Kralli, 2009, Williams, 1986) however in our study increased MtDNA levels in patients (Fig 1A) are not accompanied by increased mitochondrial encoded mRNAs (Fig. 1B–D), we see the same trend in cells, where the mitochondrial encoded mRNAs are reduced (Fig. 4C–E) whereas the MtDNA in increased (Fig. 4A). As the function of MtDNA is to encode mitochondrial mRNAs, we deduce that the increased MtDNA in not functional. The only exception is the glucose induced up-regulation of TFAM mRNA, which has been previously reported (Choi et al., 2004). Our observation is suggestive of a disconnect between the increase in MtDNA and mitochondrial transcription/translation. A similar disconnect was reported in a study investigating the mechanisms of insulin resistance in muscle, with increased MtDNA but not increased mitochondrial content in myotubes (Aguer et al., 2013). Increased MtDNA in cultured cells was reported in response to hydrogen peroxide induced oxidative stress (Lee and Wei, 2005). HG induced ROS may inhibit mitochondrial biogenesis, as has been shown in other systems (Ballinger et al., 2000) and this view is consistent with reduced/unaltered mitochondrial transcription and translation which we observe. The increase in MtDNA in these conditions could be a compensatory response to the impairment of transcription.
We measured cellular respiration in HMCs and found that oxidative metabolism can cope for several days of high stress conditions and continues to provide energy. Cells started to display altered morphology within a few hours of exposure to HG with mitochondrial fragmentation, which may be part of the initiation of an adaptive biogenesis programme to increase MtDNA content. Whilst glucose induced MtDNA increase was evident within 24 h, cells showed no detectable perturbation in their metabolic profile at this time point. Some changes were seen after 8 days, however it was only after 12 days of culture that significantly reduced ECAR and OCR were observed. To our knowledge this is the first report of the metabolic response of HMCs, although previously a meeting report on mouse mesangial cells showed similar profiles to ours (Chacko et al., 2010a). Our data agree with a study using rat retinal endothelial cells where growth in HG for 6 days led to reduced basal and maximal respiration, however unlike our study where ECAR was decreased, they found increased ECAR (Trudeau et al., 2010). The reduced ECAR in our cells is suggestive of an energy deficit and may be a consequence of a ROS induced glycolytic block (Colussi et al., 2000). This idea is supported by our finding of HG induced ROS in our cells. The importance of mitochondrial ROS in oxidative stress in the kidney is well established, and mitochondrial targeted antioxidants ameliorate certain markers of renal damage (Chacko et al., 2010b). Here we show that the increased ROS is accompanied by increased MtDNA damage and NF-κB and MYD88 mRNAs. Like bacterial DNA, MtDNA is un-methylated and can initiate immune responses via the intracellular Toll like receptor (TLR) 9 (Higginbotham et al., 2002). The inflammatory properties of MtDNA (Collins et al., 2004, Zhang et al., 2010) and disruption of the normal degradation of MtDNA in the cytosol of cardiomyocytes was shown to cause TLR9-mediated inflammation leading to a heart failure in a mouse model (Oka et al., 2012). It is well established that chronic inflammation is a key mediator in diabetic complications but the underlying mechanisms have not been elucidated (Navarro-Gonzalez et al., 2011). Our data in HMCs of increased MtDNA content, ROS, MtDNA damage, and up-regulation of MYD88 and NF-κB support the idea of MtDNA induced activation of the inflammatory TLR9 pathway in these cells and are suggestive of a novel mechanism leading to chronic inflammation in DN.
MtDNA is usually present in multiple identical copies in the cells, but the location of MtDNA close to the electron transport chain renders it more susceptible to ROS induced damage and mutations. In diabetes patients, MtDNA may be exposed to hyperglycaemia induced oxidative stress (Singh et al., 2011) resulting in random mutations in the mitochondrial genome leading to heteroplasmy, the presence of normal and mutated MtDNA in the same cell. The damaged MtDNA in such cases would only have an impact once it reached a threshold where the normal non-mutated MtDNA is no longer able to compensate for mitochondrial dysfunction (Chinnery, 2002). It has recently been demonstrated that MtDNA heteroplasmic mutations are associated with widely spread chronic diseases, including atherosclerosis and cancer (Sobenin et al., 2014, Ye et al., 2014). We used two methods to detect MtDNA damage, both of which suggest that MtDNA from diabetes samples shows increased damage, with heteroplasmy being detected only in DN samples. Furthermore, HMCs showed a significant increase in MtDNA damage after 4 days of growth in hyperglycaemic conditions, suggesting MtDNA damage in HMCs could play a role in the subsequent bioenergetic deficit observed in these cells after 8 and 12 days of hyperglycaemia.
The combination of the data presented in this paper paint a picture of severely compromised mitochondria in patients with DN. These patients have reduced MtDNA in circulation, with increased MtDNA damage, and decreased mitophagy, the functional impact of these changes is the reduced metabolic flexibility of these cells. Furthermore if we assume that the PBMCs are representative of systemic changes in the body, then it could be predicted that kidney cells in the DN patients will show a similar compromised response, which could play a major role in progression of pathology. Our results support a recent metabolomics study showing that patients with DN have reduced mitochondrial metabolites in urine (Sharma et al., 2013) and we propose that DN could be viewed as a disease of acquired mitochondrial dysfunction.
In conclusion, we have shown that metabolic dysfunction can be detected in peripheral blood samples of patients with DN. We have also shown using renal cells in-vitro that hyperglycaemia affects mitochondria, with MtDNA levels changing before other indicators of mitochondrial dysfunction. Our data suggests that the BHI formula can indicate mitochondrial dysfunction in live PBMCs from patients, and therefore could be developed into a non-invasive translational measure of mitochondrial function. It is of importance to determine if the changes we observe in clinical samples precede the onset of DN, and therefore longitudinal studies should be carried out to determine this. The potential of regular and routine monitoring of MtDNA content, integrity and BHI as indicators of DN should be further evaluated.
Acknowledgements
Special thanks to the patients, and the volunteers without whose samples this work could not have been done, we are indebted to the nursing/clinical staff at the Diabetes clinic at Guy's Hospital, especially Nurse Siew Cohen for her tireless help and constant encouragement. Thanks to Dr Sylvie Bannwarth (Laboratoire de GénétiqueMoléculaire, Nice France) for the help with the surveyor nuclease method, Dr John Harris and the Nikon Imaging Centre at KCL, Alex Liversage of Seahorse Biosciences for loaning us a Seahorse XFe96, Professor Kinya Otsu, KCL, Professor Victor Darley-Usmar (University of Alabama, USA) and Dr David Ferrick, Seahorse Biosciences, for the critical evaluation of this manuscript.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.04.002.
Appendix A. Supplementary Data
S1: baseline characteristics of healthy controls and diabetic patients.
S2: baseline characteristics of patients and healthy controls used in Seahorse subset showing a similar trend to the whole study group in Table 1.
S3: human oligonucleotide primers used in this study.
S4: raw values of data used for calculation of BHI.
S5: MtDNA content of patients by sub-dividing as type 1 and type 2 diabetes.
References
- Aguer C., Pasqua M., Thrush A.B., Moffat C., McBurney M., Jardine K., Zhang R., Beauchamp B., Dent R., McPherson R. Increased proton leak and SOD2 expression in myotubes from obese non-diabetic subjects with a family history of type 2 diabetes. Biochim. Biophys. Acta. 2013;1832:1624–1633. doi: 10.1016/j.bbadis.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Ajaz S., Czajka A., Malik A.N. Accurate measurement of circulating mitochondrial DNA content from human blood samples using real-time quantitative PCR. Methods Mol. Biol. 2014;1264 doi: 10.1007/978-1-4939-2257-4_12. [DOI] [PubMed] [Google Scholar]
- Alaveras A.E., Thomas S.M., Sagriotis A., Viberti G.C. Promoters of progression of diabetic nephropathy: the relative roles of blood glucose and blood pressure control. Nephrol. Dial. Transplant. 1997;12(Suppl. 2):71–74. [PubMed] [Google Scholar]
- Aronson D. Hyperglycemia and the pathobiology of diabetic complications. Adv. Cardiol. 2008;45:1–16. doi: 10.1159/000115118. [DOI] [PubMed] [Google Scholar]
- Ballinger S.W., Patterson C., Yan C.N., Doan R., Burow D.L., Young C.G., Yakes F.M., Van Houten B., Ballinger C.A., Freeman B.A. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ. Res. 2000;86:960–966. doi: 10.1161/01.res.86.9.960. [DOI] [PubMed] [Google Scholar]
- Bannwarth S., Procaccio V., Paquis-Flucklinger V. Rapid identification of unknown heteroplasmic mutations across the entire human mitochondrial genome with mismatch-specific Surveyor Nuclease. Nat. Protoc. 2006;1:2037–2047. doi: 10.1038/nprot.2006.318. [DOI] [PubMed] [Google Scholar]
- Brand M.D., Nicholls D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- Chacko B.K., Reily C., Srivastava A., Johnson M.S., Ye Y., Ulasova E., Agarwal A., Zinn K.R., Murphy M.P., Kalyanaraman B. Prevention of diabetic nephropathy in Ins2(+/)−(AkitaJ) mice by the mitochondria-targeted therapy MitoQ. Biochem. J. 2010;432:9–19. doi: 10.1042/BJ20100308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacko B.K., Reily C., Benavides C.A., Johnson M.S., Darley-Usmar A.V.M. 2010. Chronic Hyperglycemia-induced Attenuation of Mitochondrial Reserve Capacity Mediates Mesangial Cell Dysfunction in Diabetes. [Google Scholar]
- Chacko B.K., Kramer P.A., Ravi S., Johnson M.S., Hardy R.W., Ballinger S.W., Darley-Usmar V.M. Methods for defining distinct bioenergetic profiles in platelets, lymphocytes, monocytes, and neutrophils, and the oxidative burst from human blood. Lab. Investig. 2013;93:690–700. doi: 10.1038/labinvest.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacko B.K., Kramer P.A., Ravi S., Benavides G.A., Mitchell T., Dranka B.P., Ferrick D., Singal A.K., Ballinger S.W., Bailey S.M. The Bioenergetic Health Index: a new concept in mitochondrial translational research. Clin. Sci. (Lond.) 2014;127:367–373. doi: 10.1042/CS20140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery P.F. Modulating heteroplasmy. Trends Genet. 2002;18:173–176. doi: 10.1016/s0168-9525(01)02636-1. [DOI] [PubMed] [Google Scholar]
- Choi Y.S., Lee K.U., Pak Y.K. Regulation of mitochondrial transcription factor A expression by high glucose. Ann. N. Y. Acad. Sci. 2004;1011:69–77. doi: 10.1007/978-3-662-41088-2_8. [DOI] [PubMed] [Google Scholar]
- Clarkson M.R., Murphy M., Gupta S., Lambe T., Mackenzie H.S., Godson C., Martin F., Brady H.R. High glucose-altered gene expression in mesangial cells. Actin-regulatory protein gene expression is triggered by oxidative stress and cytoskeletal disassembly. J. Biol. Chem. 2002;277:9707–9712. doi: 10.1074/jbc.M109172200. [DOI] [PubMed] [Google Scholar]
- Collins L.V., Hajizadeh S., Holme E., Jonsson I.M., Tarkowski A. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J. Leukoc. Biol. 2004;75:995–1000. doi: 10.1189/jlb.0703328. [DOI] [PubMed] [Google Scholar]
- Colussi C., Albertini M.C., Coppola S., Rovidati S., Galli F., Ghibelli L. 2000. H2O2-induced block of glycolysis as an active ADP-ribosylation reaction protecting cells from apoptosis. [DOI] [PubMed] [Google Scholar]
- D'Aco K.E., Manno M., Clarke C., Ganesh J., Meyers K.E., Sondheimer N. Mitochondrial tRNA(Phe) mutation as a cause of end-stage renal disease in childhood. Pediatr. Nephrol. 2013;28:515–519. doi: 10.1007/s00467-012-2354-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranka B.P., Benavides G.A., Diers A.R., Giordano S., Zelickson B.R., Reily C., Zou L., Chatham J.C., Hill B.G., Zhang J. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic. Biol. Med. 2011;51:1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrick D.A., Neilson A., Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov. Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Furda A., Santos J.H., Meyer J.N., Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 2014;1105:419–437. doi: 10.1007/978-1-62703-739-6_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordin D., Waden J., Forsblom C., Thorn L.M., Rosengard-Barlund M., Heikkila O., Saraheimo M., Tolonen N., Hietala K., Soro-Paavonen A. Arterial stiffness and vascular complications in patients with type 1 diabetes: the Finnish Diabetic Nephropathy (FinnDiane) Study. Ann. Med. 2012;44:196–204. doi: 10.3109/07853890.2010.530681. [DOI] [PubMed] [Google Scholar]
- Gruden G., Thomas S., Burt D., Lane S., Chusney G., Sacks S., Viberti G. Mechanical stretch induces vascular permeability factor in human mesangial cells: mechanisms of signal transduction. Proc. Natl. Acad. Sci. U. S. A. 1997;94:12112–12116. doi: 10.1073/pnas.94.22.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman M.L., Shirihai O.S., Holbrook M., Xu G., Kocherla M., Shah A., Fetterman J.L., Kluge M.A., Frame A.A., Hamburg N.M. Relation of mitochondrial oxygen consumption in peripheral blood mononuclear cells to vascular function in type 2 diabetes mellitus. Vasc. Med. 2014;19:67–74. doi: 10.1177/1358863X14521315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., King G.L. Microvascular complications of diabetes. Endocrinol. Metab. Clin. N. Am. 2004;33:215–238. doi: 10.1016/j.ecl.2003.12.003. (xi-xii) [DOI] [PubMed] [Google Scholar]
- Higginbotham J.N., Seth P., Blaese R.M., Ramsey W.J. The release of inflammatory cytokines from human peripheral blood mononuclear cells in vitro following exposure to adenovirus variants and capsid. Hum. Gene Ther. 2002;13:129–141. doi: 10.1089/10430340152712683. [DOI] [PubMed] [Google Scholar]
- Hock M.B., Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu. Rev. Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- Hofmann M.A., Schiekofer S., Isermann B., Kanitz M., Henkels M., Joswig M., Treusch A., Morcos M., Weiss T., Borcea V. Peripheral blood mononuclear cells isolated from patients with diabetic nephropathy show increased activation of the oxidative-stress sensitive transcription factor NF-kappaB. Diabetologia. 1999;42:222–232. doi: 10.1007/s001250051142. [DOI] [PubMed] [Google Scholar]
- Hudson G., Gomez-Duran A., Wilson I.J., Chinnery P.F. Recent mitochondrial DNA mutations increase the risk of developing common late-onset human diseases. PLoS Genet. 2014;10:e1004369. doi: 10.1371/journal.pgen.1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IDF . 6th edition. International Diabetes Federation; 2013. Diabetes Atlas. [Google Scholar]
- Ihm C.G., Park J.K., Hong S.P., Lee T.W., Cho B.S., Kim M.J. Circulating factors in sera or peripheral blood mononuclear cells in patients with membranous nephropathy or diabetic nephropathy. J. Korean Med. Sci. 1997;12:539–544. doi: 10.3346/jkms.1997.12.6.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyanaraman B., Darley-Usmar V., Davies K.J., Dennery P.A., Forman H.J., Grisham M.B., Mann G.E., Moore K., Roberts L.J., II, Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med. 2012;52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick E.S., Rigby A.S., Atkin S.L. The Diabetes Control and Complications Trial: the gift that keeps giving. Nat. Rev. Endocrinol. 2009;5:537–545. doi: 10.1038/nrendo.2009.179. [DOI] [PubMed] [Google Scholar]
- Koopman W.J., Verkaart S., Visch H.J., van der Westhuizen F.H., Murphy M.P., van den Heuvel L.W., Smeitink J.A., Willems P.H. Inhibition of complex I of the electron transport chain causes O2−•-mediated mitochondrial outgrowth. Am. J. Physiol. Cell Physiol. 2005;288:C1440–C1450. doi: 10.1152/ajpcell.00607.2004. [DOI] [PubMed] [Google Scholar]
- Lee H.C., Wei Y.H. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int. J. Biochem. Cell Biol. 2005;37:822–834. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Lee J.E., Park H., Ju Y.S., Kwak M., Kim J.I., Oh H.Y., Seo J.S. Higher mitochondrial DNA copy number is associated with lower prevalence of microalbuminuria. Exp. Mol. Med. 2009;41:253–258. doi: 10.3858/emm.2009.41.4.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A.N., Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion. 2013;13(5):481–492. doi: 10.1016/j.mito.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Malik A.N., Shahni R., Iqbal M.M. Increased peripheral blood mitochondrial DNA in type 2 diabetic patients with nephropathy. Diabetes Res. Clin. Pract. 2009;86:e22–e24. doi: 10.1016/j.diabres.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Malik A.N., Shahni R., Rodriguez-de-Ledesma A., Laftah A., Cunningham P. Mitochondrial DNA as a non-invasive biomarker: accurate quantification using real time quantitative PCR without co-amplification of pseudogenes and dilution bias. Biochem. Biophys. Res. Commun. 2011;412:1–7. doi: 10.1016/j.bbrc.2011.06.067. [DOI] [PubMed] [Google Scholar]
- Maynard S., Keijzers G., Gram M., Desler C., Bendix L., Budtz-Jorgensen E., Molbo D., Croteau D.L., Osler M., Stevnsner T. Relationships between human vitality and mitochondrial respiratory parameters, reactive oxygen species production and dNTP levels in peripheral blood mononuclear cells. Aging (Albany NY) 2013;5:850–864. doi: 10.18632/aging.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzaccara C., Iafusco D., Liguori R., Ferrigno M., Galderisi A., Vitale D., Simonelli F., Landolfo P., Prisco F., Masullo M. Mitochondrial diabetes in children: seek and you will find it. PLoS One. 2012;7:e34956. doi: 10.1371/journal.pone.0034956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel S., Wanet A., De Pauw A., Rommelaere G., Arnould T., Renard P. Crosstalk between mitochondrial (dys)function and mitochondrial abundance. J. Cell. Physiol. 2012;227:2297–2310. doi: 10.1002/jcp.23021. [DOI] [PubMed] [Google Scholar]
- Moraes C.T., Shanske S., Tritschler H.J., Aprille J.R., Andreetta F., Bonilla E., Schon E.A., DiMauro S. mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am. J. Hum. Genet. 1991;48:492–501. [PMC free article] [PubMed] [Google Scholar]
- Murphy M., Godson C., Cannon S., Kato S., Mackenzie H.S., Martin F., Brady H.R. Suppression subtractive hybridization identifies high glucose levels as a stimulus for expression of connective tissue growth factor and other genes in human mesangial cells. J. Biol. Chem. 1999;274:5830–5834. doi: 10.1074/jbc.274.9.5830. [DOI] [PubMed] [Google Scholar]
- Navarro-Gonzalez J.F., Mora-Fernandez C., Muros de Fuentes M., Garcia-Perez J. Inflammatory molecules and pathways in the pathogenesis of diabetic nephropathy. Nat. Rev. Nephrol. 2011;7:327–340. doi: 10.1038/nrneph.2011.51. [DOI] [PubMed] [Google Scholar]
- Niaudet P. Mitochondrial disorders and the kidney. Arch. Dis. Child. 1998;78:387–390. doi: 10.1136/adc.78.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Hikoso S., Yamaguchi O., Taneike M., Takeda T., Tamai T., Oyabu J., Murakawa T., Nakayama H., Nishida K., Akira S., Yamamoto A., Komuro I., Otsu K. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz E., Orth S.R. Nephropathy in patients with type 2 diabetes mellitus. N. Engl. J. Med. 1999;341:1127–1133. doi: 10.1056/NEJM199910073411506. [DOI] [PubMed] [Google Scholar]
- Rudkowska I., Raymond C., Ponton A., Jacques H., Lavigne C., Holub B.J., Marette A., Vohl M.C. Validation of the use of peripheral blood mononuclear cells as surrogate model for skeletal muscle tissue in nutrigenomic studies. OMICS. 2011;15:1–7. doi: 10.1089/omi.2010.0073. [DOI] [PubMed] [Google Scholar]
- Seidowsky A., Hoffmann M., Glowacki F., Dhaenens C.M., Devaux J.P., de Sainte Foy C.L., Provot F., Gheerbrant J.D., Hummel A., Hazzan M. Renal involvement in MELAS syndrome — a series of 5 cases and review of the literature. Clin. Nephrol. 2013;80:456–463. doi: 10.5414/CN107063. [DOI] [PubMed] [Google Scholar]
- Shahni R., Czajka A., Mankoo B.S., Guvenel A.K., King A.J., Malik A.N. Nop-7-associated 2 (NSA2), a candidate gene for diabetic nephropathy, is involved in the TGFbeta1 pathway. Int. J. Biochem. Cell Biol. 2013;45:626–635. doi: 10.1016/j.biocel.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Sharma K., Karl B., Mathew A.V., Gangoiti J.A., Wassel C.L., Saito R., Pu M., Sharma S., You Y.H., Wang L. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J. Am. Soc. Nephrol. 2013;24:1901–1912. doi: 10.1681/ASN.2013020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D.K., Winocour P., Farrington K. Oxidative stress in early diabetic nephropathy: fueling the fire. Nat. Rev. Endocrinol. 2011;7:176–184. doi: 10.1038/nrendo.2010.212. [DOI] [PubMed] [Google Scholar]
- Sobenin I.A., Mitrofanov K.Y., Zhelankin A.V., Sazonova M.A., Postnov A.Y., Revin V.V., Bobryshev Y.V., Orekhov A.N. Quantitative assessment of heteroplasmy of mitochondrial genome: perspectives in diagnostics and methodological pitfalls. BioMed Res. Int. 2014;2014:292017. doi: 10.1155/2014/292017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourris K.C., Harcourt B.E., Penfold S.A., Yap F.Y., Morley A.L., Morgan P.E., Davies M.J., Baker S.T., Jerums G., Forbes J.M. Modulation of the cellular expression of circulating advanced glycation end-product receptors in type 2 diabetic nephropathy. Exp. Diabetes Res. 2010;2010:974681. doi: 10.1155/2010/974681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoves J., Lindley E.J., Barnfield M.C., Burniston M.T., Newstead C.G. MDRD equation estimates of glomerular filtration rate in potential living kidney donors and renal transplant recipients with impaired graft function. Nephrol. Dial. Transplant. 2002;17:2036–2037. doi: 10.1093/ndt/17.11.2036. [DOI] [PubMed] [Google Scholar]
- Stratton I.M., Adler A.I., Neil H.A., Matthews D.R., Manley S.E., Cull C.A., Hadden D., Turner R.C., Holman R.R. Association of glycaemia with macrovascular and microvascular complications of type 2 diabetes (UKPDS 35): prospective observational study. BMJ. 2000;321:405–412. doi: 10.1136/bmj.321.7258.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Vanuystel J., Gruden G., Rodriguez V., Burt D., Gnudi L., Hartley B., Viberti G. Vascular endothelial growth factor receptors in human mesangium in vitro and in glomerular disease. J. Am. Soc. Nephrol. 2000;11:1236–1243. doi: 10.1681/ASN.V1171236. [DOI] [PubMed] [Google Scholar]
- Trudeau K., Molina A.J., Guo W., Roy S. High glucose disrupts mitochondrial morphology in retinal endothelial cells: implications for diabetic retinopathy. Am. J. Pathol. 2010;177:447–455. doi: 10.2353/ajpath.2010.091029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuppen H.A., Blakely E.L., Turnbull D.M., Taylor R.W. Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta. 2010;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Wallace D.C. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wallace D.C., Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Med. 2013;3:a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild S., Roglic G., Green A., Sicree R., King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- Williams R.S. Mitochondrial gene expression in mammalian striated muscle. Evidence that variation in gene dosage is the major regulatory event. J. Biol. Chem. 1986;261:12390–12394. [PubMed] [Google Scholar]
- Wojtczak L., Zabłocki K. Basic mitochondria physiology in cell viability and death. In: Dykens J.A., Will Y., editors. Drug-induced Mitochondria Dysfunction. John Wiley & Sons, Inc.; New Jersey: 2008. pp. 3–36. [Google Scholar]
- Ye K., Lu J., Ma F., Keinan A., Gu Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc. Natl. Acad. Sci. U. S. A. 2014;111:10654–10659. doi: 10.1073/pnas.1403521111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi B., Hu X., Zhang H., Huang J., Liu J., Hu J., Li W., Huang L. Nuclear NF-kappaB p65 in peripheral blood mononuclear cells correlates with urinary MCP-1, RANTES and the severity of type 2 diabetic nephropathy. PLoS One. 2014;9:e99633. doi: 10.1371/journal.pone.0099633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lee A.S., Shameli A., Geng X., Finegood D., Santamaria P., Dutz J.P. TLR9 blockade inhibits activation of diabetogenic CD8 + T cells and delays autoimmune diabetes. J. Immunol. 2010;184:5645–5653. doi: 10.4049/jimmunol.0901814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1: baseline characteristics of healthy controls and diabetic patients.
S2: baseline characteristics of patients and healthy controls used in Seahorse subset showing a similar trend to the whole study group in Table 1.
S3: human oligonucleotide primers used in this study.
S4: raw values of data used for calculation of BHI.
S5: MtDNA content of patients by sub-dividing as type 1 and type 2 diabetes.