Abstract
In the present study, the methylation and protein expression of the runt-related transcription factor 3 (Runx3) gene was detected in sporadic colorectal cancer, colonic adenoma and normal colon tissue to evaluate their clinical significance in colorectal carcinogenesis. A total of 34 colonic cancer specimens, 34 colonic adenoma specimens and 34 normal colonic tissue specimens were used in the study. The CpG island methylation status of the Runx3 gene was detected by methylation-specific polymerase chain reaction and the protein expression of Runx3 was detected by immunohistochemistry. The results showed that the rates of methylation of the Runx3 gene in colonic cancer and colonic adenomas were significantly higher than that in the normal colonic tissue (23.5, 20.6 vs. 0.0%; P<0.05). There was no significant difference in the percentage of methylation of the Runx3 gene between colonic adenoma and colonic cancer (P>0.05). The positive percentage of Runx3 protein expression was significantly lower in colonic cancer compared with colonic adenoma and normal tissue (17.7 vs. 61.8, 76.5%; P<0.05). Methylation of the promoter CpG islands of the Runx3 gene is an important genetic event of colon carcinogenesis and may be associated with an altered protein level of Runx3.
Keywords: Runx3, methylation, colonic cancer
Introduction
DNA methylation is one of the most common types of epigenetic change, whose methylation is provided by S-adenosyl methionine. With the catalytic potential of DNA methyltransferase, the cytosine of DNA at the C-5 position is converted to 5-methylcytosine, occurring at the transcriptional level, and this does not change the primary structure of DNA base modifications (1,2). The methylation position is often a 5′-CpG-3′ dinucleotide. Extensive gene methylation exists in different tumors, and studies have found that hypermethylation occurs in human primary tumor-suppressor gene (3–5).
The tumor-suppressor gene runt-related transcription factor 3 (Runx3) is an classic gene in the Runx family, with all the features attributed to this family. Runx3 regulates the growth and differentiation of epithelial cells and it is located downstream of transforming growth factor-β (TGF-β). Under the guidance of the Runx proteins, Smad complexes can be transfered from the cytoplasm into the nuclei in a specific location. In cooperation with the Runx3 protein, they can induce transcription and activate target genes, so as to effect cell differentiation, cell cycle regulation, apoptosis and malignant transformation.
Runx3 is an apoptotic factor correlated with tumorigenesis and cancer progression, and research has indicated that Runx3 is involved in numerous tumorigeneses, such as gastric (6), breast (7), ovarian (8) and head and neck cancer (9). Several studies have reported Runx3 expression in colon cancer (10,11). To complement the relevant clinical data, the present study applied glycosylation methylation-specific polymerase chain reaction methods to check the CpG island methylation percentage for colon cancer, colorectal adenomas and normal colonic mucosa Runx3 gene, in order to explore the function of the CpG island methylation of the Runx3 gene in colon cancer.
Materials and methods
Tissue sample collection
The study was approved and registered by the Ethics Committee of Sichuan Provincial People's Hospital (Sichuan, China) in July 2008. The Ethics Committee approved the screening, treatment and data collection of these patients, and all the subjects provided written informed consent. All the studies were undertaken following the provisions of the Declaration of Helsinki.
In total, 68 surgical resection of colon cancer and adenomatous tissue samples were collected from Sichuan Provincial People's Hospital between October 2008 and February 2009.
The 68 patients were all first diagnosis and underwent treatment with preoperative radiotherapy, chemotherapy or immunotherapies, in which 34 cases were of colorectal adenoma and 34 cases were of colon cancer. A total of 34 healthy cases were normal tissue biopsy specimens under colonoscopy, and used as the negative control. The collection of new fresh tissue specimens was stored in liquid nitrogen within 20 min at −183°C. A total of 8 males and 16 females provided the colon cancer samples, and were aged from 38–78 years (mean age, 58±2.5 years). A total of 7 males and 17 females, aged from 28–73 years (mean age, 53±2.7 years), provided the colonic adenoma samples. The normal colon mucosa samples were provided from 18 males and 16 females, aged from 38–78 years (mean age, 58±2.2 years).
The TIANamp Genomic DNA kit was purchased from Tiangen Biotechnology Co., Ltd., (Beijing, China), and the EZ DNA Methylation-Gold™ kit was from Rimo Science and Technology Development Co., Ltd., (Beijing, China). DNA markers λDNA/HindⅢ, 50-bp DNA Labber and 2X Taq polymerase chain reaction (PCR) MasterMix were from Tiangen Technology Co., Ltd. The rabbit anti-human monoclonal antibody Runx3 was from Abcam (ab40278; Cambridge, MA, USA); and the DAB kit was from Zhongshan Golden Bridge Biological Technology Co., Ltd. (Beijing, China).
Primer synthesis and preparation
The DNA sequences were as reported in GenBank, and the PCR primers were designed according to the study by Issa (12). The primers were as follows: Runx3 methylation front guide sequences [methylated-specific forward (MF)], 5′-TTACGAGGGGCGGTCGTACGCGGG-3′ and methylated reverse primer sequence [methylated-specific reverse (MR)], 5′-AAAACGACCGACGCGAACGCCTCC-3′; Runx3 unmethylated front guide sequences (unmethylated-specific forward primers), 5′-TTATGAGGGGTGGTTGTATGTGGG-3′ and reverse primer sequence (unmethylated-specific reverse primers), 5′-AAAACAACCAACACAAACACCTCC-3′.
DNA extraction
The present study referred to Herman et al (13) for the MSP methods. The TIANamp Genomic DNA kit was used to extract tissue sample DNA. The EZ DNA Methylation-Gold kit was used for the methylation-extracted tissue. The DNA Runx3M system included 12.5 µl 2X Taq PCR MasterMix, 1 µl Runx3 MF and 1 µl Runx3 MR methylation. Following the treatment, 2.5 µL DNA and 8 µl ddH2O underwent the following Runx3 gene PCR conditions: denaturation at 94°C for 10 min, annealing at 65°C for 45 sec, and extension at 72°C for 10 min. The PCR products were applied to 1% agarose gel electrophoresis and gel images were captured using the Gel Doc 1000 imager (Bio-Rad, Hercules, CA, USA).
Immunohistochemical detection of the expression of Runx3 proteins
The Runx3 antigen retrieval underwent a high-pressure hot fix. Phosphate-buffered saline solution was used instead of a primary antibody for the negative control. A known positive plate was used as a positive control.
Criteria
Noticeable methylated-specific PCR products at 250 bp indicated positive methylation. Negative was the presence of a 240-bp unmethylated band, but no band for the methylated-specific PCR product (13). Following IHC staining, when >10% of the whole section was brown/yellow, this was deemed as positive.
Statistical analysis
Statistical analysis was used to compare the percentage of Fisher's exact probability. Statistical analysis was performed using statistical software SPSS 13.0 (SPSS, Inc., Chicago, IL, USA). Data processing used a test level of α=0.05, and P<0.05 was considered to indicate a statistically significant difference.
Results
Runx3 gene promoter CpG island methylation
The percentage of Runx3 promoter CpG island methylation positive expression in the colon carcinomas and adenomas were 23.5 (8/34) and 20.6% (7/34), and there was no detection in the normal tissue. The methylation of the Runx3 gene in the colon cancer and colon adenoma groups was significantly different from normal colon mucosa (P<0.05). However, there was no significant difference between colon cancer and colon adenoma (P>0.05, Table I).
Table I.
Percentage of Runx3 CpG island methylation.
| Runx3 CpG island methylation, % | ||
|---|---|---|
| Groups | Positive, n | Negative |
| Colon cancer | 23.5 | 76.5 |
| Adenoma | 20.6 | 79.4 |
| Normal tissue | 0.0 | 100.0 |
Runx3, runt-related transcription factor 3; negative, detection of unmethylated product, but not methylation-specific PCR product.
Runx3 protein
Runx3 protein expression percentages were 17.6 (6/34), 61.8 (21/34) and 76.5% (26/34) in the colorectal cancer, adenoma and normal groups, respectively. There was a significant difference between the colon cancer and the colon adenoma and normal groups (P<0.05, Table II, Fig. 1).
Table II.
Percentage of Runx3 protein expression.
| Runx3 protein expression, % | ||
|---|---|---|
| Groups | Positive | Negative |
| Colon cancer | 17.6 | 82.4 |
| Adenoma | 61.8 | 38.2 |
| Normal tissue | 76.5 | 23.5 |
Runx3, runt-related transcription factor 3; negative, <10% of nucleus was brown/yellow.
Figure 1.
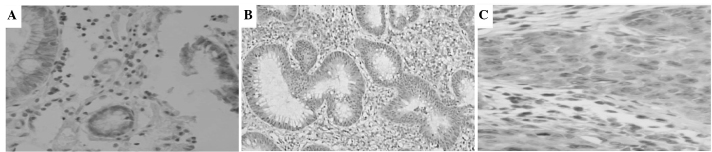
Expression level of runt-related transcription factor 3 (Runx3) in colon tissue in (A) normal mucosa adenoma tissue, (B) adenoma tissue and (C) colon cancer tissue. Immunohistochemical assay was used to detect the expression of Runx3 (magnification, x100).
Discussion
DNA methylation is closely associated with tumorigenesis, which is one of the inactivation mechanisms of the tumor-suppressor genes (14). The Runx3 gene is a newly discovered suppressor oncogene, which is the most primitive Runx alkylene type in the mammalian gene family, and it has a regulatory effect on epithelial cell growth and differentiation. It also plays an important role in the spinal ganglia development and differentiation of T cells. The Runx3 protein is involved in the TGF-β signal transduction pathway of a transcription factor, which is directly combined with Smad receptor binding. The protein guides the TGF-β1/Smad signaling pathway to activate Smad (one type of apoptotic factor). The complex moves from the cytoplasm into the nucleus of the target site, and the TGF-β1 Smad complexes and target sites combine to promote TGF-β1 signaling to mediate apoptosis in normal cells. Synergistic action of TGF-β1 on epithelial cell growth has a negative regulation, which occurs in the TGF-β1 signaling pathway and plays a key role (15). A previous study found that for the Runx3 gene colon cancer hypermethylation, the Runx3 CpG island methylation guides Runx3-induced gene inactivation, which is closely associated with colon cancer (16). Runx3 protein deficiencies can lead to the TGF-β1 signaling pathway blocking TGF-β1-induced cell growth inhibition, which reduces the sensitivity to apoptosis. β-catenin accumulates in the cytoplasm resulting in signal pathway activation, cell proliferation and apoptosis imbalance. The genetically unstable cancerous cell clonal expansion promotes the occurrence of tumors (17). Runx3 gene-knockout mice, which have decreased Runx3 protein expression, significantly increased the incidence of tumors in mice (18). Ku et al (19) applied the MSP technique for detecting methylation in human colon cancer cell lines and found that 50% of human colon cancer cell line expression of Runx3 was decreased or there was no expression. The study also found that compared with the positive expression percentage of corresponding normal tissue, the percentage of Runx3 protein expression was significantly decreased in human colon cancer, liver cell cancer, bile duct cancer, pancreatic cancer, lung cancer, esophageal cancer, endodermal sinus tumors, breast (20–27) and other tumors.
In the present study, 8 cases of Runx3 CpG island methylation were found in 34 cases of colon cancer. Methylation was therefore 23.5% (8/34), and Ku et al (19) reported that there is an 18.4% methylation rate on the Runx3 CpG island in colon cancer, while there is no Runx3 methylation in normal colon tissue. Goel et al (28) also identified 20.8% (19/91) of the presence of Runx3 CpG island methylation in colon cancer cases, and no methylation was found in the normal group; these two studies show close and similar results with our present study. The present study identified that the Runx3 protein expression percentage was 76.5% (26/34) in normal colonic mucosa, 61.8% (21/34) in the adenoma group, and 17.6% (6/34) in the colon cancer group. The colon adenoma group was significantly lower than the colon cancer group, and there was a significant difference (P<0.05). Chen et al (27) reported similar results, which suggests that Runx3 CpG island methylation may occur in normal colonic mucosa transforming into colon gland adenoma. This may be an early stage of development in the cancer sequence, and the colon cancer early onset molecular events and low expression of Runx3 protein may be associated with colon Runx3 CpG island methylation.
References
- 1.Esteller M. DNA methylation and cancer therapy: New developments and expectations. Curr Opin Oncol. 2005;17:55–60. doi: 10.1097/01.cco.0000147383.04709.10. [DOI] [PubMed] [Google Scholar]
- 2.Lewandowska J, Bartoszek A. DNA methylation in cancer development, diagnosis and therapy - multiple opportunities for genotoxic agents to act as methylome disruptors or remediators. Mutagenesis. 2011;26:475–487. doi: 10.1093/mutage/ger019. [DOI] [PubMed] [Google Scholar]
- 3.Xia Y, Zhang M, Zhang X, Liu X. A systematic review and meta-analysis of runt-related transcription factor 3 gene promoter hypermethylation and risk of gastric cancer. J Cancer Res Ther. 2014;10:310–313. doi: 10.4103/0973-1482.151539. (Suppl) [DOI] [PubMed] [Google Scholar]
- 4.Pellacani D, Kestoras D, Droop AP, Frame FM, Berry PA, Lawrence MG, Stower MJ, Simms MS, Mann VM, Collins AT, et al. DNA hypermethylation in prostate cancer is a consequence of aberrant epithelial differentiation and hyperproliferation. Cell Death Differ. 2014;21:761–773. doi: 10.1038/cdd.2013.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wojdacz TK, Windeløv JA, Thestrup BB, Damsgaard TE, Overgaard J, Hansen L. Identification and characterization of locus-specific methylation patterns within novel loci undergoing hypermethylation during breast cancer pathogenesis. Breast Cancer Res. 2014;16:R17. doi: 10.1186/bcr3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedrich MJ, Rad R, Langer R, et al. Lack of RUNX3 regulation in human gastric cancer. J Pathol. 2006;210:141–146. doi: 10.1002/path.2042. [DOI] [PubMed] [Google Scholar]
- 7.Chen LF. Tumor suppressor function of RUNX3 in breast cancer. J Cell Biochem. 2012;113:1470–1477. doi: 10.1002/jcb.24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CW, Chuang LS, Kimura S, et al. RUNX3 functions as an oncogene in ovarian cancer. Gynecol Oncol. 2011;122:410–417. doi: 10.1016/j.ygyno.2011.04.044. [DOI] [PubMed] [Google Scholar]
- 9.Tsunematsu T, Kudo Y, Iizuka S, et al. RUNX3 has an oncogenic role in head and neck cancer. PloS One. 2009;4:e5892. doi: 10.1371/journal.pone.0005892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Tan SY, Zhang J, You HX. Effects of paeonol on intracellular calcium concentration and expression of RUNX3 in LoVo human colon cancer cells. Mol Med Rep. 2013;7:1425–1430. doi: 10.3892/mmr.2013.1372. [DOI] [PubMed] [Google Scholar]
- 11.Slattery ML, Lundgreen A, Herrick JS, Caan BJ, Potter JD, Wolff RK. Associations between genetic variation in RUNX1, RUNX2, RUNX3, MAPK1 and eIF4E and risk of colon and rectal cancer: additional support for a TGF-β-signaling pathway. Carcinogenesis. 2011;32:318–326. doi: 10.1093/carcin/bgq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 13.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 15.Vogelmann R, Nguyen-Tat MD, Giehl K, Adler G, Wedlich D, Menke A. TGFbeta-induced downregulation of E-cadherin-based cell-cell adhesion depends on PI3-kinase and PTEN. J Cell Sci. 2005;118:4901–4912. doi: 10.1242/jcs.02594. [DOI] [PubMed] [Google Scholar]
- 16.Subramaniam MM, Chan JY, Soong R, Ito K, Yeoh KG, Wong R, Guenther T, Will O, Chen CL, Kumarasinghe MP, et al. RUNX3 inactivation in colorectal polyps arising through different pathways of colonic carcinogenesis. Am J Gastroenterol. 2009;104:426–436. doi: 10.1038/ajg.2008.141. [DOI] [PubMed] [Google Scholar]
- 17.Miyazono K, Maeda S, Imamura T. Coordinate regulation of cell growth and differentiation by TGF-beta superfamily and Runx proteins. Oncogene. 2004;23:4232–4237. doi: 10.1038/sj.onc.1207131. [DOI] [PubMed] [Google Scholar]
- 18.Soong R, Shah N, Peh BK, Chong PY, Ng SS, Zeps N, Joseph D, Salto-Tellez M, Iacopetta B, Ito Y. The expression of RUNX3 in colorectal cancer is associated with disease stage and patient outcome. Br J Cancer. 2009;100:676–679. doi: 10.1038/sj.bjc.6604899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ku JL, Kang SB, Shin YK, Kang HC, Hong SH, Kim IJ, Shin JH, Han IO, Park JG. Promoter hypermethylation downregulates RUNX3 gene expression in colorectal cancer cell lines. Oncogene. 2004;23:6736–6742. doi: 10.1038/sj.onc.1207731. [DOI] [PubMed] [Google Scholar]
- 20.Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/S0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 21.Park WS, Cho YG, Kim CJ, Song JH, Lee YS, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY. Hypermethylation of the RUNX3 gene in hepatocellular carcinoma. Exp Mol Med. 2005;37:276–281. doi: 10.1038/emm.2005.37. [DOI] [PubMed] [Google Scholar]
- 22.Kim WJ, Kim EJ, Jeong P, Quan C, Kim J, Li QL, Yang JO, Ito Y, Bae SC. RUNX3 inactivation by point mutations and aberrant DNA methylation in bladder tumors. Cancer Res. 2005;65:9347–9354. doi: 10.1158/0008-5472.CAN-05-1647. [DOI] [PubMed] [Google Scholar]
- 23.Sato K, Tomizawa Y, Iijima H, Saito R, Ishizuka T, Nakajima T, Mori M. Epigenetic inactivation of the RUNX3 gene in lung cancer. Oncol Rep. 2006;15:129–135. [PubMed] [Google Scholar]
- 24.Hiramatsu T, Osaki M, Ito Y, Tanji Y, Tokuyasu N, Ito H. Expression of RUNX3 protein in human esophageal mucosa and squamous cell carcinoma. Pathobiology. 2005;72:316–324. doi: 10.1159/000091329. [DOI] [PubMed] [Google Scholar]
- 25.Vogiatzi P, De Falco G, Claudio PP, Giordano A. How does the human RUNX3 gene induce apoptosis in gastric cancer? Latest data, reflections and reactions. Cancer Biol Ther. 2006;5:371–374. doi: 10.4161/cbt.5.4.2748. [DOI] [PubMed] [Google Scholar]
- 26.Kato N, Tamura G, Fukase M, Shibuya H, Motoyama T. Hypermethylation of the RUNX3 gene promoter in testicular yolk sac tumor of infants. Am J Pathol. 2003;163:387–391. doi: 10.1016/S0002-9440(10)63668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W, Salto-Tellez M, Palanisamy N, Ganesan K, Hou Q, Tan LK, Sii LH, Ito K, Tan B, Wu J, et al. Targets of genome copy number reduction in primary breast cancers identified by integrative genomics. Genes Chromosomes Cancer. 2007;46:288–301. doi: 10.1002/gcc.20411. [DOI] [PubMed] [Google Scholar]
- 28.Goel A, Arnold CN, Tassone P, Chang DK, Niedzwiecki D, Dowell JM, Wasserman L, Compton C, Mayer RJ, Bertagnolli MM, et al. Epigenetic inactivation of RUNX3 in microsatellite unstable sporadic colon cancers. Int J Cancer. 2004;112:754–759. doi: 10.1002/ijc.20472. [DOI] [PubMed] [Google Scholar]


