Abstract
Papillomaviruses are a very successful group of viruses that replicate persistently in localized regions of the stratified epithelium of their specific host. Infection results in pathologies ranging from asymptomatic infection, benign warts, to malignant carcinomas. Despite this diversity, papillomavirus genomes are small (7-8 kbp) and contain at most eight genes. To sustain the complex papillomaviral life cycle, each viral protein has multiple functions and interacts with and manipulates a plethora of cellular proteins. In this study, we use tandem affinity purification and mass spectrometry to identify host factors that interact with eleven different papillomavirus E2 proteins from diverse phylogenetic groups. The E2 proteins function in viral transcription and replication and correspondingly interact with host proteins involved in transcription, chromatin remodeling and modification, replication and RNA processing.
Keywords: E2, BRD4, HPV, papillomavirus, virus
1. Introduction
The Papillomaviridae consists of several hundred small DNA viruses that replicate in specific anatomical regions of the stratified epithelium of their specific host. Infection is persistent and results in clinical outcomes ranging from asymptomatic infection to verrucae, plantar and filiform warts, and condylomata acuminata. A subset of HPVs is associated with carcinomas of the oropharyngeal and anogenital tracts. In fact, oncogenic HPV infection is the causative agent of almost all cervical carcinomas and about 25% of head and neck cancers [1].
Papillomaviruses have a remarkable infectious cycle that depends on the development of a stratified epithelium (reviewed in [2]). The virus initially infects the lower, dividing layers of the epithelium; viral DNA is transported to the nucleus where it must escape intrinsic host defenses and establish the genome as a stable, extra-chromosomal, autonomously replicating element. Next, in the maintenance phase, genomes replicate at low copy number in concert with host DNA and are partitioned to daughter cells upon cell division. During this phase there is only low level viral gene expression, which helps the infected cells escape detection by the host immune system. Finally, as infected cells differentiate and traffic to the surface of the epithelium, high level viral DNA amplification and capsid protein synthesis is triggered to form progeny virus.
The E2 proteins play a pivotal role in the papillomavirus lifecycle (reviewed in [3]). E2 is a sequence-specific DNA binding protein that binds to consensus motifs (ACC(N)6GGT) that are contained in transcriptional regulatory regions and in the replication origin of the viral genome. E2 functions as an activator and repressor of viral transcription by binding to these sites and recruiting either positive or negative host transcription factors. E2 also functions in viral DNA replication by displacing nucleosomes, helping load the viral helicase onto the replication origin, and recruiting cellular replication proteins. The E2 proteins have additional roles in long-term genome maintenance whereby they tether viral genomes to host chromosomes; this ensures that viral DNA is efficiently partitioned to daughter cells. However, the region of host chromatin targeted by the E2 protein might also influence the infection. For example, we find that some E2 proteins bind to transcriptionally active regions of the nucleus, which can facilitate viral processes by providing a favorable environment for viral transcription [4]. We also find that the E2 protein links viral replication foci to regions of cellular chromatin undergoing replication stress; PV replication requires the host cell DNA damage and repair response and this localization likely benefits viral replication [5]. The E2 proteins have also been implicated in RNA processing [6].
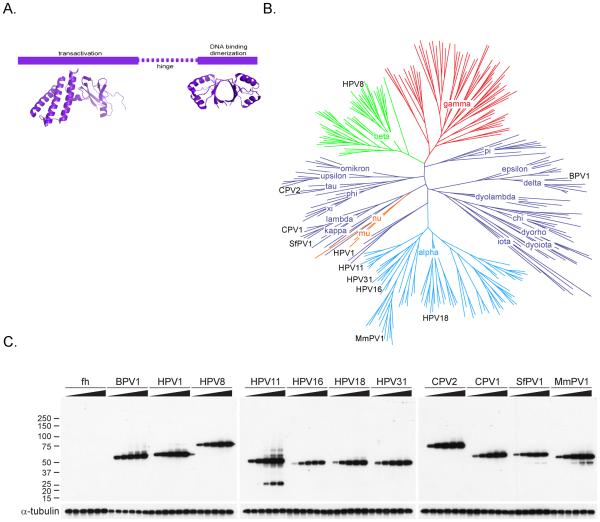
All E2 proteins have a similar structural organization with a conserved N-terminal domain of approximately 200-210 amino acids and a conserved C-terminal DNA binding and dimerization domain of about 90-100 amino acids (reviewed in [3] and shown in Figure 1A). The NMR or X-ray crystal structures of these domains have been solved for many papillomavirus types and can be found on the PaVE website http://pave.niaid.nih.gov/[7]. The polypeptide sequence between these domains is much less conserved and varies in length considerably between different E2 proteins (from about 50 to greater than 200 residues). This region has been designated as the hinge and forms an unstructured linker between the conserved domains [8]. However, despite the lack of strong conservation between E2 proteins from different genera, several genus specific functions have been mapped to the hinge regions (reviewed in [3]) and there is also some evidence that genus-specific protein interactions are mediated through this region. For example, the hinge regions of the Beta-HPV E2 proteins mediate the localization of these proteins to splicing speckles in the nucleus of interphase cells [6, 9] and to condensed chromosomes in mitotic cells [9-11]. Therefore, it is likely that many of the E2 protein specific interactions observed in this study are mediated by hinge sequences.
Figure 1.
A. Domains of the papillomavirus E2 proteins. The structure of the transactivation domain is HPV16 E2 from the pdb file, 1DTO [62]. The DNA binding domain structure is from HPV16, pdb 1BY9 [63].
B. Phylogenetic tree of papillomaviruses based on L1 sequences. The animal virus genera are shown in dark blue and the primate virus genera in red, green, cyan, and orange. The E2 proteins analyzed in this study were derived from the individual PVs indicated on the tree in black.
C. Titration of E2 protein levels. C-33A cells containing inducible expression vectors for control (pMEP4) and various E2s (BPV1, HPV1, HPV8, HPV11, HPV16, HPV18, HPV31, CPV2, CPV1, SfPV1, and MmPV1) were induced to determine optimal expression levels. The cells were stimulated by 0, 0.5, 1, 2, 4, and 8 µM CdSO4 for 4 h to induce expression of the E2 proteins and lysed in 1X LDS sample buffer (Invitrogen). The total lysates were analyzed by immunoblot analysis using an anti-HA antibody to evaluate the level of each E2 protein. As a loading control, the level of Alpha-tubulin protein was analyzed by immunoblot analysis in parallel experiments.
In this study we use Tandem Affinity Purification (TAP) followed by mass spectrometry to identify the major interacting host proteins of eleven different E2 proteins from seven different genera of the Papillomaviridae. The TAP method selects for high affinity interacting proteins and likely misses transient, low affinity interactions. Nevertheless, we have identified over two hundred different E2 interacting proteins. Here, we discuss the pros and cons of this method and use examples of some of these interacting proteins to explore approaches to further characterize and validate these interactions.
2. Materials and Methods
2.1 Plasmids
The pMEP4 expression vectors for FLAG-tagged E2 have previously been described [12]. BPV1 E2 proteins containing alanine substitutions in residues R37 and I73 were described previously [13]. The FLAG-HA tag (fh) was introduced into HindIII and blunted NotI sites of pMEP4 to generate the control plasmid, pMEP4/fh. E2 genes were sub-cloned into the blunted XhoI and SfiI sites of pMEP4/fh for HPV1 and CPV1 and blunted XhoI and BamHI of pMEP4/fh for other E2 genes. A FLAG-tagged version of HPV8 E2 containing hinge residues 216-312 fused to the C-terminal DNA binding domain (residues 404 to 498) has been described previously [9].
2.2 Antibodies
Control mouse IgG and rabbit IgG were purchased from Jackson ImmunoResearch. Anti-FLAG M2 and anti-BAZ1B antibodies were from Sigma. Antibodies for SRPK1 and SRPK2 were from BD Biosciences. Antibodies for EFTUD2 and EIF4A3 were purchased from ProteinTech Group. The polyclonal anti-BRD4 antiserum (directed against a C-terminal peptide) was described previously [13]. Horseradish peroxidase-conjugated second antibodies were from Thermo Scientific.
2.3 Establishment of E2 expressing cell lines
Stable cell lines were generated by transfecting C-33A cells with the pMEP4-E2 expression plasmids using Fugene 6 (Roche). Cells containing the extrachromosomal pMEP plasmids were selected with 80 µg/ml hygromycin B. Drug-resistant colonies were pooled after 2 weeks and cultured in complete media containing 40 µg/ml of hygromycin B. E2 protein expression was induced by cadmium sulfate induction at the concentrations indicated for 4 hours before harvest.
To express equal amounts of E2 proteins, their levels were titrated and adjusted by differential cadmium sulfate concentrations. The cells (2×105) were seeded in 24 well plates and cultured for 2 days. The pMEP metallothionein promoter was induced with different concentrations (0.0.5, 1, 2, 4, 8 μM) of cadmium sulfate for 4 h. After washing twice with cold PBS, the cells were lysed in 100 μl of 1X NuPAGE sample buffer containing 50 mM DTT, sheared by passing 5 times through a 19 gauge needle, and heated at 95°C for 5 min. Equal volumes of lysates were separated on 4-12% NuPAGE gels and immunoblotted using standard procedures using an anti-HA antibody.
2.4 Purification of E2 Complexes
To purify the protein complexes associated with the various E2 proteins, 1 × 109 C-33A cells expressing fh (tag only) and fh-tagged E2s (BPV1, HPV1, HPV8, HPV11, HPV16, HPV18, HPV31, CPV2, CPV1, SfPV1, and MmPV1) were induced with the appropriate concentration of cadmium sulfate, previously determined by calibration of cadmium sulfate concentration. The induced cells were pelleted, resuspended in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 1 mM PMSF, and Complete protease inhibitor cocktail), incubated on ice for 10 min, and disrupted by 20 strokes of a Dounce homogenizer. After centrifugation, nuclear pellets were vigorously mixed with the equal volumes of 2X buffer C (20 mM HEPES, pH 7.9, 0.42 M KCl, 1.5 mM MgCl2, 25% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 1 mM PMSF, and Complete protease inhibitor cocktail) at 4°C for 30 min, followed by centrifugation at 25,000 g for 20 min. The supernatants were dialyzed against buffer D (20 mM HEPES, pH 7.9, 20% glycerol, 150 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, and 1 mM PMSF) for 3 hours. The lysate was clarified by centrifugation at 25,000 g for 20 min and the resulting nuclear extracts (~50 mg) were subjected to immunoprecipitation with 0.25 ml (packed volume) of anti-FLAG M2 agarose beads (Sigma). After washing, bound proteins were eluted twice with 0.25 ml of FLAG peptide (0.1 mg/ml in buffer D; Sigma). The eluates were further immunoprecipitated with 50 μl of anti-HA agarose (Sigma) for 2 hours. After extensive washing with buffer D containing 0.1% NP-40, HA bound proteins were eluted with 100 μl HA peptide (0.1 mg/ml in TBS; Sigma) or 100 μl of 2% sarcosyl. The eluates were separated on 4-12% NuPAGE gel and stained with colloidal blue or silver stain.
2.5 Co-immunoprecipitation and Immunoblot analysis
To verify E2 protein interactors, nuclear extracts (~400 μg) were prepared from specific E2-expressing cells as described above and incubated with anti-FLAG M2 agarose for 16 hours or overnight at 4°C. After extensive washing with buffer D containing 0.1% NP-40, bound complexes were eluted with FLAG peptides or 2% sarcosyl, as indicated. Eluates were separated on 4-12% NuPAGE gels and immunoblotted using standard procedures.
2.6 Mass Spectrometric Analysis of E2 Protein Complexes
Mass spectrometry was performed in the Proteomics and Mass Spectrometry Facility of National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. E2 protein complexes were concentrated with 6 volumes of cold acetone, resuspended in 1X NuPAGE sample buffer and separated on 4-12% NuPAGE gel and stained with Colloidal Blue staining kit (Invitrogen). After destaining, the gel was separated into individual bands or zones. Gel slices were transferred to a 96-well plate and subjected to in-gel digestion with an automated fluid transfer robot (Massprep, Waters). Extracted samples were concentrated by vacuum to a volume of 2-5 μl. 20 μl 0.1% formic acid/1% acetonitrile was added and the samples were reconcentrated to 15-20 μl as judged visually with a control vial. Samples were flash frozen and stored at –80 C being thawed just prior to analysis .Samples were submitted to the queue of an LC/MS system comprising of a micro-HPLC system (NanoAcquity, Waters) and a hybrid mass spectrometer (LTQ-FTultra, Finnigan). The LC system was configured in the vented configuration with a 20mm × 180μm C18 trap column (Waters 1860003514) and a 250mm × 75μm analytical column (Waters 186003545). After the analytical column the flow entered a conductive union mounted on a Proxeon Nanospray source (pre-Thermo ES240) which held a fused silica emitter (New Objective, FS360-50-10-CE-20) cut to about 2.0cm and held about 4mm from the inlet capillary of the mass spectrometer. Initially, with the vent open, a 10μL injection was made and the the 20μL loop and trap column received a flow of 10μL/minute for 6 minutes. Then the vent was closed, the loop taken off-line and the flow rate dropped to 0.3μL/minute. After 2 minutes of flow at 1% acetonitrile a linear gradient was run from 1% to 40% acetonitrile over 30 minutes, from 40% to 80% acetonitrile in another 10 minutes, held at 80% for five minutes and then returned to 1% acetonitrile for 10 minutes. A separate sample was run for each band or zone with corresponding control bands from control lanes in the gel collected before those from the experiment. Hybrid data sets were collected with a single parent peptide mass spectrum (contour) from 400 to 1800 M/Z collected using the FT-ICR with the 100K resolution setting followed by up to 12 MS/MS (centroid) fragmentation spectra. The fill target for the FTICR was 1E6 and that for the Ion Trap was 1E4 with maximum injection times of 200ms and 100ms respectively. Charge state recognition was used to select ions with no effective intensity threshold. The success of the charge state recognition seeming to be a useful bar for fragmentation targeting. Peptides with known charge states of 2 and above were selected for fragmentation with dynamic exclusion used with two repeats and a time constant of 30 seconds. Previewed spectra were used which effectively ruled out fragmentation of very high charge state peptides.
A number of analysis campaigns were carried out over more than a year. To standardize comparisons all experiments were analyzed using the same final parameter set. Collected data was abstracted to .mgf format (using the extractMsn.exe available with Xcalibur 2.0) and submitted to the NIH Mascot cluster (running Mascot 2.2.0). Parent mass tolerances of 0.05 Daltons and peptide mass tolerances of 0.25 Daltons were used. The MSDB data base (version 20060831, 3239079 entries) was searched without species specificity (as a conservative measure) with variable modification of oxidation on methionine and fixed modification of carbamidomethyl on cysteines specified with up to 3 missed sites for trypsin. Mascot flat files from the searches were then used as input to the Scaffold program (2.06.02) which created an output which was then used by the primary experimenter to cull identifications with the following criteria; homo sapiens proteins represented by ≥ 2 peptides.
Due to the low complexity of each sample expectation values have little value and the 95% probability of the accuracy of the mascot result is only a rough approximation. The supplementary table lists the peptides used for the identification of each protein together with each peptide’s Mascot score. A subset of proteins were used in antibody based assays of protein complexes discussed in the manuscript and these assays imply a second level of validation of these identifications.
2.7 Purification of HPV8 E2 complexes in presence of RNase
The C-33A cells expressing fh (tag only) or fh-tagged HPV8 E2 (1×108) were treated as described above and resuspended in buffer A in presence of RNase A (40 µg/ml). After disrupting and removing cytosolic fractions, isolated nuclear pallets were resuspended in 2X buffer C for 30 min in presence of RNase A. The extracted nuclear fractions were dialyzed against buffer D for 3 h and subjected to immunoprecipitation using anti-FLAG M2 agarose (50 µl) for overnight. The precipitants were extensively washed with buffer D containing 0.1% NP-40 and eluted with 100 µl FLAG peptide (0.1mg/ml in TBS), followed by an elution with 100 µl 2% sarcosyl. One tenth of FLAG eluates were analyzed by immunoblotting for components of HPV8 E2 complexes.
2.8 Fractionation of E2 protein complexes using gel filtration column chromatography
Nuclear extracts were prepared from C-33A cells as described above and subjected to dialysis against buffer D for 3 h. Two milligrams of the nuclear extracts were fractionated into 0.3 ml volumes by Superose 6 10/300 GL column (GE Healthcare) chromatography using a running buffer (20 mM HEPES pH 7.9, 20% glycerol, 150 mM KCl, 0.2 mM EDTA). The fractions were analyzed by immunoblotting using anti-FLAG M2 antibody and antibodies for components of the E2 protein complexes. Gel filtration standards (Bio-Rad) were separated in parallel to independently assess the sizes of the complexes.
3. Results and Discussion
3.1 Selection of E2 Proteins
To identify cellular targets of papillomavirus E2 proteins, 11 different E2s were expressed in human C-33A (HPV-negative cervical carcinoma) cells. The E2 proteins were derived from papillomaviruses from Alpha (HPV11, HPV16, HPV18, HPV31 and MmPV1), Beta (HPV8), delta (BPV1), mu (HPV1), kappa (SfPV1), lambda (CPV1) and tau (CPV2) genera. These viruses infect a wide range of species (human, cow, dog, rabbit and rhesus monkey) and anatomical sites (oral and genital mucosa and several distinct cutaneous regions). Alpha HPV E2 proteins were obtained from both “low oncogenic risk” and “high oncogenic risk” types. Figure 1B shows a phylogenetic tree of 260 papillomavirus types, with the viruses used in this study indicated.
3.2 Expression System
Analysis of cellular targets of the E2 proteins during infection is difficult for a number of reasons. The papillomavirus life cycle takes place in a stratified tissue with different temporal stages taking place in different cell layers over a period of weeks. Furthermore, papillomavirus genomes are small (<8kb) and are packed with essential overlapping genes and regulatory elements that make it very difficult to epitope tag individual viral proteins. Therefore, we decided to stably express the E2 proteins in an HPV-negative cervical carcinoma cell line, C-33A, that we have previously shown can tolerate low levels of E2 expression [12].
The E2 genes were cloned in the pMEP4 plasmid, an EBV-derived episomal vector that can be used to efficiently establish stable cell lines by virtue of an encoded hygromycin-resistance gene. Each E2 protein contained tandem FLAG and HA epitope tags fused to the N-terminus. This allows purification of the E2 proteins and associated proteins by tandem affinity purification. This method was first described by Rigaut and colleagues [14] and has proven useful for isolating complexes containing chromatin modifiers [15]. The advantage of this technique is that sequential affinity purification greatly reduces the background of non-specific proteins; however it also selects for stable protein-protein interactions and can miss transient interactions. The E2 proteins were expressed from a metallothionein promoter and titration of cadmium sulfate concentration ensured that relatively low, but equal amounts of each E2 protein were expressed. When analyzed by immunofluorescence, the selected levels gave a distinctive nuclear, but nucleolar excluded, pattern of E2 staining. Our previous studies have shown these levels to be comparable to those expressed in wart tissue [16]. Notably, all five E2 proteins from the Alpha-genus had to be at least partially codon-modified to achieve expression levels comparable to those of E2 proteins from other genera [12]. Figure 1C shows the relative level of each E2 protein after induction with 0, 0.5, 1, 2, 4, and 8 µM CdSO4.
3.3 Extraction, Purification and Identification of Protein Complexes
The choice of extraction method is critical in the purification of protein complexes, especially those that are tightly bound to cellular components such as chromatin. Gentle extraction methods can preserve protein complexes but chromatin bound proteins are often difficult to extract from the nucleus. In this study we chose to use the classical Dignam nuclear extract procedure, which selectively extracts transcriptional components that are functional in vitro [17]. The E2 proteins are nuclear proteins involved in processes such as transcription, replication, splicing and chromatin binding [3]; thus, the Dignam nuclear extraction protocol is ideal for removing contaminating cytoplasmic components and preserving functional nuclear complexes. One disadvantage with this method is that prolonged dialysis after high salt extraction could promote the formation of complexes in vitro that were not present in the unextracted cells. However, careful validation of interacting proteins can overcome this limitation. Overall, the extraction of functional complexes greatly outweighs this potential drawback.
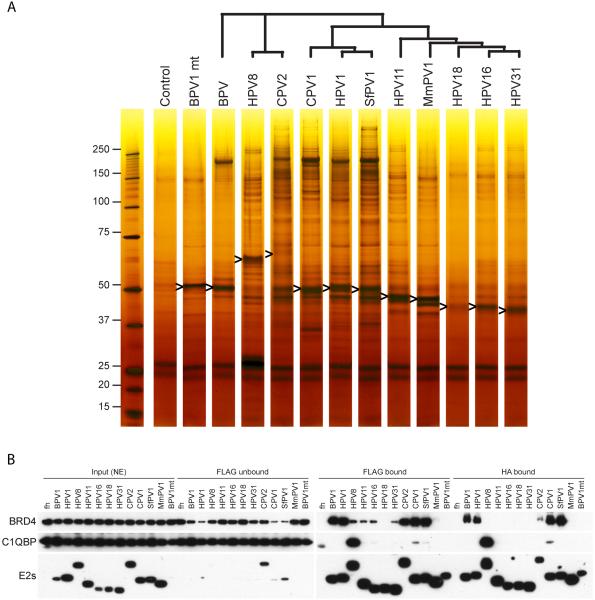
E2 containing complexes were initially purified from cell extracts by virtue of the N-terminal FLAG tag. These complexes were eluted from the affinity beads with a FLAG peptide and repurified using beads coated with antibodies to the second, HA tag. Protein complexes were then eluted with either HA peptide or sarcosyl. Figure 2A shows a silver stained polyacrylamide gel of the resulting protein complexes. The E2 proteins, ranging in size from 45 to 75kD, are indicated by arrows. The original gel and coomassie blue stained version of the gel are shown in Supplementary Figure 1. In general, the Alpha-PV E2 proteins did not have any highly prominent interacting partners. This correlates with previous studies showing that Alpha-PV E2 proteins were not tightly associated with the nucleus compared to non-Alpha-PV E2 proteins [18]. The non-Alpha-PV E2 proteins had varying numbers of strong interacting proteins, visible by coomassie staining. After destaining, individual bands or zones were excised from the coomassie blue stained gel and proteins extracted from the gel slices were identified by LC/MS, as described in Methods. Three of the most prominent bands are boxed in Supplementary Figure 1. The bands in boxes #1 contain the BRD4 chromatin binding protein; boxes # 2 contain nucleophosmin (NPM1); and the major band in box 3 represents C1QBP, the P32 subunit of the pre-mRNA splicing factor SRSF1. Supplementary Table 1 contains lists of additional proteins identified by this screen. Several additional TAP experiments followed by mass spectrometry were carried out on BPV1 and HPV8 E2 proteins, because these had been well characterized as chromatin binding proteins in our laboratory [10, 19]. Complexes associated with each of the 11 E2 proteins were isolated by smaller scale TAP for use in comparative immunoblot analysis.
Figure 2.
A. Silver stained gel of E2 protein complexes
C-33A cells containing the various E2 expression vectors were induced with the appropriate concentration of CdSO4 for 4 h to give equivalent levels of E2 expression. Protein complexes were extracted from the nucleus and purified by immunopurification with anti-FLAG M2 and anti-HA antibodies as described in Materials and Methods. The final bound proteins were eluted from anti-HA agarose with 2% sarcosyl. The purified E2 complexes were analyzed on a NuPAGE gel and stained with SilverXpress silver staining kit (Invitrogen). An band for each E2 protein is marked with an arrowhead. The tree above the lanes shows the phylogenetic relationship among the different E2 proteins.
B. BRD4 and C1QBP have varying affinities for different E2 proteins
The various E2 protein complexes were purified as described in Materials and Methods and Figure 2A. During the preparation of E2 protein complexes, portions of each fraction were collected from nuclear extracts (NE), unbound fractions from the first IP using anti-FLAG (FLAG unbound), eluates from the first IP (FLAG bound), and eluates from the second IP using anti-HA (HA bound). The fractions were analyzed by immunoblot analysis using anti-BRD4, anti-C1QBP, and anti-HA antibodies to detect the E2 protein.
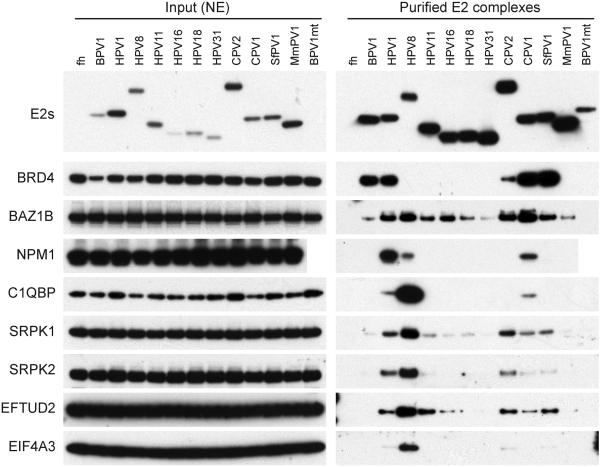
The E2 interacting proteins function mostly in transcription, replication, histone modification, chromatin recognition and remodeling (see Table 1), consistent with their role in the Papillomavirus lifecycle. In addition, the Beta HPV8 E2 protein complex contained many proteins from the splicesome, indicating a role in RNA processing. This interaction is accordant with the detection of the Beta HPV E2 proteins in nuclear speckles [9, 20]. Notable interactors will be discussed individually below in Section 3.4.
Table 1.
Categories of cellular proteins interacting with one or more PV E2 Protein
| Category | Examples of E2 Interacting Proteins |
|---|---|
| Nuclear Matrix | MATR3; HNRNPU; HNRNPA1 |
| Post translational Modification |
CSNK2A1, CSNK2A2, CSNK2B; SRPK1/2; PRMT5; PPP2CA/CB/R1A, BAZ1B |
| DNA Replication/Repair | RFC 2-5; ATAD5; WICH (BAZ1B, SMARCA5) |
| Transcription Factor | BNC2; YY1 |
| RNA Processing | Splicesome factors; C1QBP |
| Chromosome Structure | SMC5/6 |
| Histone Acetylases | TTRAP/TIP60 (INO80, p400, TRRAP) |
| Histone Deacetylases | NCOR1, HDAC, SAP18, |
| Histone Demethylases | KDM1B |
| Histone Methyltransferase | WDR5; WHSC (WHSC1/WHSCL1, SALL4); PRMT5; EHMT1 |
| Chromatin Binding | BRD4; Polycomb (E2F6, PCGF6, RNF2) |
| Chromatin Remodeling | SWI/SNF (SMARCA4); NuRD (CHD4, SMARCA5, HDAC1/2, MTA1/2); WICH (BAZ1B, SMARCA5); TRRAP/TIP60 (INO80, EP400, TRRAP) |
3.4 Notable E2 Interacting Proteins
3.4.1 Chromatin Binders, Modifiers and Remodelers
3.4.1.1 BRD4
The most significant E2 interacting protein for many E2 proteins was the double bromodomain protein BRD4 (see Figure 2A and Supplementary Figure 1). BRD4 is a cellular chromatin adaptor protein first identified as a significant binding partner for the HPV16 and BPV1 E2 proteins [21, 22]. Most notable is that the BRD4 protein was the major binding factor for Delta, Lambda, Kappa, Mu and Tau PV E2 proteins. Many of the other proteins found in the BPV1 E2 complexes are BRD4-associated factors. Examples of these are ATAD5 [23], the RFC complex [24], WHSC1 [25] and WHSC1L1 (NSD3) [23]. Furthermore, a specific mutation in BPV1 E2 that eliminates interaction with BRD4 (R37A/I73A) also disrupts association in our TAP experiments (see Figure 2A and Supplementary Figure 1). Notably, and as shown previously [26], missing from the E2-BRD4 complexes are components of the transcriptional elongation factor, pTEF-b, likely because E2 and pTEF-b compete for binding to the C-terminus of the BRD4 protein [26].
However, BRD4 was not present in high abundance in the Alpha and Beta E2 protein complexes. This correlates well with the observation that the Alpha-PV E2 proteins do not stabilize the interaction of BRD4 with host chromatin [13], while the HPV8 E2 protein binds to the rDNA loci on acrocentric chromosomes in a BRD4-independent manner [10, 12]. It also correlates well with previous studies that showed that the Alpha-PV E2 proteins bind BRD4 with low affinity compared to other E2 proteins [18].
However, we have shown previously that HPV8 E2 (a Beta-PV) binds to BRD4 with high affinity in vitro [18] and we suspect that the lack of associated BRD4 in the TAP studies presented here is because HPV8 E2 has so many competing binding factors.
The different E2 immune complexes were analyzed for the presence of the BRD4 protein at each stage of purification. As shown in Figure 2B, precipitation of HPV1, BPV1, CPV1 and SfPV1 E2 proteins depleted much of the BRD4 protein in the lysate, indicating that the majority of BRD4 in the cell was in complex with these proteins. While C1QBP remains tightly bound through both anti-FLAG and anti-HA immunoprecipitation, a significant portion of associated BRD4 is lost in the second anti-HA immunoprecipitation. Figure 2 shows the amounts of BRD4 and C1QBP associated with E2 at each step of the tandem affinity purification.
3.4.1.2 Chromatin Remodelers
The E2 proteins interact with several chromatin remodeling complexes such as SWI/SNF (SMARCA4); NuRD (CHD4, SMARCA5, HDAC1/2, MTA1/2); WICH (BAZ1B, SMARCA5); and TRRAP/TIP60 (INO80, EP400, TRRAP). Of particular interest for HPV8 E2 is the WICH complex (BAZ1B and SMARCA5), a chromatin remodeling complex that is involved in the transcription of rDNA and might also function to maintain ribosomal chromatin structure, a region bound by HPV8 E2. Also bound by HPV8 E2 are components of the nucleosome remodeling and deacetylase complex, NuRD. This complex is targeted by cytomegalovirus to promote accumulation of immediate early RNAs [27]. Tip60 has been shown to contribute to E2 regulation of the major viral early promoter in Alpha-PVs [28].
3.4.1.3 Histone Modifiers
The E2 proteins interact with several histone modifiers such as acetyl transferases, TRRAP/TIP60 (INO80, p400, TRRAP, TIP60), deacetyl transferases NCOR1/HDAC, demethylase KDM1B and methylases WDR5, WHSC (WHSC1, WHSC1L1, SALL4) and PRMT5. These factors could function in many of the roles that E2 plays in the viral life cycle.
3.4.2 Structural Maintenance of Chromosomes SMC5/6
SMC5 and 6 belong to the Structural Maintenance of Chromosome protein Family. These proteins have architectural similarity to condensin and cohesion (SMC 1-4). Compared to the other family members, the function of SMC5 and SMC6 are less well understood but they have been implicated in chromosome replication, segregation, and repair (reviewed in [29, 30].These are all functions related to the role of E2 in the viral life cycle. SMC5 and 6 have previously been identified in complexes of HPV11 E2 purified from 293 cells [31]. Here we confirm this observation and preliminary observations show that all E2 proteins tested are in complex with SMC5/6 (data not shown).
3.4.3 RNA Processing Proteins and components of the splicesome: EFTUD2, EIF4A3, SRPK1, SRPK2
Many of the E2-interacting proteins identified have highest affinity for the HPV8 E2 protein (Figures 2 and 3). Uniquely among the E2 proteins studied here, Beta HPV8 E2 (and the related HPV5 E2) colocalize with splicing speckles [6, 9] and this is reflected in many of the co-precipitating proteins. These include SRPK1/SRPK2, EFTUD2, EIF4A3 (Figure 3) and many other proteins involved in mRNA processing (Table 1). The HPV8 E2 hinge has multiple SR/RS dipeptide motifs that are usually found in the SR protein family involved in RNA splicing processes. Other E2 proteins also bind to these proteins but with less apparent affinity, and correspondingly they contain only one or more SR motifs. Notably, many of these proteins are also bound to the EBNA1 protein of Epstein-Barr virus (EBV) [32].
Figure 3.
Immunoblot analysis of interacting proteins purified from complexes from all E2 proteins.
E2 protein complexes were purified as described in Materials and Methods and Figure 3. The E2 complexes were eluted with 2% sarcosyl (HA bound) and analyzed by immunoblot analysis using various antibodies against components of the E2 protein complexes - E2 proteins (HA), BRD4, BAZ1B, NPM1, C1QBP, SRPK1, SRPK2, EFTUD2, and EIF4A3
3.4.4 C1QBP
C1QBP (TAP/P32) was originally described as a protein that interacted with the splicing factor ASF/SF2 [33, 34]. It binds to many viral proteins such as EBV EBNA1, adenovirus core protein V, herpesvirus IE63, and herpesvirus saimiri orf73 [35-38]. Somewhat disparately, C1QBP also interacts with the plasma complement component C1q [39] and is observed localized primarily to mitochondria [40]. C1QBP has both structural and functional similarities to histone chaperones NAP-1, nucleoplasmin and nucleophosmin [41]. Notably, NAP-1 and nucleophosmin are also E2 interacting proteins (this study and [42, 43]) and C1QBP has also been shown to interact with HPV5 E2 [6, 44]. Recently, C1QBP, NPM1, and NAP1 have been shown to promote the dissociation of protamine and association of histones in sperm chromatin [45]. C1QBP has been described as a frequent contaminant in TAP methods [46] but because of the association with many other viral proteins, and its specificity for HPV8 E2, it remains under consideration as a valid E2 interacting protein.
3.4.5 Nucleophosmin
Nucleophosmin (NPM1/ B23) is a nucleolar protein that is transported to the nucleoplasm in response to stress. It interacts with adenovirus basic core proteins, functioning as a histone chaperone and stimulating adenovirus replication [47]. Nucleophosmin also contributes to the transcriptional activation function of the EBV EBNA1 protein [48]. As shown below, HPV8 E2 exists in a complex with NPM1 and C1QBP.
3.4.6 Replication Factors
The E2 proteins are in complex with several proteins related to Replication Factor C (RFC2, 3, 4, 5 and ATAD5). The RFC complex functions as a clamp loader to load PCNA onto the replication fork; ATAD5 acts in concert with RFC to unload the clamp [49]. BRD4 also interacts with these proteins and likely mediates their association [23, 24].
HPV8 E2 interacts with BAZ1B and SMARCA5, which are components of the WICH complex. This chromatin remodeling complex regulates RNA polymerase I, II and III mediated transcription but also maintains chromatin structure at the replication fork and regulates the DNA damage response [50]. BAZ1B also localizes to heterochromatic replication foci [51].
3.4.7 Factors involved in post-translational modification
SRPK1/2 are serine protein kinases that specifically phosphorylate SR proteins [52]. We have previously shown that the hinge of HPV8 E2 is highly phosphorylated, and the majority of this phosphorylation is likely to be of the many SR/RS dipeptides located here [11]. SRPK1 can phosphorylate the hinge region of HPV5 and 8 E2 proteins in vitro [6, 53]. Prescott et al. have recently shown that SRPK1 can also phosphorylate HPV1 E2 in vitro, even though it only contains three SR/RS dipeptides [54] . Furthermore, the E1^E4 proteins of many HPVs bind SRPK1 and inhibit its ability to phosphorylate E2 [54]. In our study, the observed interaction of the different E2 proteins with SRPK1/2 correlates somewhat with the number of SR/RS dipeptides in the hinge region (HPV8 has 28; others have 0-5). In general, the Alpha-E2 proteins do not contain many SR/RS dipeptides, and correspondingly do not interact strongly with SRPK1/2 (Figures 3).
The BPV1 E2 hinge region is also phosphorylated by CK2 (casein kinase 2) and this modification promotes proteosomal degradation of E2 [16, 55, 56]. Notably, CSNK2A1 and/or CSNK2A2 is associated with BPV1 E2 and other PV E2 proteins in our TAP purification.
PRMT5, a protein arginine methyl transferase was a strong interacting partner of HPV8 and CPV2 E2. HPV8 and CPV2 contain may RG motifs, which are the preferred target of PRMT5. Significantly, the EBNA 1 protein of EBV (which has many functions analogous to E2) also contains multiple RG motifs that are methylated by PRMT5 [57].
3.5 The interaction HPV8 E2 with RNA binding proteins is not mediated indirectly by RNA
mRNA is associated with RNA binding proteins from the onset of transcription until, as processed messages they are translated in the cytoplasm. As described above, Beta-HPV8 E2 interacts with many proteins involved in RNA processing. Each of these proteins is an RNA binding protein and HPV8 E2 (as well as other Beta-PV E2 proteins) have many glycine-arginine rich sequences in the hinge region, reminiscent of the RGG RNA binding motifs [58]. Thus it is possible that the interaction of HPV8 E2 with cellular RNA binding proteins is mediated by mutual binding to RNA.
To investigate this further, complexes were extracted from the nucleus and purified in the presence of RNase (Figure 4). Co-precipitation of NPM1, C1QBP, EFTUD2, SRPK2, and EIF4A3 were monitored by immunoblot analysis. However, RNAse treatment increased, rather than decreased, the copurification of EFTUD2, SRPK2, and EIF4A3 with HPV8 E2. There was also a small increase in the level of background binding of these factors (precipitation in the absence of E2) suggesting that they might become more accessible after RNAse treatment. The interaction of E2 with nucleophosmin was decreased, but not eliminated, after RNAse treatment. This could be due to the observation that nucleophosmin becomes displaced from host rDNA chromatin after RNAse treatment [59]. We have previously shown that HPV8 E2 binds to rDNA [10] and the decrease in coprecipitation of E2 and nucleophosmin after RNAse treatment could reflect a specific disruption of the complex in this location. Finally, the interaction of C1QBP with E2 was completely unaffected by RNase treatment.
Figure 4.
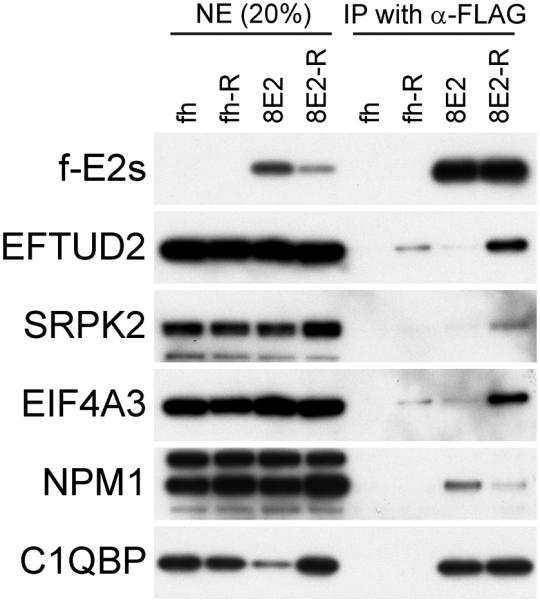
RNase treatment of E2 complexes
FLAG-tagged HPV8 E2 protein was expressed in C-33A cells and purified with anti-FLAG M2 agarose without adding RNase A (fh and 8E2) or in presence of RNase A (fh-R and 8E2-R). The purified E2 complexes (FLAG eluates) were analyzed by immunoblot analysis using various antibodies against components of the E2 protein complexes - E2 proteins (FLAG), EFTUD2, SRPK2, EIF4A3, NPM1, and C1QBP.
3.6 Analysis of E2 complexes by Gel filtration analysis
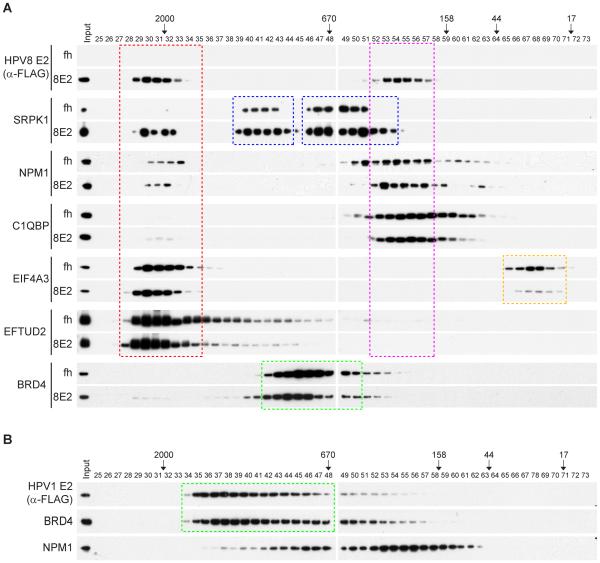
Many of the cellular proteins copurified with the E2 proteins are part of large multi-subunit complexes. Coprecipitation of E2 with multiple subunits from a particular complex increases the validity of the interaction but also indicates that each interacting protein might not necessarily interact directly with E2. One way to analyze the components of these complexes directly is by gel filtration. HPV1 and HPV8 E2 protein complexes were further characterized by gel filtration and western blot analysis and are shown in Figure 5.
Figure 5.
Fractionation of E2 protein complexes by Superose 6 size exclusion column chromatography.
(A) Nuclear extracts from C-33A cells expressing FLAG-HA-tag only (fh) and FLAG-HA-tagged HPV8 E2 (8E2) were fractionated by Superose 6 10/300 GL column chromatography. The fractions were analyzed by immunoblotting using various antibodies against components of the E2 protein complexes – HPV8 E2 protein (FLAG), SRPK1, NPM1, C1QBP , EIF4A3, EFTUD2 and BRD4. Gel filtration standards (Bio-Rad) were run in parallel to independently assess the size of the complexes and are indicated in kilodaltons. (B) Nuclear extract from C-33A cells expressing FLAG-HA-tagged HPV1 E2 was fractionated by Superose 6 10/300 GL column chromatography. The fractions were analyzed by immunoblotting using anti-FLAG M2 (E2), anti-BRD4, anti-NPM1 antibodies. Gel filtration standards (Bio-Rad) were run in parallel to independently assess the sizes of the complexes and are indicated in kilodaltons.
When separated by size exclusion chromatography, HPV8 E2 is found in two fractions corresponding to apparent molecular masses of about ~350KD and approximately 2MD. Immunoblot analysis of E2 interacting proteins SRPK1, NPM1, C1QBP, E1F4A3 and ETUD2 shows that the smaller complex contains NPM1 and C1QBP. As described above, NPM1 and C1QBP have common roles as histone chaperones, suggesting a possible function for this complex. The larger HPV8 E2 protein corresponds to fractions containing E1F4A3 and ETUD2, which are both components of the spliceosome. The spliceosome is a very large, dynamic complex of 200-300 proteins that rivals the size of ribosomes [60] and it is likely that this complex is at the limit of resolution of Superose 6 size exclusion. HPV8 E2 does not dramatically effect the size distribution of E1F4A3 and ETUD2, but its presence greatly enhances the incorporation of SRPK1 into the large putative spliceosome complex.
In contrast, HPV1 E2 is present primarily in a distinct 1400KD complex with BRD4. Small amounts of nucleophosmin are found recruited into the lower molecular weight fractions of the broad E2-BRD4 peak.
3.7 Validation of Protein Interaction by Colocalization
Another useful method to validate protein-protein interactions is to determine whether they colocalize within a cell. We have previously analyzed a number of the E2 interactors for their intracellular localization in the presence of the E2 proteins. A classic example is the chromatin binding protein BRD4. Expression of E2 proteins that bind BRD4 with high affinity (eg BPV1 and HPV1 E2) stabilizes binding of BRD4 to host chromatin in punctate speckles that contain the E2 protein [13]. The Alpha-PV E2 proteins do not stabilize BRD4 binding, but in the presence of the viral E1 replication protein, all three proteins colocalize in replication factories [61].
We have examined the colocalization of HPV8 E2 with NPM1 and with C1QBP, however it is very difficult to detect significant colocalization. NPM1 localizes primarily to the nucleoli and C1QBP to the mitochondria and there is no dramatic relocalization in the presence of E2 expression (data not shown). It is possible that at a different stage of the viral life cycle, or in the presence of additional viral proteins, colocalization might be more evident.
HPV8 E2 localizes to RNA splicing speckles as identified by co-immunofluorescence with the SC35 protein. This correlates well with the interaction with the plethora of spliceosome associated proteins shown here to bind HPV8 E2.
4. Concluding Remarks
In this study we have identified the major host proteins that are in complex with papillomavirus E2 proteins in nuclear extracts. Several of these proteins have been identified previously, and we have been able to augment the characterization of these interactions. Others are novel; but almost all are compatible with the known functions of E2 in viral gene expression and viral replication.
There are dramatic differences in the number of host proteins that strongly interact with E2 proteins from different genera; most notably all Alpha-PV E2 proteins have very few major, strong interacting proteins. Conversley, some other PV types have many strong interactors. This could be due to differences in amino acid sequence (particularly of the divergent hinge sequences of the different E2 proteins). In other cases, a second viral protein (such as E1) or process (such as replication) could strengthen the interaction between E2 and host factors.
Supplementary Material
Acknowledgments
We are grateful to Caleb McKinney and Katherine Hussman for comments on the manuscript. This research was supported by the Intramural Research Program of the NIAID, NIH (grant number ZIAAI000713).
Abbreviations
- BPV1-bovine
papillomavirus type 1
- CPV1–canine
papillomavirus type 1
- SfPV1-Sylvilagus
floridanus Papillomavirus 1
- MmPV1-Macaca
mulata Papillomavirus 1
- TAP-Tandem
Affinity Purification
Footnotes
The authors have declared no conflict of interest
References
- [1].Crow JM. HPV: The global burden. Nature. 2012;488:S2–S3. doi: 10.1038/488S2a. [DOI] [PubMed] [Google Scholar]
- [2].Doorbar J, Quint W, Banks L, Bravo IG, et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- [3].McBride AA. The Papillomavirus E2 proteins. Virology. 2013;445:57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jang MK, Kwon D, McBride AA. Papillomavirus E2 proteins and the host BRD4 protein associate with transcriptionally active cellular chromatin. J Virol. 2009;83:2592–2600. doi: 10.1128/JVI.02275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jang MK, Shen K, McBride AA. Papillomavirus Genomes Associate with BRD4 to Replicate at Fragile Sites in the Host Genome. PLoS pathogens. 2014;10:e1004117. doi: 10.1371/journal.ppat.1004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lai MC, Teh BH, Tarn WY. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J Biol Chem. 1999;274:11832–11841. doi: 10.1074/jbc.274.17.11832. [DOI] [PubMed] [Google Scholar]
- [7].Van Doorslaer K, Tan Q, Xirasagar S, Bandaru S, et al. The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res. 2013;41:D571–578. doi: 10.1093/nar/gks984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gauthier JM, Dillner J, Yaniv M. Structural analysis of the human papillomavirus type 16-E2 transactivator with antipeptide antibodies reveals a high mobility region linking the transactivation and the DNA-binding domains. Nucleic.Acids.Res. 1991;19:7073–7079. doi: 10.1093/nar/19.25.7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sekhar V, Reed SC, McBride AA. Interaction of the betapapillomavirus E2 tethering protein with mitotic chromosomes. J Virol. 2010;84:543–557. doi: 10.1128/JVI.01908-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Poddar A, Reed SC, McPhillips MG, Spindler JE, McBride AA. The human papillomavirus type 8 E2 tethering protein targets the ribosomal DNA loci of host mitotic chromosomes. J.Virol. 2009;83:640–650. doi: 10.1128/JVI.01936-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sekhar V, McBride AA. Phosphorylation regulates binding of the human papillomavirus type 8 E2 protein to host chromosomes. J Virol. 2012;86:10047–10058. doi: 10.1128/JVI.01140-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Oliveira JG, Colf LA, McBride AA. Variations in the association of papillomavirus E2 proteins with mitotic chromosomes. Proc Natl Acad Sci U S A. 2006;103:1047–1052. doi: 10.1073/pnas.0507624103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].McPhillips MG, Ozato K, McBride AA. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J Virol. 2005;79:8920–8932. doi: 10.1128/JVI.79.14.8920-8932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rigaut G, Shevchenko A, Rutz B, Wilm M, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- [15].Ogawa H, Ishiguro K.-i., Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F-and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- [16].Penrose KJ, McBride AA. Proteasome-mediated degradation of the papillomavirus E2-TA protein is regulated by phosphorylation and can modulate viral genome copy number. Journal of virology. 2000;74:6031–6038. doi: 10.1128/jvi.74.13.6031-6038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic acids research. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J Virol. 2006;80:9530–9543. doi: 10.1128/JVI.01105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Skiadopoulos MH, McBride AA. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J Virol. 1998;72:2079–2088. doi: 10.1128/jvi.72.3.2079-2088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bodaghi S, Jia R, Zheng ZM. Human papillomavirus type 16 E2 and E6 are RNA-binding proteins and inhibit in vitro splicing of pre-mRNAs with suboptimal splice sites. Virology. 2009;386:32–43. doi: 10.1016/j.virol.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- [22].Baxter MK, McPhillips MG, Ozato K, McBride AA. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J Virol. 2005;79:4806–4818. doi: 10.1128/JVI.79.8.4806-4818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rahman S, Sowa ME, Ottinger M, Smith JA, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Maruyama T, Farina A, Dey A, Cheong J, et al. A Mammalian bromodomain protein, Brd4, interacts with replication factor C and inhibits progression to S phase. Mol.Cell Biol. 2002;22:6509–6520. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sarai N, Nimura K, Tamura T, Kanno T, et al. WHSC1 links transcription elongation to HIRA-mediated histone H3. 3 deposition. The EMBO journal. 2013;32:2392–2406. doi: 10.1038/emboj.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yan J, Li Q, Lievens S, Tavernier J, You J. Abrogation of the Brd4-positive transcription elongation factor B complex by papillomavirus E2 protein contributes to viral oncogene repression. J Virol. 2010;84:76–87. doi: 10.1128/JVI.01647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Terhune SS, Moorman NJ, Cristea IM, Savaryn JP, et al. Human cytomegalovirus UL29/28 protein interacts with components of the NuRD complex which promote accumulation of immediate-early RNA. PLoS pathogens. 2010;6:e1000965. doi: 10.1371/journal.ppat.1000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Smith JA, Haberstroh FS, White EA, Livingston DM, et al. SMCX and components of the TIP60 complex contribute to E2 regulation of the HPV E6/E7 promoter. Virology. 2014;468:311–321. doi: 10.1016/j.virol.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Murray JM, Carr AM. Smc5/6: a link between DNA repair and unidirectional replication? Nat Rev Mol Cell Biol. 2008;9:177–182. doi: 10.1038/nrm2309. [DOI] [PubMed] [Google Scholar]
- [30].Kegel A, Sjogren C. The Smc5/6 complex: more than repair? Cold Spring Harbor symposia on quantitative biology. 2010;75:179–187. doi: 10.1101/sqb.2010.75.047. [DOI] [PubMed] [Google Scholar]
- [31].Wu SY, Lee AY, Hou SY, Kemper JK, et al. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20:2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Malik-Soni N, Frappier L. Proteomic profiling of EBNA1-host protein interactions in latent and lytic Epstein-Barr virus infections. J Virol. 2012;86:6999–7002. doi: 10.1128/JVI.00194-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- [34].Petersen-Mahrt SK, Estmer C, Öhrmalm C, Matthews DA, et al. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. The EMBO journal. 1999;18:1014–1024. doi: 10.1093/emboj/18.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ito S, Ikeda M, Kato N, Matsumoto A, et al. Epstein–Barr virus nuclear antigen-1 binds to nuclear transporter karyopherin α1/NPI-1 in addition to karyopherin α2/Rch1. Virology. 2000;266:110–119. doi: 10.1006/viro.1999.0054. [DOI] [PubMed] [Google Scholar]
- [36].Matthews D, Russell W. Adenovirus core protein V interacts with p32--a protein which is associated with both the mitochondria and the nucleus. Journal of General Virology. 1998;79:1677–1685. doi: 10.1099/0022-1317-79-7-1677. [DOI] [PubMed] [Google Scholar]
- [37].Chen M-R, Yang J-F, Wu C-W, Middeldorp JM, Chen J-Y. Physical association between the EBV protein EBNA-1 and P32/TAP/hyaluronectin. Journal of biomedical science. 1998;5:173–179. doi: 10.1007/BF02253466. [DOI] [PubMed] [Google Scholar]
- [38].Bryant HE, Matthews DA, Wadd S, Scott JE, et al. Interaction between herpes simplex virus type 1 IE63 protein and cellular protein p32. Journal of virology. 2000;74:11322–11328. doi: 10.1128/jvi.74.23.11322-11328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ghebrehiwet B, Lim BL, Peerschke EI, Willis AC, Reid KB. Isolation, cDNA cloning, and overexpression of a 33-kD cell surface glycoprotein that binds to the globular "heads" of C1q. J Exp Med. 1994;179:1809–1821. doi: 10.1084/jem.179.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Soltys BJ, Kang D, Gupta RS. Localization of P32 protein (gC1q-R) in mitochondria and at specific extramitochondrial locations in normal tissues. Histochemistry and cell biology. 2000;114:245–255. doi: 10.1007/s004180000191. [DOI] [PubMed] [Google Scholar]
- [41].Dutta S, Akey IV, Dingwall C, Hartman KL, et al. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Molecular cell. 2001;8:841–853. doi: 10.1016/s1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- [42].Muller M, Jacob Y, Jones L, Weiss A, et al. Large scale genotype comparison of human papillomavirus E2-host interaction networks provides new insights for e2 molecular functions. PLoS Pathog. 2012;8:e1002761. doi: 10.1371/journal.ppat.1002761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rehtanz M, Schmidt HM, Warthorst U, Steger G. Direct interaction between nucleosome assembly protein 1 and the papillomavirus E2 proteins involved in activation of transcription. Mol Cell Biol. 2004;24:2153–2168. doi: 10.1128/MCB.24.5.2153-2168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Muller M, Demeret C. The HPV E2-Host Protein-Protein Interactions: A Complex Hijacking of the Cellular Network. Open Virol J. 2012;6:173–189. doi: 10.2174/1874357901206010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Emelyanov AV, Rabbani J, Mehta M, Vershilova E, et al. Drosophila TAP/p32 is a core histone chaperone that cooperates with NAP-1, NLP, and nucleophosmin in sperm chromatin remodeling during fertilization. Genes & development. 2014;28:2027–2040. doi: 10.1101/gad.248583.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chen GI, Gingras AC. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods. 2007;42:298–305. doi: 10.1016/j.ymeth.2007.02.018. [DOI] [PubMed] [Google Scholar]
- [47].Samad MA, Okuwaki M, Haruki H, Nagata K. Physical and functional interaction between a nucleolar protein nucleophosmin/B23 and adenovirus basic core proteins. FEBS Lett. 2007;581:3283–3288. doi: 10.1016/j.febslet.2007.06.024. [DOI] [PubMed] [Google Scholar]
- [48].Malik-Soni N, Frappier L. Nucleophosmin contributes to the transcriptional activation function of the Epstein-Barr virus EBNA1 protein. J Virol. 2014;88:2323–2326. doi: 10.1128/JVI.02521-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ulrich HD. New insights into replication clamp unloading. Journal of molecular biology. 2013;425:4727–4732. doi: 10.1016/j.jmb.2013.05.003. [DOI] [PubMed] [Google Scholar]
- [50].Xiao A, Li H, Shechter D, Ahn SH, et al. WSTF regulates the H2A. × DNA damage response via a novel tyrosine kinase activity. Nature. 2008;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 2002;21:2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gui JF, Tronchere H, Chandler SD, Fu XD. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci U S A. 1994;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wang WS, Lee MS, Tseng CE, Liao IH, et al. Interaction between human papillomavirus type 5 E2 and polo-like kinase 1. J Med Virol. 2009;81:536–544. doi: 10.1002/jmv.21404. [DOI] [PubMed] [Google Scholar]
- [54].Prescott EL, Brimacombe CL, Hartley M, Bell I, et al. Human Papillomavirus Type 1 E1^ E4 Protein Is a Potent Inhibitor of the Serine-Arginine (SR) Protein Kinase SRPK1 and Inhibits Phosphorylation of Host SR Proteins and of the Viral Transcription and Replication Regulator E2. Journal of virology. 2014;88:12599–12611. doi: 10.1128/JVI.02029-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].McBride AA, Bolen JB, Howley PM. Phosphorylation sites of the E2 transcriptional regulatory proteins of bovine papillomavirus type 1. J.Virol. 1989;63:5076–5085. doi: 10.1128/jvi.63.12.5076-5085.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Penrose KJ, Garcia-Alai M, Prat-Gay G, McBride AA. CK2 phosphorylation-induced conformational switch triggers degradation of the papillomavirus E2 protein. J.Biol.Chem. 2004;279:22430–22439. doi: 10.1074/jbc.M314340200. [DOI] [PubMed] [Google Scholar]
- [57].Shire K, Kapoor P, Jiang K, Hing MN, et al. Regulation of the EBNA1 Epstein-Barr virus protein by serine phosphorylation and arginine methylation. J.Virol. 2006;80:5261–5272. doi: 10.1128/JVI.02682-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Thandapani P, O’Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Molecular cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- [59].Hisaoka M, Ueshima S, Murano K, Nagata K, Okuwaki M. Regulation of nucleolar chromatin by B23/nucleophosmin jointly depends upon its RNA binding activity and transcription factor UBF. Molecular and cellular biology. 2010;30:4952–4964. doi: 10.1128/MCB.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- [61].Sakakibara N, Chen D, Jang MK, Kang DW, et al. Brd4 Is Displaced from HPV Replication Factories as They Expand and Amplify Viral DNA. PLoS Pathog. 2013;9:e1003777. doi: 10.1371/journal.ppat.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Antson AA, Burns JE, Moroz OV, Scott DJ, et al. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature. 2000;403:805–809. doi: 10.1038/35001638. [DOI] [PubMed] [Google Scholar]
- [63].Hegde RS, Androphy EJ. Crystal structure of the E2 DNA-binding domain from human papillomavirus type 16: implications for its DNA binding-site selection mechanism. Journal of molecular biology. 1998;284:1479–1489. doi: 10.1006/jmbi.1998.2260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.