Abstract
Imprinted genes, which are monoallelically expressed by virtue of an epigenetic process initiated in the germline, are known to play key roles in regulating fetal growth and placental development. Numerous studies are investigating the expression of these imprinted genes in the human placenta in relation to common complications of pregnancy such as fetal growth restriction and preeclampsia. This study aimed to determine whether placental sampling protocols or other factors such as fetal sex, gestational age and mode of delivery may influence the expression of imprinted genes predicted to regulate placental signalling.
Methods
Term placentas were collected from Caucasian women delivering at University Hospital of Wales or Royal Gwent Hospital within two hours of delivery. Expression of the imprinted genes PHLDA2, CDKN1C, PEG3 and PEG10 was assayed by quantitative real time PCR. Intraplacental gene expression was analysed (N = 5). Placental gene expression was compared between male (N = 11) and female (N = 11) infants, early term (N = 8) and late term (N = 10) deliveries and between labouring (N = 13) and non-labouring (N = 21) participants.
Results
The paternally expressed imprinted genes PEG3 and PEG10 were resilient to differences in sampling site, fetal sex, term gestational age and mode of delivery. The maternally expressed imprinted gene CDKN1C was elevated over 2-fold (p < 0.001) in placenta from labouring deliveries compared with elective caesarean sections. In addition, the maternally expressed imprinted gene PHLDA2 was elevated by 1.8 fold (p = 0.01) in samples taken at the distal edge of the placenta compared to the cord insertion site.
Conclusion
These findings support the reinterpretation of existing data sets on these genes in relation to complications of pregnancy and further reinforce the importance of optimising and unifying placental collection protocols for future studies.
Keywords: Placenta, Imprinted genes, Sampling, Delivery mode
Highlights
-
•
77% variation in expression of PHLDA2 between distal and proximal sampling sites.
-
•
2.4-fold increase in placental expression of CDKN1C induced by labour.
-
•
May explain conflicting links between pregnancy pathologies and imprinted genes.
1. Introduction
Imprinted genes display parent of origin specific gene expression [1] and play a key role in regulating placental development and controlling fetal growth (reviewed in Ref. [2]). Recent studies have identified several imprinted genes that regulate the endocrine lineages of the mouse placenta to modulate placental hormone production (reviewed in Ref. [3]). Imprinting of four of these genes, PLECKSTRIN HOMOLOGY-LIKE DOMAIN FAMILY A MEMBER 2 (PHLDA2), CYCLIN-DEPENDENT KINASE INHIBITOR 1C (CDKN1C), PATERNALLY EXPRESSED GENE 3 (PEG3) and PATERNALLY EXPRESSED GENE 10 (PEG10), is conserved in the human placenta [4]. While a number of studies have examined human placental imprinted gene expression focusing primarily on fetal growth restriction (FGR) as an adverse outcome [4–9], the potential for aberrant placental signalling presents additional implications for human pregnancies.
Gene expression in the placenta can be influenced by many factors including differences in sampling site, fetal sex, gestational age and mode of delivery (reviewed in Ref. [10]). Variation in gene expression may occur in different regions of the placenta due to differences in placental architecture or blood supply and therefore consistency in placental sampling is of key importance [10]. A number of studies have demonstrated differences in intraplacental gene expression depending on sampling site [11–17] but the possibility of intraplacental variation in expression of PHLDA2, CDKN1C, PEG3 or PEG10 has not been investigated.
Sexual dimorphism in human placental gene expression (of both autosomal and sex chromosome linked genes) has also previously been demonstrated [17,18] which has been proposed to underlie sex differences in fetal growth [19]. As sexual dimorphism in placental imprinted gene expression has been demonstrated in an animal model [20] and for the imprinted H19 gene in human placenta [9], controlling for fetal sex is important to studies of imprinted gene expression in the human placenta.
Infant morbidity and mortality [21–23] as well as placental pathology [24] have been observed to vary in response to gestational age in term pregnancies. It has therefore been proposed that infants should be distinguished as early term (37 0/7 to 38 6/7 weeks) and full term (39 0/7 to 41 6/7) [23]. While it has been demonstrated that placental expression of PHLDA2 varies between preterm and term deliveries [7], it has not yet been established whether expression of PHLDA2, CDKN1C, PEG3 or PEG10 varies between early term and full term placentas. This is of particular relevance to the study of pregnancy complications such as FGR, as these growth restricted infants may require earlier delivery [25].
Importantly, a number of studies have reported differences in gene expression between labouring and non-labouring placentas [26–29], which may arise from a decrease in placental oxygen supply during contractions [10] and/or differences in exposure to hormones associated with labour [30]. PHLDA2 is known to be responsive to hypoxia [31–33] but it is not known whether placental expression of PHLDA2, CDKN1C, PEG3 or PEG10 is altered in response to labour.
To address this lack of information, this study examined placental expression of the maternally expressed imprinted genes PHLDA2 and CDKN1C and the paternally expressed imprinted genes PEG3 and PEG10 to determine whether their expression varied in relation to sampling site, fetal sex, gestational age or mode of delivery.
2. Methods
2.1. Study participants
Study participants (N = 40) were recruited prior to delivery at University Hospital Wales, Cardiff and Royal Gwent Hospital, Newport. Written informed consent was obtained and the study was approved by the South East Wales Research Ethics Committee (REC number: 10/WSE02/10). Healthy Caucasian women with singleton pregnancies and no known medical disorders were recruited and all infants were delivered normal birth weight at term (≥37 weeks). Information on birth outcomes were obtained from the maternal medical notes.
In order to determine the effect of fetal sex on placental imprinted gene expression, gene expression was compared between male (N = 11) and female (N = 11) placentas delivered by elective C-section. Gene expression was also compared between early term (N = 8) and full term (N = 10) placentas delivered by elective C-section to determine whether expression varies with gestational age at term. Finally, to determine whether labour effects placental imprinted gene expression, gene expression was compared between placentas delivered by elective C-section in the absence of labour (N = 21) and by vaginal (N = 9) or emergency C-section delivery following a period of labour (N = 4). Allocation of participants to each group and participant characteristics is summarized in Supplementary Table 1.
2.2. Placental dissection
Placentas were weighed and dissected within 2 h of delivery. Villous trophoblast samples were obtained at five random sites on the maternal surface of the placenta, midway between the cord and distal edge. Samples were thoroughly rinsed in ice cold phosphate buffered saline to remove excess blood. Placental samples were then stored in RNAlater (Sigma–Aldrich, Dorset, UK) at 4 °C prior to long term storage at −80 °C.
To examine intraplacental variation in imprinted gene expression, three placental samples (close to the cord insertion, middle and distal edge) were taken from each of the fetal, middle and maternal layers of the placenta as described by Wyatt et al. (2005) (16). This was carried out for five term placentas (2 male, 3 female) from elective C-sections. Average placental gene expression (fetal, middle and maternal layers of the placenta) was compared between sampling sites close to the cord insertion and at distal sites. Similarly, average placental gene expression (cord insertion, middle and distal edge) was compared between basal and chorionic surface sampling sites.
2.3. Gene expression analysis
Total RNA was extracted from the placental tissue samples using GenElute Mammalian Total RNA Miniprep Kit (Sigma–Aldrich, Dorset, UK) with an on-column DNase digestion (Sigma–Aldrich, Dorset, UK). 5 μg of RNA was reverse transcribed using M-MuLV reverse transcriptase (Promega, Southampton, UK) with random hexamers, according to manufacturer's instructions.
Quantitative RT-PCR was performed using Chromo 4 Four Colour Real Time Detector (MJ Research) in a 20 μl reaction containing 5 μl of cDNA (diluted 1 in 50), 1X Buffer 2 mM MgCl2, 2 mM dNTPs, 0.65 Units Taq (Fermentas (Thermo), Loughborough, UK), 1 μM of each primer (Sigma–Aldrich, Dorset, UK) and 0.12X Sybr Green (Invitrogen, Paisley, UK). All samples were run in triplicate and duplicate plates were performed. Conditions for amplification were: 1) 15 min at 94 °C, 2) 30 s at 94 °C, 3) 30 s at 60 °C, 4) 30 s at 72 °C and 5) 30 s 75 °C, with steps 2–5 repeated for a total of 40 cycles. Melt Curve was performed from 70 °C to 94 °C, reading every 0.5 °C and holding for 2 s.
Primer sequences were as follows: YWHAZ forward: TTCTTGATCCCCAATGCTTC and reverse: AGTTAAGGGCCAGACCCAGT, PHLDA2 forward: GAGCGCACGGGCAAGTA and reverse: CAGCGGAAGTCGATCTCCTT [6], CDKN1C forward: CCCATCTAGCTTGCAGTCTCTT and reverse: CAGACGGCTCAGGAACCATT [4], PEG3 forward: CTCACAACACAATCCAGGAC and reverse: TAGACCTCGACTGGTGCTTG, PEG10 forward: AAATTGCCTGACATGAAGAGGAGTCTA and reverse: AAGCCTAGTCACCACTTCAAAACACACTAAA [4].
Gene expression data is presented as the ΔCT (target gene expression relative to the housekeeping gene YWHAZ) and as the fold change in expression, calculated using the 2−ΔΔCT [34] where the ΔΔCT is the target gene expression relative to expression in the control group. The housekeeping gene YWHAZ was chosen as this gene has been demonstrated in a number of studies to be stably expressed in the human placenta of normal pregnancies [35–37] and in pregnancies complicated by growth restriction [38]. In the current study there was no significant effect of fetal sex (p = 0.56), early term delivery (p = 0.44) or labour (p = 0.74) on placental YWHAZ gene expression.
2.4. Statistical analysis
All statistical analysis was carried out using IBM SPSS Statistics for Windows (version 20.0, 2011). A Shapiro–Wilk test was used to test for normal distribution of the data. All gene expression data was normally distributed and therefore differences in gene expression were analysed using an independent samples T-test.
3. Results
A summary of participant demographics is shown in Table 1.
Table 1.
Characteristics of the study participants (N = 40). Mean (SD)/range or number (%) is shown.
| Study Participants (N = 40) | |
|---|---|
| Maternal Characteristic | |
| Ethnicity: | |
| Caucasian | 40 (100%) |
| Parity | 1 (0.87)/0–3 |
| Maternal age | 30 (6.12)/19–40 |
| Maternal BMI | 27 (5.91)/17 - 38 |
| Birth Outcome | |
| Mode of Delivery: | |
| Vaginal | 9 (22%) |
| Elective C section | 27 (68%) |
| Emergency C section | 4 (10%) |
| Birth weight (g) | 3417 (303)/2680 - 4030 |
| Gestational age (weeks) | 39 (1.19)/37 - 42 |
| Placental weight (g) | 676 (120)/434 - 891 |
| Gender | |
| Male | 18 (45%) |
| Female | 22 (55%) |
3.1. Sampling site variation
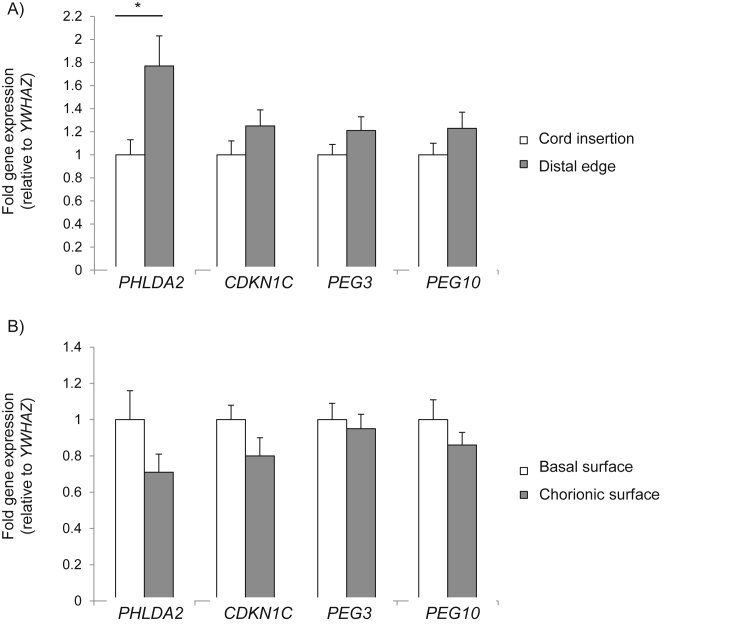
Placental CDKN1C, PEG3 and PEG10 expression did not differ significantly between cord insertion and distal sampling sites (Fig. 1A) or between the basal and chorionic surface (Fig. 1B). Placental PHLDA2 expression was significantly increased, by 77%, in samples taken at distal sampling sites compared with sampling sites close to the cord insertion (Fig. 1A). There was no significant difference in PHLDA2 expression between sampling sites on the basal and chorionic placental surface (Fig. 1B).
Fig. 1.
Effect of sampling site on placental imprinted gene expression. Gene expression is shown relative to sampling site near the cord insertion (A) and on the basal surface (B) of the placenta. Mean fold gene expression is shown (±SEM). Three samples were taken at each site for a total of five placentas (N = 15 samples per site). Differences in gene expression were analysed using an independent samples t-test. *p < 0.05.
3.2. Fetal sex
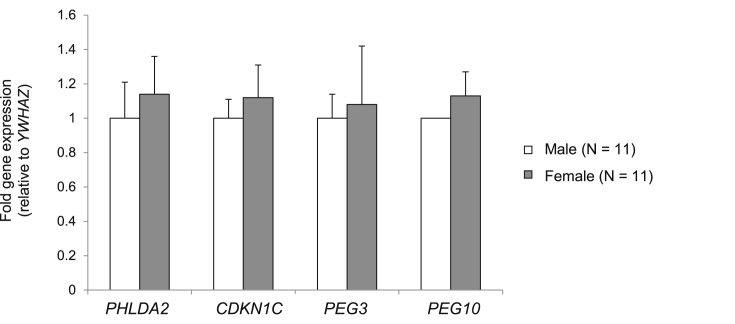
There was no significant difference in mean placental weight (663 g vs. 726 g; p = 0.23), birth weight (3381 g vs. 3601 g; p = 0.07) or gestational age (273 days v 274 days; p = 0.85) between male and female infants. There was no significant difference in placental PHLDA2, CDKN1C, PEG3 or PEG10 expression between male and female placentas (Fig. 2).
Fig. 2.
Sex differences in placental imprinted gene expression. Mean fold gene expression is shown (±SEM) relative to expression in male placentas. Differences in gene expression were not statistically significant using an independent samples t-test.
3.3. Gestational age
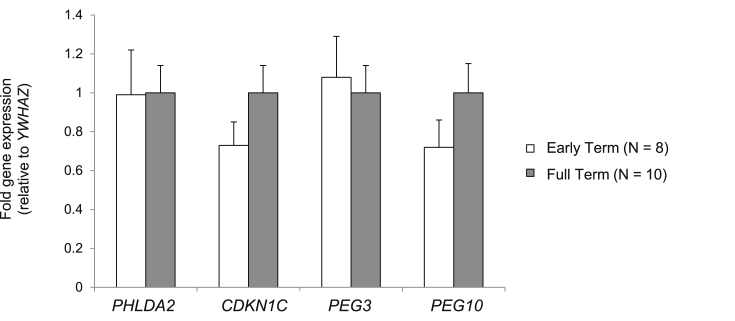
Early term infants were delivered between 37 and 38 weeks (mean gestational age 266 days) and full term infants between 39 and 40 weeks (mean gestational age 277 days). There was no significant difference in mean placental weight (648 g vs. 717 g; p = 0.23), birth weight (3445 g vs. 3392 g; p = 0.07) between early term and full term infants. The proportion of male and female infants was also not significantly different between early (4 Male, 4 Female) and full term (4 Male and 6 Female) participants (p = 0.67). PHLDA2, CDKN1C, PEG3 and PEG10 expression was not significantly altered in early term compared with full term placentas (Fig. 3).
Fig. 3.
Early term delivery and placental imprinted gene expression. Mean fold gene expression is shown (±SEM) relative to expression in full term placentas. Differences in gene expression were not statistically significant using an independent samples T-test.
3.4. Labour
There was no significant difference in mean placental weight (708 g vs. 661 g; p = 0.31), birth weight (3470 g vs. 3451 g; p = 0.85) or gestational age (273 days v 276 days; p = 0.49) between labouring and non-labouring participants. There was also no significant difference in the proportion of male and female infants between labouring (6 Male, 7 Female) and non-labouring (11 Male, 10 Female) participants (p = 0.72).
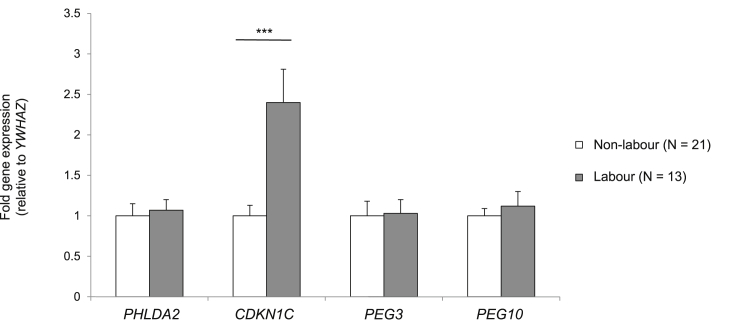
There was no significant effect of labour on placental PHLDA2, PEG3 or PEG10 expression (Fig. 4). Placental CDKN1C was increased, by 140%, in labouring compared with non-labouring participants (Fig. 4). This increase in placental CDKN1C expression remained significant when labouring participants were divided into those delivering vaginally (115% increase, p = 0.001, n = 9) and those delivering by emergency C-section (197% increase, p = 0.001, n = 4).
Fig. 4.
Effect of labour on placental imprinted gene expression. Mean fold gene expression is shown (±SEM) relative to expression in placentas from elective C section deliveries. Differences in gene expression were analysed using an independent samples T-test. ***p < 0.001.
4. Discussion
This study demonstrates that both the site of sampling and the mode of delivery can influence the quantitative assessment of imprinted gene expression in the human placenta.
Our first key finding was that the expression of PHLDA2 varied with site of sampling. PHLDA2 is a maternally expressed imprinted gene whose elevated placental expression has been reported in relation to fetal growth restriction in several human studies [39]. In mice we have shown that transgenically elevated Phlda2 drives a late and asymmetric fetal growth restriction [40] suggesting a causal role for PHLDA2 in human fetal growth restriction. In this current study, we found that placental PHLDA2 expression differed significantly with sampling site, with increased expression at distal sampling sites compared with sites close to the umbilical cord. Although no previous study has examined placental PHLDA2 expression in relation to sampling site, a number of studies have similarly reported significant intraplacental variation in gene expression [12–14,16,17]. Given that placental PHLDA2 expression is altered in response to hypoxia [31–33], it is possible that differences in perfusion between distal sites and sites close to the umbilical cord [14,16] could underlie the differences in expression observed. The finding of intraplacental variation in PHLDA2 expression is novel and has implications for interpretation of previous studies and for future studies with consistency in placental sampling of key importance.
A second key finding in this study was the variation in CDKN1C in relation to mode of delivery. In humans, loss of expression of CDKN1C has been linked to the childhood overgrowth disorder Beckwith Weidemann Syndrome while microduplications spanning the gene and alterations in the PCNA domain of CDKN1C have been linked to the growth restriction disorders Silver-Russell syndrome (SRS) and IMAGe syndrome, respectively [41]. Mouse studies demonstrate that Cdkn1c functions early in life to limit fetal growth and to regulate placental development [42–45] consistent with a role for this gene in these disorders.
Loss of function of CDKN1C may also play a role in preeclampsia/HELLP syndrome [46] which has also been partially supported by animal studies [47]. However, while one study reported significantly reduced CDKN1C expression in preeclamptic placentas [48] two other studies reported significantly elevated expression [49,50]. In the study by Kawasaki et al. [48], placental villous samples were obtained immediately after caesarean section in the absence of labour for both controls and preeclampsia pregnancies that would exclude mode of delivery as a confounder. In the study by Enquobahrie et al. [50] preeclamptic pregnancies had a higher caesarean delivery rate but a similar rate of labour with controls while in the study by Unek et al. [49] placenta were sampled after caesarean delivery without stating whether labour was initiated. Similarly, studies examining placental CDKN1C expression in relation to fetal growth restriction have reported conflicting results with both unaltered [4], increased [5] and decreased [51] expression reported in FGR placentas which may result from a failure to control for mode of delivery in these analyses.
In this study, the expression of CDKN1C varied in response to mode of delivery with both vaginally delivered and emergency caesarean placenta expressing CDKN1C at significantly higher levels than placenta obtained by elective caesarean in which labour was not initiated. This is consistent with previous reports of significantly increased CDKN1C expression in the mouse uterus and human myometrium in association with labour [52]. In light of these new findings, it will be important to re-examine the data both from studies reporting a link between placental CDKN1C and preeclampsia or fetal growth restriction and those studies finding no association.
No significant difference was observed in placental PHLDA2, PEG3 or PEG10 expression in response to labour, and no intraplacental variation was observed in placental CDKN1C, PEG3 or PEG10 expression at different sampling sites. None of the four genes showed a significant difference in expression between early and late term deliveries and there was no significant difference in relation to fetal sex confirming the findings of Moore et al. [9]. However, sexual dimorphism exists in the human fetal response to an adverse environment (reviewed in Ref. [19]) and differential imprinted gene expression has been reported in male and female mouse placentas in response to maternal diet alteration [53]. Thus, a sex-specific response in human placental imprinted gene expression to an adverse intrauterine environment merits further investigation.
In conclusion, we have found that the expression of PHLDA2 varied with sampling site and that expression of CDKN1C varied with mode of delivery. These types of study further highlight the importance of developing uniform collection protocols with details on mode of delivery as well as birth outcomes [10].
Acknowledgements
With thanks to the participants at University Hospital Wales and Newport Gwent Hospital who kindly donated their placenta anonymously for this study. ABJ was funded by BBSRC doctoral training grant BB/F016557/1. SJT was funded by BBSRC grant BB/J015156.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.placenta.2015.06.011.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Surani M.A. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93(3):309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- 2.Lim A.L., Ferguson-Smith A.C. Genomic imprinting effects in a compromised in utero environment: implications for a healthy pregnancy. Semin. Cell Dev. Biol. 2010;21(2):201–208. doi: 10.1016/j.semcdb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 3.John R.M. Epigenetic regulation of placental endocrine lineages and complications of pregnancy. Biochem. Soc. Trans. 2013;41(3):701–709. doi: 10.1042/BST20130002. [DOI] [PubMed] [Google Scholar]
- 4.Diplas A.I., Lambertini L., Lee M.-J., Sperling R., Lee Y.L., Wetmur J.G. Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics. 2009;4(4):235–240. doi: 10.4161/epi.9019. [DOI] [PubMed] [Google Scholar]
- 5.McMinn J., Wei M., Schupf N., Cusmai J., Johnson E.B., Smith A.C. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta. 2006;27(6–7):540–549. doi: 10.1016/j.placenta.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Apostolidou S., Abu-Amero S., O'Donoghue K., Frost J., Olafsdottir O., Chavele K. Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. J. Mol. Med. 2007;85(4):379–387. doi: 10.1007/s00109-006-0131-8. [DOI] [PubMed] [Google Scholar]
- 7.Kumar N., Leverence J., Bick D., Sampath V. Ontogeny of growth-regulating genes in the placenta. Placenta. 2012;33(2):94–99. doi: 10.1016/j.placenta.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 8.Demetriou C., Abu-Amero S., Thomas A.C., Ishida M., Aggarwal R., Al-Olabi L. Paternally expressed, imprinted insulin-like growth factor-2 in chorionic villi correlates significantly with birth weight. PLoS One. 2014;9(1):e85454. doi: 10.1371/journal.pone.0085454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore G.E., Ishida M., Demetriou C., Al-Olabi L., Leon L.J., Thomas A.C. The role and interaction of imprinted genes in human fetal growth. Philos. Trans. R. Soc. Lond B Biol. Sci. 2015;370(1663):20140074. doi: 10.1098/rstb.2014.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton G.J., Sebire N.J., Myatt L., Tannetta D., Wang Y.L., Sadovsky Y. Optimising sample collection for placental research. Placenta. 2014;35(1):9–22. doi: 10.1016/j.placenta.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Brameld J.M., Hold R., Pipkin F.B. Regional variation in angiotensin converting enzyme activity in the human placenta. Placenta. 2011;32(11):906–908. doi: 10.1016/j.placenta.2011.07.085. [DOI] [PubMed] [Google Scholar]
- 12.Tzschoppe A.A., Struwe E., Dorr H.G., Goecke T.W., Beckmann M.W., Schild R.L. Differences in gene expression dependent on sampling site in placental tissue of fetuses with intrauterine growth restriction. Placenta. 2010;31:178–185. doi: 10.1016/j.placenta.2009.12.002. England: 2009 Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- 13.Pidoux G., Gerbaud P., Laurendeau I., Guibourdenche J., Bertin G., Vidaud M. Large variability of trophoblast gene expression within and between human normal term placentae. Placenta. 2004;25:469–473. doi: 10.1016/j.placenta.2003.10.016. England. [DOI] [PubMed] [Google Scholar]
- 14.Hempstock J., Bao Y.P., Bar-Issac M., Segaren N., Watson A.L., Charnock-Jones D.S. Intralobular differences in antioxidant enzyme expression and activity reflect the pattern of maternal arterial bloodflow within the human placenta. Placenta. 2003;24:517–523. doi: 10.1053/plac.2002.0955. England. [DOI] [PubMed] [Google Scholar]
- 15.Avila L., Yuen R.K., Diego-Alvarez D., Penaherrera M.S., Jiang R., Robinson W.P. Evaluating DNA methylation and gene expression variability in the human term placenta. Placenta. 2010;31:1070–1077. doi: 10.1016/j.placenta.2010.09.011. England: 2010 Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- 16.Wyatt S.M., Kraus F.T., Roh C.R., Elchalal U., Nelson D.M., Sadovsky Y. The correlation between sampling site and gene expression in the term human placenta. Placenta. 2005;26:372–379. doi: 10.1016/j.placenta.2004.07.003. England. [DOI] [PubMed] [Google Scholar]
- 17.Sood R., Zehnder J.L., Druzin M.L., Brown P.O. Gene expression patterns in human placenta. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5478–5483. doi: 10.1073/pnas.0508035103. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buckberry S., Bianco-Miotto T., Bent S.J., Dekker G.A., Roberts C.T. Integrative transcriptome meta-analysis reveals widespread sex-biased gene expression at the human fetal-maternal interface. Mol. Hum. Reprod. 2014;20:810–819. doi: 10.1093/molehr/gau035. England: The Author 2014. Published by Oxford University Press on behalf of the European Society of Human Reproduction and Embryology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clifton V.L. Review: Sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta. 2010;31:S33–S39. doi: 10.1016/j.placenta.2009.11.010. England: Crown 2010. Published by Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- 20.Faisal M., Kim H., Kim J. Sexual differences of imprinted genes' expression levels. Gene. 2014;533(1):434–438. doi: 10.1016/j.gene.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crump C., Sundquist K., Winkleby M.A., Sundquist J. Early-term birth (37–38 weeks) and mortality in young adulthood. Epidemiology. 2013;24(2):270–276. doi: 10.1097/EDE.0b013e318280da0f. [DOI] [PubMed] [Google Scholar]
- 22.Engle W.A., Kominiarek M.A. Late preterm infants, early term infants, and timing of elective deliveries. Clin. Perinatol. 2008;35(2):325–341. doi: 10.1016/j.clp.2008.03.003. vi. [DOI] [PubMed] [Google Scholar]
- 23.Fleischman A.R., Oinuma M., Clark S.L. Rethinking the definition of term pregnancy. Obstet. Gynecol. 2010;116(1):136–139. doi: 10.1097/AOG.0b013e3181e24f28. [DOI] [PubMed] [Google Scholar]
- 24.Stanek J. Comparison of placental pathology in preterm, late-preterm, near-term, and term births. Am. J. Obstet. Gynecol. 2014;210(3)(234):e1–6. doi: 10.1016/j.ajog.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Galan H.L. Timing delivery of the growth-restricted fetus. Semin. Perinatol. 2011;35(5):262–269. doi: 10.1053/j.semperi.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Cindrova-Davies T., Yung H.W., Johns J., Spasic-Boskovic O., Korolchuk S., Jauniaux E. Oxidative stress, gene expression, and protein changes induced in the human placenta during labor. Am. J. Pathol. 2007;171:1168–1179. doi: 10.2353/ajpath.2007.070528. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sitras V., Paulssen R.H., Gronaas H., Vartun A., Acharya G. Gene expression profile in labouring and non-labouring human placenta near term. Mol. Hum. Reprod. 2008;14:61–65. doi: 10.1093/molehr/gam083. England. [DOI] [PubMed] [Google Scholar]
- 28.Lee K.J., Shim S.H., Kang K.M., Kang J.H., Park D.Y., Kim S.H. Global gene expression changes induced in the human placenta during labor. Placenta. 2010;31:698–704. doi: 10.1016/j.placenta.2010.05.006. England. [DOI] [PubMed] [Google Scholar]
- 29.Peng H.H., Kao C.C., Chang S.D., Chao A.S., Chang Y.L., Wang C.N. The effects of labor on differential gene expression in parturient women, placentas, and fetuses at term pregnancy. Kaohsiung J. Med. Sci. 2011;27:494–502. doi: 10.1016/j.kjms.2011.06.012. China Republic : 1949-: 2011. Published by Elsevier B.V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss G. Endocrinology of parturition. J. Clin. Endocrinol. Metab. 2000;85(12):4421–4425. doi: 10.1210/jcem.85.12.7074. [DOI] [PubMed] [Google Scholar]
- 31.Kim H.S., Roh C.R., Chen B., Tycko B., Nelson D.M., Sadovsky Y. Hypoxia regulates the expression of PHLDA2 in primary term human trophoblasts. Placenta. 2007;28(2–3):77–84. doi: 10.1016/j.placenta.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 32.Roh C.R., Budhraja V., Kim H.S., Nelson D.M., Sadovsky Y. Microarray-based identification of differentially expressed genes in hypoxic term human trophoblasts and in placental villi of pregnancies with growth restricted fetuses. Placenta. 2005;26(4):319–328. doi: 10.1016/j.placenta.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Tomlinson T.M., Garbow J.R., Anderson J.R., Engelbach J.A., Nelson D.M., Sadovsky Y. Magnetic resonance imaging of hypoxic injury to the murine placenta. Am. J. Physiol – Regul. Integr. Comp. Physiol. 2010;298(2) doi: 10.1152/ajpregu.00425.2009. R312–R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Meller M., Vadachkoria S., Luthy D.A., Williams M.A. Evaluation of housekeeping genes in placental comparative expression studies. Placenta. 2005;26:601–607. doi: 10.1016/j.placenta.2004.09.009. England. [DOI] [PubMed] [Google Scholar]
- 36.Cleal J.K., Day P., Hanson M.A., Lewis R.M. Measurement of housekeeping genes in human placenta. Placenta. 2009;30:1002–1003. doi: 10.1016/j.placenta.2009.09.002. England. [DOI] [PubMed] [Google Scholar]
- 37.Cleal J.K., Day P.L., Hanson M.A., Lewis R.M. Sex differences in the mRNA levels of housekeeping genes in human placenta. Placenta. 2010;31:556–557. doi: 10.1016/j.placenta.2010.03.006. England. [DOI] [PubMed] [Google Scholar]
- 38.Murthi P., Fitzpatrick E., Borg A.J., Donath S., Brennecke S.P., Kalionis B. GAPDH, 18S rRNA and YWHAZ are suitable endogenous reference genes for relative gene expression studies in placental tissues from human idiopathic fetal growth restriction. Placenta. 2008;29:798–801. doi: 10.1016/j.placenta.2008.06.007. England. [DOI] [PubMed] [Google Scholar]
- 39.Jensen A.B., Tunster S.J., John R.M. The significance of elevated placental PHLDA2 in human growth restricted pregnancies. Placenta. 2014;35(8):528–532. doi: 10.1016/j.placenta.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 40.Tunster S.J., Van De Pette M., John R.M. Isolating the role of elevated PHLDA2 in asymmetric late fetal growth restriction in mice. Dis. Model Mech. 2014;7:1185–1191. doi: 10.1242/dmm.017079. England: 2014. Published by The Company of Biologists Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eggermann T., Binder G., Brioude F., Maher E.R., Lapunzina P., Cubellis M.V. CDKN1C mutations: two sides of the same coin. Trends Mol. Med. 2014;20(11):614–622. doi: 10.1016/j.molmed.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi K., Kobayashi T., Kanayama N. p57(Kip2) regulates the proper development of labyrinthine and spongiotrophoblasts. Mol. Hum. Reprod. 2000;6(11):1019–1025. doi: 10.1093/molehr/6.11.1019. [DOI] [PubMed] [Google Scholar]
- 43.Andrews S.C., Wood M.D., Tunster S.J., Barton S.C., Surani M.A., John R.M. Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7. BMC Dev. Biol. 2007;7:53. doi: 10.1186/1471-213X-7-53. England. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang P., Liegeois N.J., Wong C., Finegold M., Hou H., Thompson J.C. Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith–Wiedemann syndrome. Nature. 1997;387(6629):151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 45.Yan Y., Frisen J., Lee M.H., Massague J., Barbacid M. Ablation of the CDK inhibitor p57Kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes. Dev. 1997;11(8):973–983. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- 46.Romanelli V., Belinchon A., Campos-Barros A., Heath K.E., Garcia-Minaur S., Martinez-Glez V. CDKN1C mutations in HELLP/preeclamptic mothers of Beckwith-Wiedemann Syndrome (BWS) patients. Placenta. 2009;30:551–554. doi: 10.1016/j.placenta.2009.03.013. England. [DOI] [PubMed] [Google Scholar]
- 47.Kanayama N., Takahashi K., Matsuura T., Sugimura M., Kobayashi T., Moniwa N. Deficiency in p57Kip2 expression induces preeclampsia-like symptoms in mice. Mol. Hum. Reprod. 2002;8(12):1129–1135. doi: 10.1093/molehr/8.12.1129. [DOI] [PubMed] [Google Scholar]
- 48.Kawasaki K., Kondoh E., Chigusa Y., Ujita M., Murakami R., Mogami H. Reliable pre-eclampsia pathways based on multiple independent microarray data sets. Mol. Hum. Reprod. 2015;21(2):217–224. doi: 10.1093/molehr/gau096. [DOI] [PubMed] [Google Scholar]
- 49.Unek G., Ozmen A., Mendilcioglu I., Simsek M., Korgun E.T. The expression of cell cycle related proteins PCNA, Ki67, p27 and p57 in normal and preeclamptic human placentas. Tissue Cell. 2014;46(3):198–205. doi: 10.1016/j.tice.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Enquobahrie D.A., Meller M., Rice K., Psaty B.M., Siscovick D.S., Williams M.A. Differential placental gene expression in preeclampsia. Am. J. Obstet. Gynecol. 2008;199 doi: 10.1016/j.ajog.2008.04.020. United States 566 e1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajaraman G., Murthi P., Pathirage N., Brennecke S.P., Kalionis B. Downstream targets of homeobox gene HLX show altered expression in human idiopathic fetal growth restriction. Am. J. Pathol. 2010;176:278–287. doi: 10.2353/ajpath.2010.090187. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bethin K.E., Nagai Y., Sladek R., Asada M., Sadovsky Y., Hudson T.J. Microarray analysis of uterine gene expression in mouse and human pregnancy. Mol. Endocrinol. 2003;17(8):1454–1469. doi: 10.1210/me.2003-0007. [DOI] [PubMed] [Google Scholar]
- 53.Radford E.J., Isganaitis E., Jimenez-Chillaron J., Schroeder J., Molla M., Andrews S. An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming. PLoS Genet. 2012;8(4):e1002605. doi: 10.1371/journal.pgen.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.