Abstract
Background
New biomarkers are needed to identify the stage of hepatitis C virus (HCV)-infected diseases in order to reduce the mortality rates. Herein, we investigated whether serum 3β-hydroxysterol Δ24-reductase antibody (DHCR24 Ab) may serve as a prognostic marker for hepatitis C infection progression to hepatocellular carcinoma (HCC).
Methods
Serum DHCR24 Abs from 395 HCV-positive patients, including 133 chronic hepatitis (CHC), 85 liver cirrhosis (LCC), and 177 HCC (HCC-C) patients; 232 hepatitis B virus (HBV)-positive patients, including 103 chronic hepatitis (CHB), 56 liver cirrhosis (LCB), and 73 HCC (HCC-B) patients; and 24 healthy controls, were measured using enzyme-linked immunosorbent assay.
Results
The serum DHCR24 Ab levels were significantly higher in patients with CHC than in healthy controls, in LCC than in CHC, and in LCC than in HCC-C (P < 0.0001 for all). The concentration of serum DHCR24 Ab in HCC-B patients showed no significant difference compared to CHB and LCB patients (P = 0.1247). The DHCR24 Ab levels were significantly higher in early HCC-C than CHC or LCC patients and in late HCC-C compared to early HCC-C patients. The sensitivity of the DHCR24 Ab for HCC-C detection (70.6%) was higher than that of alpha-fetoprotein (AFP; 54.8%) and protein induced by vitamin K absence or antagonist-II (PIVKA-II; 42 · 5%). Moreover, DHCR24 was up-regulated in HCV-positive, but not HBV-positive tissues or HBV-negative, HCV-negative HCC specimens.
Conclusions
DHCR24 auto-antibody represents a potential noninvasive biomarker for HCV-related liver disease and may facilitate the diagnosis of PIVKA-II and AFP-negative HCC.
Keywords: 3β-Hydroxysterol Δ24-reductase antibody, Serum autoantibody, Liver cancer, Biomarker, Hepatitis C virus
Highlights
-
•
The DHCR24 Ab level was found to be increased during hepatitis C virus (HCV) infection disease progression.
-
•
The measurement of DHCR24 Ab in the serum may facilitate the diagnosis of PIVKA-II and AFP-negative HCC.
-
•
The measurement of DHCR24 Ab in the serum may improve the detection rate of HCC-C.
-
•
The level of DHCR24 Ab could not distinguish HBV-related-HCC from HBV-related chronic hepatitis or liver cirrhosis.
-
•
TThe DHCR24 was up-regulated in HCV-positive, but not HBV-positive, tissues.
1. Introduction
Hepatitis C virus (HCV) infection has a high prevalence, incidence, and pathogenicity, and may evolve into cirrhosis and hepatocellular carcinoma (HCC) (Castello et al., 2010). HCV infection is considered a chronic liver disease, likely resulting from immune evasion by HCV quasi-species generated from high rates of replication errors (Farazi and DePinho, 2006). Since HCV does not integrate into the host genome, HCV-related HCC (HCC-C) is mainly induced through indirect pathways, including chronic inflammation, cirrhosis, cell death, and proliferation (Lorusso and Ruegg, 2008). A hallmark of HCV infection is perturbed lipid metabolism; HCV exploits host lipids for replication and further infection. The 3β-hydroxysterol Δ24-reductase (DHCR24) protein catalyzes the conversion of desmosterol to cholesterol; Bae and Paik, 1997 this reaction appears to be remarkable in HCV infection (Schaefer and Chung, 2013). Previously, we found that HCV up-regulated DHCR24 expression in human hepatoblastoma-derived RzM6-LC cells and that DHCR24 overexpression impaired p53 activity by suppressing acetylation and increasing interactions with the MDM2 proto-oncogene. In turn, this suppressed the hydrogen peroxide-induced apoptotic response in hepatocytes (Nishimura et al., 2009, Tsukiyama-Kohara, 2012, Saito et al., 2012).
The development of novel biomarkers and surveillance strategies in high-risk populations will permit earlier discovery of HCC and improve the survival outcomes. Currently, the most commonly used biomarker for early-stage HCC is alpha-fetoprotein (AFP) (Zhang et al., 2004). However, the AFP levels remain normal in 15–20% of advanced-stage HCC patients, and only 10–20% of early-stage HCC patients show abnormal AFP values (Monsour et al., 2013). Therefore, additional HCC biomarkers, including protein induced by vitamin K absence or antagonist-II (PIVKA-II; des-gamma-carboxy-prothrombin), have been developed (Makuuchi et al., 2008). Nonetheless, the need for novel biomarkers for early detection of HCC remains an important issue. Moreover, no biomarker corresponding to the disease progression of HCV-infected diseases, such as chronic hepatitis, cirrhosis, and HCC, has been established. Accordingly, this study aimed to evaluate the levels of serum DHCR24 auto-antibody (DHCR24 Ab) in a broad spectrum of chronic hepatitis diseases.
2. Materials and Methods
2.1. Subjects
The subjects were prospectively enrolled at the Tokyo Metropolitan Komagome Hospital, Showa University Fujigaoka Hospital, and Kanazawa University Hospital, Japan. This study was approved by the ethics committee of each hospital and conducted in accordance with the Helsinki Declaration. All patients provided informed consent.
Frozen liver samples (cancerous and noncancerous) were obtained from five HCC-HCV, five HCC-HBV and five HCC-nonBnonC (NBNC) patients that were obtained from 15 HCC patients at the Liver Unit of the Tokyo Metropolitan Komagome Hospital, Japan and used for Western blotting and immunohistochemistry.
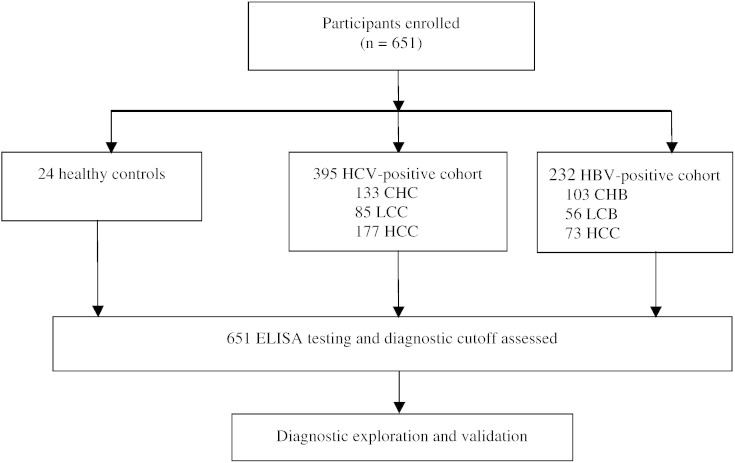
Six hundred fifty-one serum samples, collected from September 2007 to November 2014, were obtained from 395 HCV-positive patients, including 133 moderate chronic hepatitis C (CHC), 85 liver cirrhosis (LCC), and 177 HCC (HCC-C) patients; 232 HBV patients, including 103 chronic HBV (CHB), 56 liver cirrhosis (LCB), and 73 HCC (HCC-B) patients; and 24 healthy controls (Table 1).
Table 1.
Baseline demographic and disease characteristics.
| Healthy controls (N = 24) | Hepatitis C virus group (N = 395) | Hepatitis B virus group (N = 232) | |
|---|---|---|---|
| Mean age ± SD, years | 45.08 ± 16.84 | 68.32 ± 11.40 | 58.31 ± 12.90 |
| Gender (male/female) | 9/15 | 185/210 | 151/81 |
| ALT (IU/L), mean ± SD | 21.50 ± 10.10 | 54.55 ± 44.80 | 63.01 ± 159 |
| AST (IU/L), mean ± SD | 21.92 ± 6.47 | 58.32 ± 36.70 | 79.25 ± 450 |
| AFP (ng/mL), mean ± SD | 4.50 ± 3.23 | 233.31 ± 18823 | 1850 ± 180900 |
| PIVKA-II (mIU/mL), mean ± SD | – | 5991.35 ± 75342.65 | 471.60 ± 3736.78 |
| Clinical status | |||
| Moderate chronic hepatitis (n) | – | 133 | 103 |
| Liver cirrhosis (n) | – | 85 | 56 |
| HCC (n) | – | 177 | 73 |
Abbreviations: SD, standard deviation; ALT, alanine aminotransferase; AST, aspartate aminotransferase; AFP, alpha-fetoprotein; PIVKA-II, protein induced by vitamin K absence or antagonist-II; HCC, hepatocellular carcinoma.
Chronic HCV infection was defined as detectable serum anti-HCV antibodies and HCV RNA. Liver cirrhosis was by presence of ascites and/or gastroesophageal varices and defined by the aspartate transaminase (AST) to platelet ratio index (APRI) and Fibrosis-4 index. APRI (cutoff value ≧ 1.00, sensitivity 89%, specificity 74%, PPV38%, NPV 98%) was calculated as follows: (AST [IU / L] / upper normal limit) × 100/platelets (109/L) (Wai et al., 2003). The Fibrosis-4 index (cutoff value ≧ 1.45, sensitivity 70%, specificity 74%, PPV42%, NPV 90%) was calculated by: age (years) × AST (IU / L) / (platelets [109/L] × [alanine aminotransferase (ALT; IU/L)]½) (Sterling et al., 2006). HCC was diagnosed by ultrasonography and computed tomography and confirmed by liver biopsy, and classified into early and late HCCs according to the pathological criteria of the international consensus group for hepatocellular neoplasia. Early-stage HCC was defined as a single lesion between 2 and 5 cm, or less than or equal to three lesions each ≤ 3 cm. Late-stage HCC was defined by a single lesion > 5 cm, or greater than three lesions (International Consensus Group for Hepatocellular Neoplasia and The International Consensus Group for Hepatocellular N, 2009).
The diagnosis of chronic HBV infection was confirmed by the presence of HBV surface antigen in the blood for greater than six months, HBV core antibodies, and HBV-DNA detection by real-time polymerase chain reaction. The healthy controls had no medical history of liver disease, were negative for viral hepatitis markers, and had normal ALT and AST levels. Table 1 summarizes the main characteristics of the study patients.
2.2. Western Blotting
Fifteen frozen liver tissue specimens were homogenized on ice using a type-A glass homogenizer (Wheaton Science Products, Millville, NJ, USA) in RIPA buffer (1% sodium dodecyl sulfate, 0.5% NP40, 0.15 M NaCl, 10 mM Tris pH 7 · 4, 5 mM ethylenediaminetetraacetic acid [EDTA], and 1 mM dithiothreitol). Proteins were transferred to Immobilon P polyvinylidene fluoride membranes (Merck Millipore, Jaffrey, NH, USA). After blocking with 5% Block Ace™ powder (Dainippon Pharmaceutical Co., Osaka, Japan) in TBST (0.1% Tween 20, 20 mM Tris-Cl pH 7.6, and 137 mM NaCl) at room temperature for 2 h, the membranes were incubated for 2 h with 1-μg/mL of primary antibody in TBST containing 1% Block Ace. After washing, the membranes were incubated with 1:2000 horseradish peroxidase-conjugated rabbit anti-mouse antibody (Dako, Glostrup, Denmark) in TBST containing 1% Block Ace. Western blotting was performed using DHCR24 monoclonal antibody (MoAb) 2-152a6 as the primary antibody. Actin (polyclonal antibody; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used for normalization.
2.3. Immunohistochemistry of Liver Tissues
Frozen sections of cancerous and noncancerous liver tissues from HCC patients were prepared in optimal cutting temperature compound (Ted Pella Inc., Redding, CA, USA), placed on glass slides, thawed, washed with phosphate-buffered saline (PBS), and fixed with 1% paraformaldehyde for 10 min in PBS. After blocking for 1 h with PBS containing 1% bovine serum albumin and 1 mM EDTA, the slides were washed with PBS and incubated overnight at 4 °C with the primary monoclonal Abs (5 μg/mL). Next, the slides were washed with PBS thrice and incubated for 30 min with 1:1000 Alexa 488-conjugated goat anti-mouse immunoglobulin G (IgG) (Fab′)2 fragment or Alexa 568-conjugated goat anti-rabbit IgG (Fab′)2 fragment (Molecular Probes, Eugene, OR, USA) in PBS containing 0.05% Tween-20. Subsequently, the slides were washed thrice with PBS and cover-slipped using Vector-shield (Vector Laboratories Inc., Burlingame, CA, USA) containing 10-μg/mL of Topro-3 (Molecular Probes). The slides were observed using conventional fluorescent or confocal microscopy (510 meta; Zeiss, Oberkochen, Germany).
2.4. Purification of DHCR24 Protein
DHCR24 protein was purified from HuH-7 hepatocellular carcinoma cell lysate. HuH-7 cells (1 × 108 cells) were solubilized in 1 mL of lysis buffer C (1% Triton X-100, 20 mM HEPES pH 7 · 5, 1 mM EDTA, 1 mM dithiothreitol, and 1 mM diisopropylfluorophosphate), sonicated, and centrifuged at 20,400 g for 30 min. The supernatant was incubated at 4 °C for 4 h with 500 μg of DHCR24 MoAb 2-152a IgG immobilized on a HiTrap N-hydroxysuccinimide-activated high-performance column (Amersham Bioscience, Amersham, UK) according to the manufacturer's protocol. The resin was washed with buffer C, and bound proteins were eluted with 0.2 M sodium bicarbonate (pH 8 · 3) containing 0.5 M NaCl, and immediately neutralized with an equal amount of ice-cold 1 M Tris pH 8 · 5.
2.5. Enzyme-linked Immunosorbent Assay (ELISA)
ELISA was performed in flat-bottom polystyrene plates (Greiner Bio-One GmbH, Frickenhausen, Germany) precoated with 50 μl of purified antigen diluted to 5 μg/mL in 0.05 M sodium carbonate Na2CO3 pH 9.0, overnight at 4 °C. Next, the plates were washed with PBS 0.1% Tween 20 and incubated in blocking buffer (1% Block Ace/PBS-0.1% Tween 20) at 37 °C for 1–2 h. After washing, the samples (diluted with blocking buffer to 1:100), positive controls (mouse polyclonal anti-DHCR24 sera diluted to 0.5, 1, and 2 μg/mL), and blanks (assays without sample) were added and the plates were incubated at 37 °C for 1–2 h, followed by washing. Subsequently, the secondary Abs (1:1000; peroxidase conjugate polyclonal rabbit anti-human IgG [Dako] for samples, and peroxidase conjugate polyclonal rabbit anti-Mouse IgG [Dako] for positive controls) were added and incubated at 37 °C for 1 h. Finally, the plates were washed to remove the secondary Abs, tetramethylbenzidine/H2O2-TMB (Bio-Rad, Hercules, CA, USA) was added, and the plates were incubated at 37 °C for 30 min. The reaction was stopped with 1 N of H2SO4. The absorbance values were read using a plate spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) at 450 nm. The assay was done in blind and interpreted by tree researchers. In addition, the assay was repeated for approximately 3% of the total samples, and no discrepancy was observed.
2.6. Statistical Analysis
Descriptive data are presented as numbers and mean ± SD, as appropriate. Differences in continuous variables were compared using the Student's t-test, Mann–Whitney U test, and one-way analysis of variance (ANOVA). For categorical variables, the Chi-square test was used. All P-values were two-sided and P < 0.05 was considered significant. The area under the receiver operating characteristic curves (AUC of ROC) and their 95% confidence intervals (CIs), were used to evaluate the diagnostic value of the DHCR24 Ab and to test the hypothesis that the AUC is 0.5. Additionally, positive and negative predictive values and their 95% confidence intervals were calculated. All correlations were assessed using Spearman's correlation test. The survival rates were estimated using Kaplan–Meier curves and compared using two-sided log-rank tests. All statistical analyses were performed using GraphPad PRISM version 6.0e (GraphPad Software, San Diego, CA, USA) or MedCalc statistical software.
3. Results
3.1. Setup of ELISA
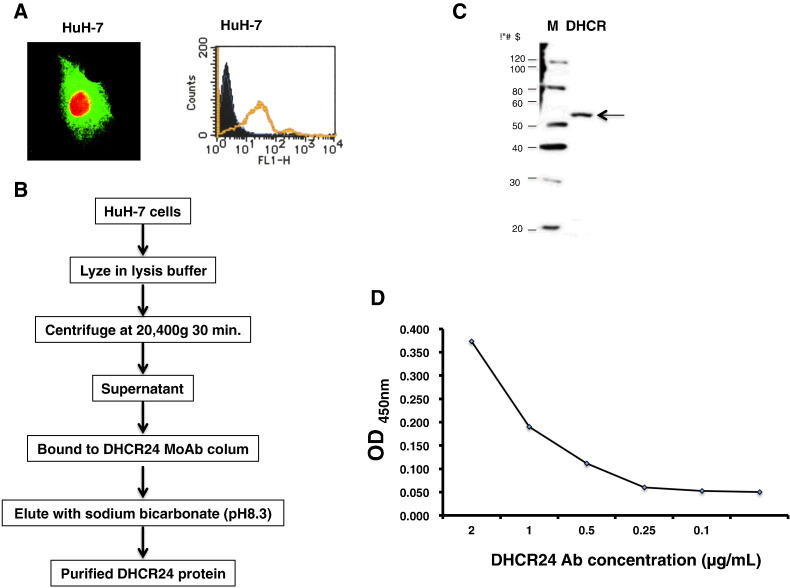
We recruited 651 participants from September 2007 to November 2014 (Fig. 1). The main clinical and demographic characteristics of the study population are summarized in Table 1. We screened the DHCR24 Ab levels in the patient sera using ELISA. The antigen for ELISA was purified from HuH-7 cells (Fig. 2), as the recombinant DHCR24 expressed in E. coli did not show proper reactivity (data not shown). The concentration of DHCR24 Ab levels was determined using mouse anti-DHCR24 as a standard. The dose-response relationship between DHCR24 Ab concentration and optical density was very good (range, 0–2 μg/mL), and the lowest detectable concentration was 0.05 μg/mL.
Fig. 1.
Study profile.
Fig. 2.
Standard concentration curve (OD450) for DHCR24 Ab. (A) Fluorescence-activated cell sorting analysis of DHCR24 expression in HuH-7 cells. DHCR24 is expressed on the surface of HuH-7 cells and is not secreted. (B) Procedure for purification of DHCR24 protein using a DHCR24 monoclonal Ab (MoAb) 2-152a column. (C) Characterization of DHCR24 protein purified from HuH-7 cells by Western blotting (arrow). The left lane contains a molecular weight marker. (D) Reactivity of MoAb 2-152a with purified DHCR24 antigen from HuH-7 cells.
3.2. DHCR24 Ab Levels in HCV-infected Patients
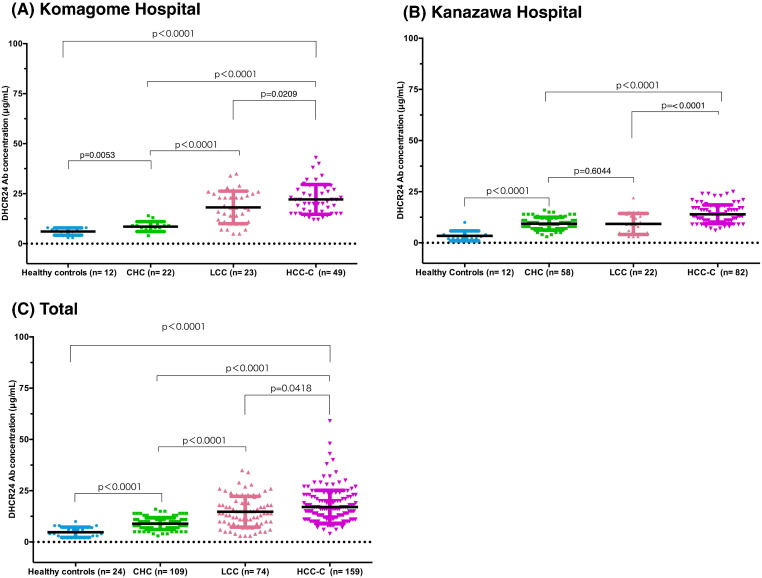
The concentration of DHCR24 Ab was significantly higher in CHC patients compared to in healthy subjects (P < 0.0001), but significantly lower compared to in HCC-C patients (P < 0.0001) in all hospitals (Fig. 3A, B, C, Supplementary Fig. 1). Overall, the DHCR24 Ab level was significantly higher in HCC-C patients (mean: 16.46 ± 0.63 μg/mL) compared to CHC (8.69 ± 0.25 μg/mL, P < 0.0001) and LCC (13.82 ± 0.81 μg/mL, P = 0.0418) patients and healthy controls (4.75 ± 0.51 μg/mL, P < 0.0001; Fig. 3C). One-way ANOVA revealed significant differences between HCC-C and LCC and CHC patients (P < 0.0001).
Fig. 3.
Serum DHCR24 Ab relative concentrations in hepatitis C virus (HCV)-infected patients with chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma, and in healthy controls. Serum levels of DHCR24 Ab in hepatitis C patients and healthy controls in (A) Komagome hospital, (B) Kanazawa hospital, and (C) Plotted serum levels of DHCR24 Ab in hepatitis C patients and healthy controls (all hospitals). Black horizontal lines indicate means, and the error bars indicate the standard deviations. CHC, chronic hepatitis C virus infection; LCC, liver cirrhosis with HCV infection; HCC-C, hepatocellular carcinoma with HCV infection.
ROC curves were plotted to identify the optimal cutoff values to distinguish HCC from HCV chronic carriers (Supplementary Fig. 2). The sensitivity and specificity of serum DHCR24 Ab for the diagnosis of HCC-C were 70.06% (95% CI: 62.73–76.70%) and 84.21% (95% CI: 76.88–89.95%), respectively (cutoff, 11.5 μg/mL). The corresponding values for AFP were 53.49% (95% CI: 45.74–61.12%) and 97.66% (95% CI: 93.30–99.51%), respectively (cutoff, 20 ng/mL), and those of PIVKA-II Ab were 42.51% (95% CI: 34.91–50.39%) and 100% (95% CI: 96.38–100%), respectively (cutoff, 40 mIU/mL). Additionally, a comprehensive assessment of the diagnostic accuracy of DHCR24 Ab was performed for HCC-C (Table 2).
Table 2.
Assessment of the diagnostic accuracy of DHCR24 Ab in HCV-related HCC.
| HCC-C vs. healthy | HCC-C vs. CHC | HCC-C vs. LCC | |
|---|---|---|---|
| Sensitivity (%), (95% CI) | 70.06 (62.73–76.70) | 70.06 (62.73–76.70) | 70.06 (62.73–76.70) |
| Specificity (%), (95% CI) | 100 (85.62–100) | 84.21 (76.88–89.95) | 42.35 (31.70–53.55) |
| Positive likelihood ratio | 50.44 | 4.44 | 1.22 |
| Negative likelihood ratio | 0.30 | 0.36 | 0.71 |
| Positive predictive value (%), (95% CI) | 100 (97.04–100) | 94.66 (91–98.32) | 71.68 (64.34–78.25) |
| False discovery rate (%) | 0 | 5.34 | 28.32 |
| Negative predictive value (%), (95% CI) | 31.17 | 41.29 (37.47–45.11) | 40.45 (30.17–51.38) |
| False omission rate (%) | 68.83 | 58.71 | 59.55 |
| Area under the ROC curve, (95% CI) | 0.98 (0.96–1) | 0.86 (0.82–0.91) | 0.60 (0.51–0.67) |
Abbreviations: DHCR24, 3β-hydroxysterol Δ24-reductase antibody; HCV, hepatitis C virus; HCC-C, HCV-related hepatocellular carcinoma; CHC, HCV-related chronic hepatitis; LCC, HCV-related liver cirrhosis; CI, confidence interval; ROC, receiver operating characteristics.
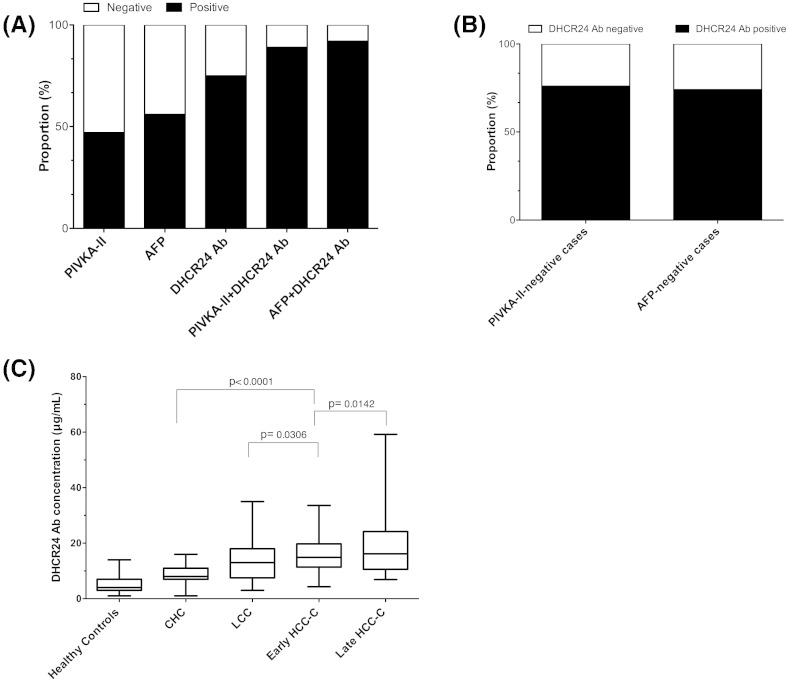
DHCR24 Ab showed an AUC of 0.86 (95% CI: 0.82–0.91) in distinguishing HCC-C from CHC patients, which was compared with AFP (0.84, 95% CI: 0.82–0.91) and PIVKA-II (0.78, 95% CI: 0.73–0.84) (Supplementary Fig. 2). A greater proportion of patients with HCC-C were positive for DHCR24 Ab (125/177, 71%) than for AFP (93/172, 54%) and PIVKA-II (73/167, 44%) (P < 0.0001; Fig. 4A). Combined serum DHCR24 Ab and PIVKA-II or AFP levels increased the positive rates of HCC cases to 87% and 88%, respectively (Fig. 4A). Furthermore, 46% (79/172) of patients were considered AFP-negative (< 20 ng/mL). Among them, the serum DHCR24 Ab levels were elevated in 73.4% (58/79) (Fig. 4B). Moreover, 56.3% (94/167) were considered PIVKA-II-negative (< 40 mIU/mL). Among them, the serum DHCR24 Ab levels were elevated in 75.5% (71/94) (Fig. 4B). Additionally, DHCR24 Ab could significantly differentiate between early HCC and cirrhosis (P = 0.0306) and early HCC-and late HCC (P = 0.0142) in chronic hepatitis C patients (Fig. 4C).
Fig. 4.
The performance of serum markers in the discrimination between hepatitis C virus-related hepatocellular carcinoma (HCC-C) and chronic hepatitis (CHC). (A) Rates of positive results for combinations of PIVKA-II, AFP, and DHCR24 Ab in patients with HCC-C. (B) Performance of DHCR24 AB in PIVKAII-negative and AFP-negative cases. (C) Serum levels of DHCR24 Ab among healthy, chronic hepatitis C, LCC, and early and late HCC-C patients. AUC, area under the curve; CI, confidence interval.
We further evaluated the performance of DHCR24 Ab in discriminating CHC vs. LCC; ROC curves showed an AUC of 0.74 (95% CI: 0.66–0.83), which was similar to that of AFP (0.68, 95% CI: 0.66–0.77) and significantly better than that of PIVKA-II (0.56, 95% CI: 0.43–0.62) (Supplementary Fig. 3).
Spearman correlation analyses between DHCR24 Ab and AFP or PIVKA-II levels showed no clear correlations (rs = − 0.15, 95% CI: − 0.30–0.01, P = 0.0668, R2 = 0.0076; and rs = 0.08, 95% CI: 0.24–0.083, P = 0.3149, R2 = 0.0161, respectively (Supplementary Fig. 4).
3.3. DHCR24 Ab Levels in HBV-infected Patients
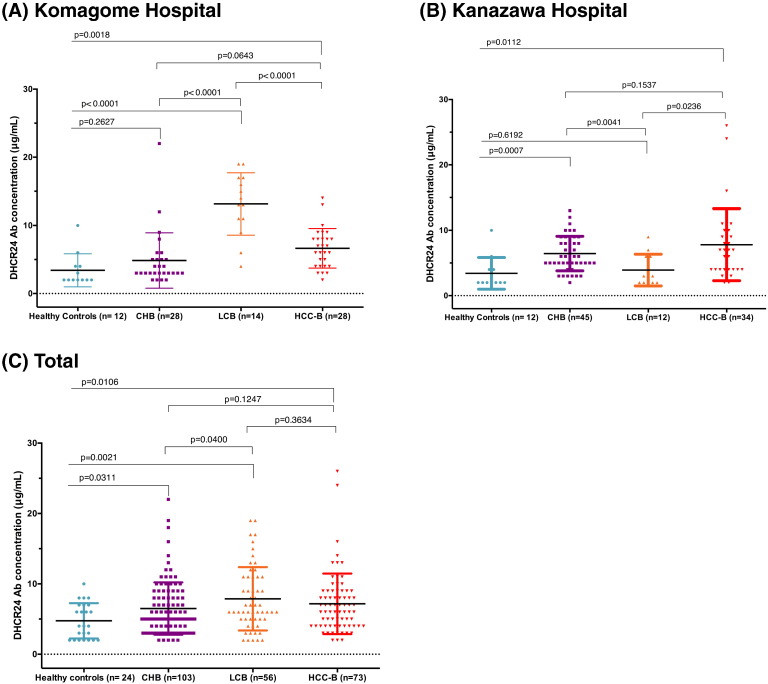
Next, we investigated the serum levels of DHCR24 Ab from patients with CHB, LCB, and HCC-B (Fig. 5, Supplementary Fig. 5). Stratification analysis by hospital showed that the DHCR24 Ab levels were higher in CHB patients compared to in healthy controls (Fig. 5A–C). Overall, the concentration of DHCR24 Ab in HCC-B (mean: 7.16 ± 0.50 μg/mL) showed no significant differences compared to CHB (6.49 ± 0.37 μg/mL) and LCB patients (7.87 ± 0.60 μg/mL) (P = 0.1247). However, it was significantly higher in patients with chronic hepatitis B (CHB, LCB, or HCC-B) than in healthy controls (P < 0.05) (Fig. 5C). The AUC for distinguishing between HCC-B and CHB for DHCR24 Ab (0.50, 95% CI: 0.41–0.60) was not superior to that of PIVKA-II (0.73, 95% CI: 0.64–0.82) or AFP (0.77, 95% CI: 0.71–0.83) (Supplementary Fig. 6), indicating that DHCR24 Ab is not a good biomarker for HCC-B.
Fig. 5.
Serum levels of DHCR24 Ab in hepatitis B virus (HBV)-infected patients with chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma, and in healthy controls. Serum levels of DHCR24 Ab in hepatitis B patients and healthy controls in (A) Komagome Hospital and (B) Kanazawa hospital. (C) Plotted serum levels of DHCR24 Ab in hepatitis B patients and healthy controls (all hospitals). Black horizontal lines indicate means and error bars indicate standard deviations. CHB, chronic hepatitis B virus infection; LCB, liver cirrhosis with HBV infection; HCC-B, hepatocellular carcinoma with HBV infection.
3.4. DHCR24 is Differentially Expressed in Liver Cancer Tissues
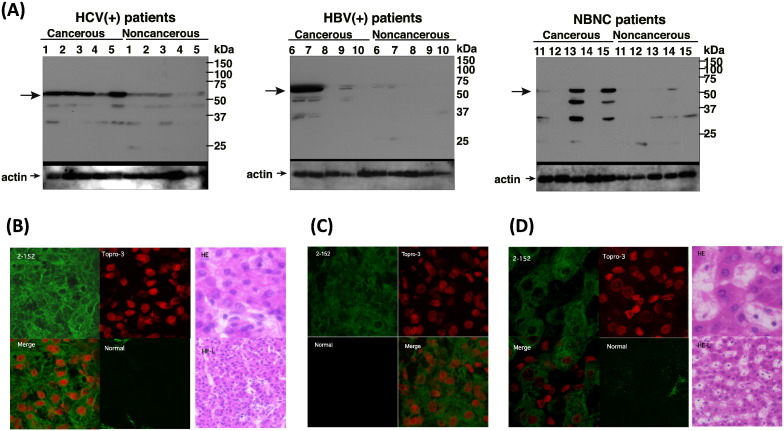
To determine whether HCV infection induces DHCR24 protein expression in HCC, 15 primary human HCC tissues and adjacent non-tumor tissues were subjected to Western blot analysis using DHCR24 monoclonal antibody 2-152a. DHCR24 was overexpressed in all HCV-positive tumor tissues compared to the adjacent non-tumor tissues (Fig. 6A), as well as in two out of five HBV-related, or HBV-negative, HCV-negative HCC specimens (Fig. 6A). Furthermore, immunohistochemical analyses revealed that the DHCR24 protein expression was increased in cancerous HCV-tissues (Fig. 6B) and reduced in cancerous HBV-positive (Fig. 5C) and HBV-negative, HCV-negative-tissues (Fig. 6D). Taken together, these data indicate that DHCR24 is more frequently up-regulated in HCV-cancerous than in HBV-cancerous tissues.
Fig. 6.
The DHCR24 expression is up-regulated in hepatitis C virus (HCV)-tumor tissues, and association between DHCR24 Ab levels with tumor progression, and transcatheter arterial chemoembolization (TACE). (A) Western blot showing DHCR24 expression in cancerous and non-cancerous regions in the livers of 10 hepatocellular (HCC) patients. Five patients were infected with HCV (HCV +), five were infected with hepatitis B virus (HBV +), and five were not infected with HBV and HCV (NBNC). Immunofluorescence and histological images of (B) HCV-infected cancerous tissues, (C) HBV-infected cancerous tissues, and (D) NBNC tissues obtained using immunohistochemistry. DHCR24 protein expression was detected using the 2-152a monoclonal Ab. Topro-3 was used for counterstaining of the nuclei. HE, hematoxylin and eosin.
3.5. Relationship Between DHCR24 Ab Levels and Clinicopathologic Features in HCC-C Patients
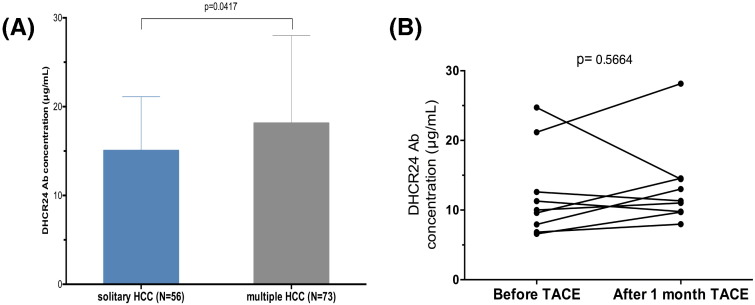
Solitary HCC is thought to be de novo occurrence and multiple HCC includes de novo and intrahepatic metastases. We compared the DHCR24 Ab levels between in solitary HCC-C and multiples HCC. In fact, the concentration of DHCR24 Ab was significantly higher among patients with multiple HCC (18.15 ± 1.15 μg/mL) than in those with solitary HCC (15.07 ± 0.81 μg/mL) (P = 0.0417; Fig. 7A). Moreover, the DHCR24 Ab levels were compared before and after treatment in patients who underwent transcatheter arterial chemoembolization for HCC (n = 8). However, the DHCR24 Ab levels did not differ before and after the treatment (P = 0.5664; Fig. 7B).
Fig. 7.
Association between 3β-hydroxysterol Δ24-reductase antibody (DHCR24 Ab) levels with tumor progression, and transcatheter arterial chemoembolization (TACE). (A) DHCR24 Ab level comparison ` solitary HCC and multiple HCCs in hepatitis C virus (HCV)-related hepatocellular carcinoma (HCC-C) patients. (B) Changes in the DHCR24 levels before and after TACE of HCC-C patients.
4. Discussion
In the serum, Abs are stable and the Ab response is enduring (Heo et al., 2012), and overexpressed, mutated, misfolded, or aberrantly degraded proteins can become immunogenic, thus resulting in auto-Ab production; consequently, the detection of these antibodies in patient sera can be exploited for prognostic and/or diagnostic purposes.
In this study, we found that serum DHCR24 Ab levels correlated with HCV-mediated chronic hepatitis, cirrhosis, and HCC, but not with HBV-related diseases, suggesting that the concentration of serum DHCR24 Abs may be useful for the diagnosis and/or prognosis in HCV-positive patients. Moreover, the sensitivity of the DHCR24 Ab in discriminating CHC from HCC-C patients was superior to serum AFP and PIVKA-II levels, and high DHCR24 Ab levels demonstrated a high positive predictive value for HCC-C. Additionally, a combination of DHCR24 Ab with AFP or PIVKA-II further increased the detection rates of HCC in HCV-positive patients. Conversely, in chronic HBV infection, the DHCR24 Ab levels did not differ between CHB, LCB, and HCC-B (P = 0.1247).
Previously, we have shown that DHCR24 is overexpressed in HCV-infected cells in vitro and in vivo, and that HCV silencing by siRNA down-regulated DHCR24 expression (Nishimura et al., 2009, Saito et al., 2012, Takano et al., 2011). However, HBV infection had no significant effect on DHCR24 expression (Makuuchi et al., 2008). DHCR24 up-regulation in HCV infection may be due to increased Sp1 phosphorylation by ataxia telangiectasia mutated as a result of HCV-induced oxidative stress, which promotes transcriptional activation of DHCR24 (Tsukiyama-Kohara, 2012). Thus, chronic HCV infection could induce the augmentation of DHCR24. Also, this augmentation of DHCR24 expression by HCV suppresses p53 activity by blocking nuclear p53 acetylation and increasing the interaction between p53 and MDM2 in the cytoplasm, potentially mediated by inhibition of p53 degradation (Tsukiyama-Kohara, 2012). Consequently, impaired apoptotic signaling upon p53 activity suppression may facilitate hepatocarcinogenesis. Moreover, DHCR24 has been shown to inhibit caspase-3 activation, one of the major proteins involved in apoptosis, suggesting a role of DHCR24 in the regulation of cell survival and death (Greeve et al., 2000, Benvenuti et al., 2008). Additionally, we found that the MoAb 2-152a clone can bind DHCR24 and suppress HCV replication (Satoh et al., 2011), suggesting that the DHCR24 Ab has antiviral effects, and that high levels of DHCR24 Ab in HCC patients may lead to lower HCV RNA levels in the cancerous tissues. DHCR24 is reportedly more abundantly expressed on the surface of liver cancer cells than normal hepatocytes Satoh et al., 2011. Accordingly, Tanaka et al. observed that non-cancerous tissues of patients with HCC and HCV mono-infection displayed virus levels 10- to 1000-fold higher than cancerous tissues, and this may be one of the reasons for the higher amount of DHCR24 Ab seen in HCC patients (Tanaka et al., 2004).
The DHCR24 gene has been mapped to chromosome 1p32.3, a region commonly amplified in HCC and which may contribute to carcinogenesis (Yuan et al., 2003, Endo et al., 2009). Recently, two somatic mutations, Q88L and R311H, were identified in the DHCR24 gene in Korean HCC patients (Ahn et al., 2014). Further, overexpression of DHCR24 is a hallmark of prostate cancer, with high levels observed in low-grade prostate cancer, but diminishing as the cancer progresses (Bonaccorsi et al., 2008), and it is also overexpressed during non-muscle-invasive urothelial carcinoma progression (Lee et al., 2014).
In conclusion, DHCR24 Ab levels might represent a novel marker for advanced liver disease. The increase in DHCR24 Abs from moderate chronic hepatitis to advanced-stage disease provides a potential novel diagnostic system for early-stage disease progression and suggests that serum DHCR24 Ab elevation may reflect the progression of autoimmunity-related inflammation to HCC in HCV-positive patients. However, further studies are required to clarify the mechanism of serum DHCR24 Ab induction during chronic HCV infection.
5. Conclusions
A thorough Medline and PubMed search without date restrictions, for original research articles, using the search terms “DHCR24 antibody OR Seladin-1 antibody AND (cancer OR tumor OR neoplasm) AND diagnosis”, and “DHCR24 Ab OR Seladin-1 Ab AND (hepatocellular carcinoma OR liver cancer)”. However, we found no studies that had assessed the diagnostic relevance of serum 3β-hydroxysterol Δ24-reductase antibody (DHCR24 Ab) in hepatocellular carcinoma (HCC) or other human cancers.
Herein, for the first time, we measured and comprehensively assessed the diagnostic accuracy of DHCR24 Ab in HCC in a large-scale, multicenter investigation. The DHCR24 Ab level was found to be increased during hepatitis C virus (HCV) infection disease progression. The sensitivity and specificity of the DHCR24 Ab were high, and the predictive values and likelihood ratios were satisfactory for the diagnosis of HCV-related-HCC and in patients with alpha-fetoprotein (AFP)-negative or protein induced by vitamin K absence or antagonist-II (PIVKA-II)-negative status. Therefore, measurement of DHCR24 Ab in the serum may facilitate the diagnosis of PIVKA-II and AFP-negative HCC and improve the detection rate of HCC-C. However, the test could not distinguish hepatitis B virus (HBV)-related-HCC from HBV-related chronic hepatitis or liver cirrhosis, and we found that DHCR24 was up-regulated in HCV-positive, but not HBV-positive, tissues.
Our data indicate that the DHCR24 Ab shows potential as a prognostic factor for HCV chronic infection and reflect the autoimmune response, and should be investigated further.
Conflicts of Interest
Authors have no conflict of interest.
Author Contributions
S.E., T.N., M.K., and K.T.-K. designed and performed experiments, K.K., K.I., H.S., and S.K., collected clinical samples and validated data, T.H. performed pathological study, and S.E. and K.T.-K. wrote the manuscript.
Funding
This work was supported by grants from the Ministry of Health Science H24-B-014, H25-009 and Welfare and the Ministry of Education, Science and Culture, Japan, 23590547.
The funders of the study had no role in the study design, data collection, analysis, interpretation, or writing of the paper. All authors had access to the raw data.
Acknowledgment
The authors acknowledge Kousuke Tanaka and Yuri Kasama for their technical support. Sayeh Ezzikouri is supported by a Japan Society for the Promotion of Science (JSPS) Fellowship for Foreign Researchers.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.04.007.
Contributor Information
Michinori Kohara, Email: kohara-mc@igakuken.or.jp.
Kyoko Tsukiyama-Kohara, Email: kkohara@vet.kagoshima-u.ac.jp.
Appendix A. Supplementary data
Supplementary figures.
References
- Ahn S.M., Jang S.J., Shim J.H. A genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60(6):1972–1982. doi: 10.1002/hep.27198. [DOI] [PubMed] [Google Scholar]
- Bae S.H., Paik Y.K. Cholesterol biosynthesis from lanosterol: development of a novel assay method and characterization of rat liver microsomal lanosterol delta 24-reductase. Biochem. J. 1997;326(Pt 2):609–616. doi: 10.1042/bj3260609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuti S., Luciani P., Cellai I. Thyroid hormones promote cell differentiation and up-regulate the expression of the seladin-1 gene in in vitro models of human neuronal precursors. J. Endocrinol. 2008;197(2):437–446. doi: 10.1677/JOE-07-0324. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi L., Luciani P., Nesi G. Androgen receptor regulation of the seladin-1/DHCR24 gene: altered expression in prostate cancer. Lab. Invest. 2008;88(10):1049–1056. doi: 10.1038/labinvest.2008.80. [DOI] [PubMed] [Google Scholar]
- Castello G., Scala S., Palmieri G., Curley S.A., Izzo F. HCV-related hepatocellular carcinoma: from chronic inflammation to cancer. Clin. Immunol. 2010;134(3):237–250. doi: 10.1016/j.clim.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Endo M., Yasui K., Nakajima T. Infrequent amplification of JUN in hepatocellular carcinoma. Anticancer Res. 2009;29(12):4989–4994. [PubMed] [Google Scholar]
- Farazi P.A., DePinho R.A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer. 2006;6(9):674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- Greeve I., Hermans-Borgmeyer I., Brellinger C. The human DIMINUTO/DWARF1 homolog seladin-1 confers resistance to Alzheimer's disease-associated neurodegeneration and oxidative stress. J. Neurosci. 2000;20(19):7345–7352. doi: 10.1523/JNEUROSCI.20-19-07345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo C.K., Bahk Y.Y., Cho E.W. Tumor-associated autoantibodies as diagnostic and prognostic biomarkers. BMB Rep. 2012;45(12):677–685. doi: 10.5483/BMBRep.2012.45.12.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Consensus Group for Hepatocellular Neoplasia, The International Consensus Group for Hepatocellular Neoplasia Pathologic diagnosis of early hepatocellular carcinoma: a report of the international consensus group for hepatocellular neoplasia. Hepatology. 2009;49(2):658–664. doi: 10.1002/hep.22709. [DOI] [PubMed] [Google Scholar]
- Lee G.T., Ha Y.S., Jung Y.S. DHCR24 is an independent predictor of progression in patients with non-muscle-invasive urothelial carcinoma, and its functional role is involved in the aggressive properties of urothelial carcinoma cells. Ann. Surg. Oncol. 2014;21 Suppl 4:S538–545. doi: 10.1245/s10434-014-3560-6. [DOI] [PubMed] [Google Scholar]
- Lorusso G., Ruegg C. The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochem. Cell Biol. 2008;130(6):1091–1103. doi: 10.1007/s00418-008-0530-8. [DOI] [PubMed] [Google Scholar]
- Makuuchi M., Kokudo N., Arii S. Development of evidence-based clinical guidelines for the diagnosis and treatment of hepatocellular carcinoma in Japan. Hepatol. Res. 2008;38(1):37–51. doi: 10.1111/j.1872-034X.2007.00216.x. [DOI] [PubMed] [Google Scholar]
- Monsour H.P., Jr., Asham E., Robert S., McFadden R.S., Victor D.W., III, Muthuswamy B., Zaheer I. Hepatocellular carcinoma: the rising tide from east to west—a review of epidemiology, screening and tumor markers. Transl. Cancer Res. 2013;2(6):492–506. [Google Scholar]
- Nishimura T., Kohara M., Izumi K. Hepatitis C virus impairs p53 via persistent overexpression of 3beta-hydroxysterol Delta24-reductase. J. Biol. Chem. 2009;284(52):36442–36452. doi: 10.1074/jbc.M109.043232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Kohara M., Tsukiyama-Kohara K. Hepatitis C virus promotes expression of the 3beta-hydroxysterol delta24-reductase through Sp1. J. Med. Virol. 2012;84(5):733–746. doi: 10.1002/jmv.23250. [DOI] [PubMed] [Google Scholar]
- Satoh M., Saito M., Takano T. Monoclonal antibody 2-152a suppresses hepatitis C virus infection through betaine/GABA transporter-1. J. Infect. Dis. 2011;204(8):1172–1180. doi: 10.1093/infdis/jir501. [DOI] [PubMed] [Google Scholar]
- Schaefer E.A., Chung R.T. HCV and host lipids: an intimate connection. Semin. Liver Dis. 2013;33(4):358–368. doi: 10.1055/s-0033-1358524. [DOI] [PubMed] [Google Scholar]
- Sterling R.K., Lissen E., Clumeck N. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43(6):1317–1325. doi: 10.1002/hep.21178. [DOI] [PubMed] [Google Scholar]
- Takano T., Tsukiyama-Kohara K., Hayashi M. Augmentation of DHCR24 expression by hepatitis C virus infection facilitates viral replication in hepatocytes. J. Hepatol. 2011;55(3):512–521. doi: 10.1016/j.jhep.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Inoue K., Hayashi Y. Virological significance of low-level hepatitis B virus infection in patients with hepatitis C virus associated liver disease. J. Med. Virol. 2004;72(2):223–229. doi: 10.1002/jmv.10566. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K. Role of oxidative stress in hepatocarcinogenesis induced by hepatitis C virus. Int. J. Mol. Sci. 2012;13(11):15271–15278. doi: 10.3390/ijms131115271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai C.T., Greenson J.K., Fontana R.J. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2003;38(2):518–526. doi: 10.1053/jhep.2003.50346. [DOI] [PubMed] [Google Scholar]
- Yuan B.Z., Zhou X., Zimonjic D.B., Durkin M.E., Popescu N.C. Amplification and overexpression of the EMS 1 oncogene, a possible prognostic marker, in human hepatocellular carcinoma. J. Mol. Diagn. 2003;5(1):48–53. doi: 10.1016/S1525-1578(10)60451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.H., Yang B.H., Tang Z.Y. Randomized controlled trial of screening for hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2004;130(7):417–422. doi: 10.1007/s00432-004-0552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.