Summary
Human indoleamine 2,3-dioxygenase (hIDO), a monomeric heme enzyme, catalyzes the oxidative degradation of L-tryptophan (L-Trp) and other indoleamine derivatives. Its activity follows typical Michaelis–Menten behavior only for L-Trp concentrations up to 50 µM; a further increase in the concentration of L-Trp causes a decrease in the activity. This substrate inhibition of hIDO is a result of the binding of a second L-Trp molecule in an inhibitory substrate binding site of the enzyme. The molecular details of the reaction and the inhibition are not yet known. In the following, we summarize the present knowledge about this heme enzyme.
Keywords: hemeproteins, enzyme mechanisms, FTIR spectroscopy, temperature derivative spectroscopy, substrate
INTRODUCTION
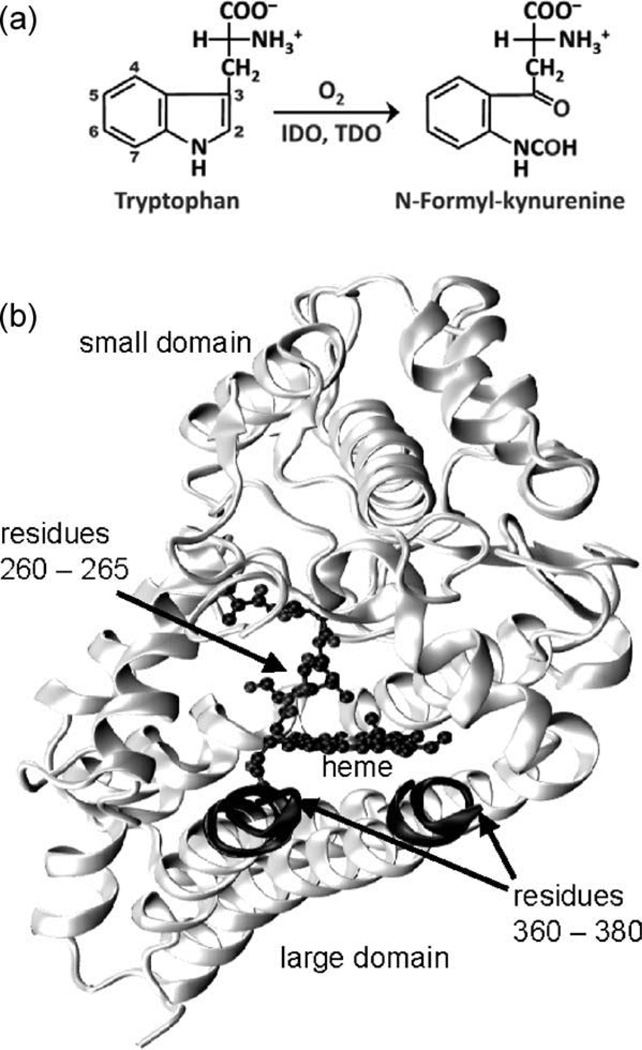
Heme proteins perform a diverse variety of tasks, including electron transfer, storage and transport of small molecules, enzymatic functions and gas sensing, owing to the remarkable chemical properties of the heme prosthetic group (1, 2). Tryptophan 2,3-dioxygenase (TDO) and indoleamine 2,3-dioxygenase (IDO) are both heme enzymes that catalyze the first and rate-limiting step in the oxidative degradation pathway of L-tryptophan (L-Trp), by inserting both atoms of dioxygen (O2) into the C2=C3 bond of the indole ring to yield N-formylkynurenine (NFK) (Fig. 1a) (3, 4). L-Trp is the scarcest amino acid in mammals. The majority of our dietary L-Trp is metabolized by TDO along the kynurenine pathway in the liver (5, 6). It ultimately leads to the biosynthesis of NAD. In contrast to the hepatic TDO, IDO is distributed in all tissues other than liver. It is thought to be responsible for providing immune protection to developing mammalian fetal tissue during gestation by decreasing local L-Trp concentrations and/or by producing cytotoxic metabolites, thereby suppressing the maternal T-cell-based immune response (7). IDO is also overexpressed in most solid tumors, its role in the immune escape of tumors has been proposed (8). An increased level of IDO expression in tumor cells is correlated with a poor prognosis for survival in several cancer types (9, 10) making IDO an appealing drug target for cancer theraphy (11, 12) and calling for the de novo design of inhibitors selective for human indoleamine 2,3-dioxygenase (hIDO).
Figure 1.
(a) The dioxygenase reaction catalyzed by IDO and TDO. (b) Cartoon representation of hIDO.
THE hIDO ACTIVE SITE
The human isoform of IDO, hIDO, is a heme enzyme of ~45 kDa. Its polypeptide chain is folded into two distinct domains (13), with the heme sandwiched in between (Fig. 1b). The large, all-α-helical domain carries the heme prosthetic group, covalently anchored via the proximal histidine, His346. The small domain covers the top of the heme pocket. A loop compromised of residues 250–267 connects the two domains and extends over the sixth coordination site of the heme (Fig. 1b). The indole ring of the substrate is assumed to be packed against the loop through hydrophobic interactions (14). Interestingly, the catalytic site of hIDO is essentially devoid of polar residues, with Ser167 being the only exception. This residue is, however, neither involved in substrate recognition nor in catalytic activity (15). Residues 360–380 form another flexible loop that could not be resolved in the crystal structure. It is thought to become ordered and close off the heme pocket once the substrate binds (16).
Vibrational spectroscopy is known to be a sensitive technique to explore structural details of the active sites in heme proteins that are too subtle for X-ray crystallography (17). In particular, the infrared stretching bands of heme-bound CO have been recognized as excellent reporters for the fine structure and heterogeneity of the active sites due to their exquisite sensitivity to local electric fields created by amino acid residues surrounding the active sites (18–21). The CO stretching bands can be studied selectively by using photolysis-induced difference spectroscopy, which involves measurement of IR spectra before and after photolysis. The difference of the two spectra contains only absorption features due to photodissociation of the ligand from the heme iron. This technique yields IR spectra of heme-bound and photodissociated ligands with exquisite sensitivity and precision. The stretching bands are fine tuned by interactions of the ligand with its environment, which provide a rich source of information on active-site conformations, effects of mutation and ligand docking sites. At ambient temperature, however, the difference spectrum decays rapidly after photolyzing illumination due to fast ligand rebinding to the heme iron. Therefore, we use Fourier transform infrared (FTIR) spectroscopy at cryogenic temperatures, which takes advantage of the fact that kinetic processes in proteins are thermally activated. Ligand rebinding in heme proteins becomes sufficiently slow near 0 K, that IR spectra of photodissociated species can be measured on a steady-state spectrometer. One could argue that processes observed at low temperature may not be relevant for the functioning of a protein at physiological temperature. However, evidence has accumulated over the years that this objection is unfounded (17, 22, 23).
In CO-ligated myoglobin (MbCO), the distal histidine, HisE7, is the main determinant of active site heterogeneity sensed by the CO stretching vibration (18, 20, 21). Two major conformations, called A substates, are observed at neutral pH, associated with the dominant A1 (1,945 cm−1) and the weaker A3 (1,933 cm−1) bands (23–25). In both A substates, a neutral HisE7 imidazole side chain resides in the distal pocket close to the heme iron, placing a positive partial charge from the Nε2 hydrogen near the CO (26, 27). Removal of the HisE7 imidazole from the CO proximity, which can be achieved by lowering the pH, gives rise to an A0 band at ~1,967 cm−1. Upon protonation, the HisE7 imidazole rotates and swings out of the heme pocket (26, 27). The A substates not only differ in structural details but also in their ligand binding properties (28).
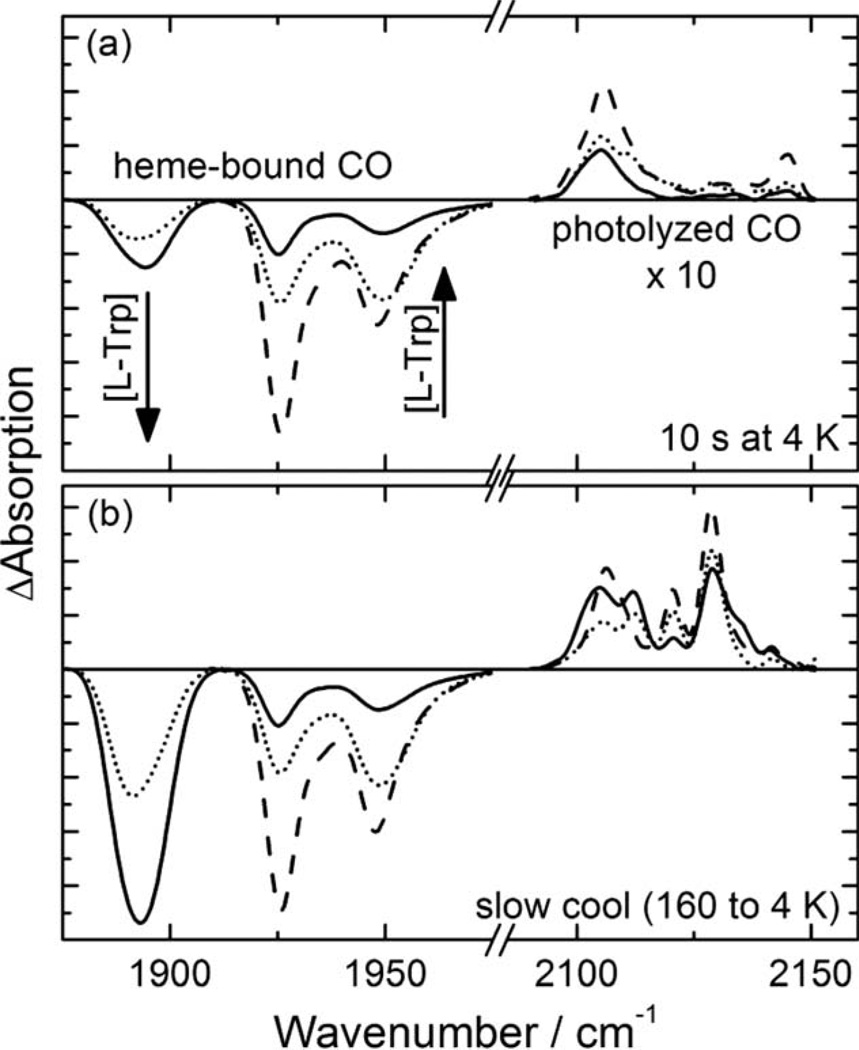
The FTIR spectrum of hIDO-CO determined at 4 K shows three different conformations at neutral pH, A1925, A1937, and A1947 (Fig. 2) (29). Rather low stretching frequencies indicate an interaction between the CO ligand and a positive partial charge (18, 19), consistent with the reported resonance Raman results (30). As there is no polar amino acid at the hIDO active site (13), we suggest that the positive partial charge originates from a backbone amine. In the 3D structure of hIDO-CN, the distance between the ligand N and the backbone amide N of Ala264 is 2.9 Å (13) and thus sufficiently short to establish an interaction. Quantum mechanics (QM)/molecular mechanics (MM) studies have proposed an interaction with Gly265 (31). Both Ala264 and Gly265 are part of the flexible loop that connects the two hIDO domains (13) and, therefore, might adopt slightly different orientations with respect to the bound ligand that result in measurable frequency differences.
Figure 2.
4-K FTIR photolysis difference spectra of substrate-free (dashed lines) and L-Trp-bound (dotted, solid) hIDO-CO determined after (a) 10-sec illumination at 4 K and (b) slow cooling from 160 to 4 K under continuous illumination (rate 0.3 K/min). Stretching bands of heme-bound CO missing after photolysis are plotted with negative amplitude. Photoproduct bands that emerge during photolysis have positive amplitudes.
Increasing concentrations of L-Trp at the active site are reflected in an A state band at 1,895 cm−1 (ASubstrate, AS) that emerges at the expense of the A bands of substrate-free hIDO-CO, in the following subsumed as ASF (Fig. 2) (29). The frequency of this band is again consistent with the resonance Raman result (30). Its shape is slightly asymmetric and might be a superposition of two bands with very similar peak positions. The strong red shift suggests that the L-Trp substrate has established a hydrogen bond to the heme-bound CO. A combination of classical molecular dynamics and hybrid quantum-classical (QM/MM) methodologies suggests that L-Trp can be accommodated in hIDO-CO in two conformations (31). In the first conformation, the indoleamine group of L-Trp is close to the CO, but the perpendicular positions of CO and L-Trp with respect to the heme plane do not allow a proper orientation for a hydrogen bonding interaction. In the second conformation, the L-Trp indole moiety is oriented almost parallel to the heme plane, and a hydrogen bond is found between the indole-NH and the ligand. This conformation might be represented by the AS band.
Ligand Migration in hIDO
FTIR-Temperature Derivative Spectroscopy (TDS) has found wide-spread application in the study of CO migration and binding in heme proteins (17, 32–35). The technique is a convenient method for the investigation of thermally activated rate processes that are characterized by distributed enthalpy barriers. TDS is a relaxation method in which a nonequilibrium state is created by perturbation of a sample at a temperature at which approach to equilibrium is slow. In heme protein studies, the sample is perturbed by ligand photolysis. Subsequently, the temperature, T, is ramped up linearly in time in the dark at a constant rate. During the temperature ramp, IR spectra are taken continuously (1 spectrum/K). They reveal the response of the sample after the perturbation, including ligand rebinding to the heme iron. At cryogenic temperatures (T < 200 K in our cryosolvent), rebinding is characterized by a static distribution of rebinding enthalpy barriers due to the absence of large-scale conformational motions (36–38). Protein molecules with small barriers rebind their ligand first, and the continuous increase in temperature causes molecules to rebind sequentially with respect to their barrier heights. TDS is a so-called “rate window method,” which allows all processes to occur with a characteristic time scale, which is ~100 s at the heating rate used in our experiments. The data are presented as two-dimensional contour plots, in which the absorbance changes between successive spectra are plotted versus temperature and wave number. Contours are usually spaced logarithmically, which emphasizes weak features, and solid and dotted lines depict gain and loss of absorption with temperature, respectively.
We use especially designed illumination protocols to selectively populate transient docking sites in heme proteins. To measure rebinding from states that can be populated at the lowest temperatures (“site B,” “primary docking site”), the sample is illuminated for 1–10 sec at 4 K. To sample recombination from sites with higher recombination barriers (“remote” or “secondary docking sites,” “sites C and D”), the slow cool protocol is applied. Here, the laser light is kept on while the sample is cooled from 160 to 4 K.
In a typical oxygen-storage protein such as Mb, transient ligand docking sites inside the protein matrix modulate the ligand association and dissociation kinetics (23, 39–42). The so-called primary docking site B located close to the binding site (A site) plays a key role, as it is the only site through which bond formation can occur. As long as this site is available, ligands entering the protein will be efficiently captured. In their study of oxygen binding in a large variety of mutant Mbs, Scott et al. (43) suggested that the globin acts like a baseball glove that “catches” and then traps incoming ligands long enough to allow bond formation with the heme iron. A recent comparison of different heme-containing globins has revealed that ligand migration between discrete docking sites is a general phenomenon in this class of proteins (17). Indeed, without these preformed sites, globins could not efficiently bind O2 at a ferrous heme iron (23, 35, 40, 43), which has to be located in the protein interior for protection against oxidation. By contrast, nitrophorin 4, an NO transporter, does not need protection against oxidation because its function relies on a ferric heme (44).
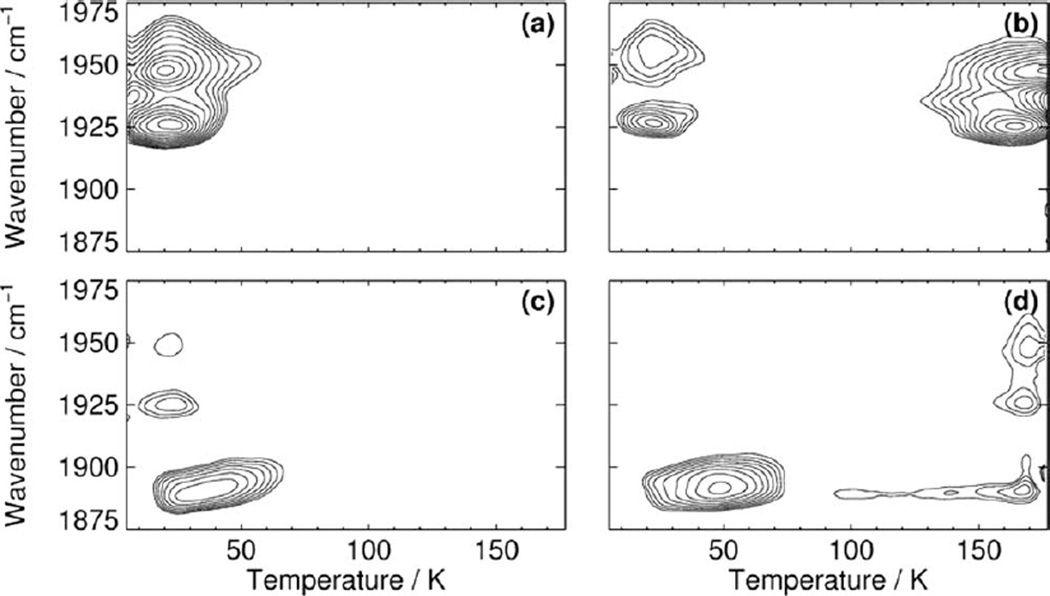
Transient docking sites exist also in the enzyme hIDO as can be inferred from the narrow bands in range 2,090–2,160 cm−1 associated with photolyzed CO (Fig. 2). Band amplitudes and positions depend on the temperature, at which the sample was illuminated, and on the illumination period, which implies that multiple sites are accessible (Fig. 2). The A state TDS contour map of substrate-free hIDO-CO illustrates rebinding in ASF from a primary docking site B at ~20 K (Fig. 3a). For comparison, rebinding from site B in the A0, A1, and A3 conformational substates of Mb peaks at ~30, 50, and 70 K, respectively (32, 45). The different temperatures have been explained by various degrees of steric hindrance against rebinding exerted by the HisE7 side chain, with minimal hindrance in A0 because the side chain is rotated away from the binding site. Similarly, we can assume that CO ligands face essentially no steric hindrance against recombination in the ASF conformations of hIDO, thus placing the site very close to the binding site.
Figure 3.
TDS contour maps of (a, b) hIDO-CO and (c, d) L-Trp bound hIDO-CO obtained after (a, c) 10-sec illumination at 4 K, and (b, d) slow cooling under continuous illumination from 160 to 4 K. Shown are the absorption changes in the bands of hemebound CO. Contours are spaced logarithmically.
Recombination from a more remote site D extends from ~140–200 K (Fig. 3b). The average rebinding temperature (~170 K) is significantly higher than that required in wild-type Mb [~120 K, (23)]. As yet, there is no structural information on where the site is located inside the protein.
The Effect of L-T on Ligand Migration and Binding
The photoproduct spectra of L-Trp bound hIDO-CO are different from those of substrate-free hIDO-CO, especially after extended illumination at higher temperatures (Fig. 2b), suggesting that L-Trp binding markedly affects CO migration and rebinding. The low amplitude of the AS band recorded after 10-sec illumination at 4 K (Fig. 3c) illustrates that only ~25% of all CO ligands in L-Trp-bound hIDO molecules have been trapped at site B. Although the photolysis quantum yield for CO is ~1 (46), most ligands are not observed to rebind in the TDS experiment because they rebind too fast even at the lowest temperature. Apparently, the accessibility of site B is significantly reduced as compared to substrate-free hIDO, possibly because the site is partially blocked by the substrate. Those few ligands that do dock at site B in AS encounter higher enthalpy barriers against rebinding as compared to those from ASF; the average recombination temperature is increased from 20 to ~45 K (Fig. 3c).
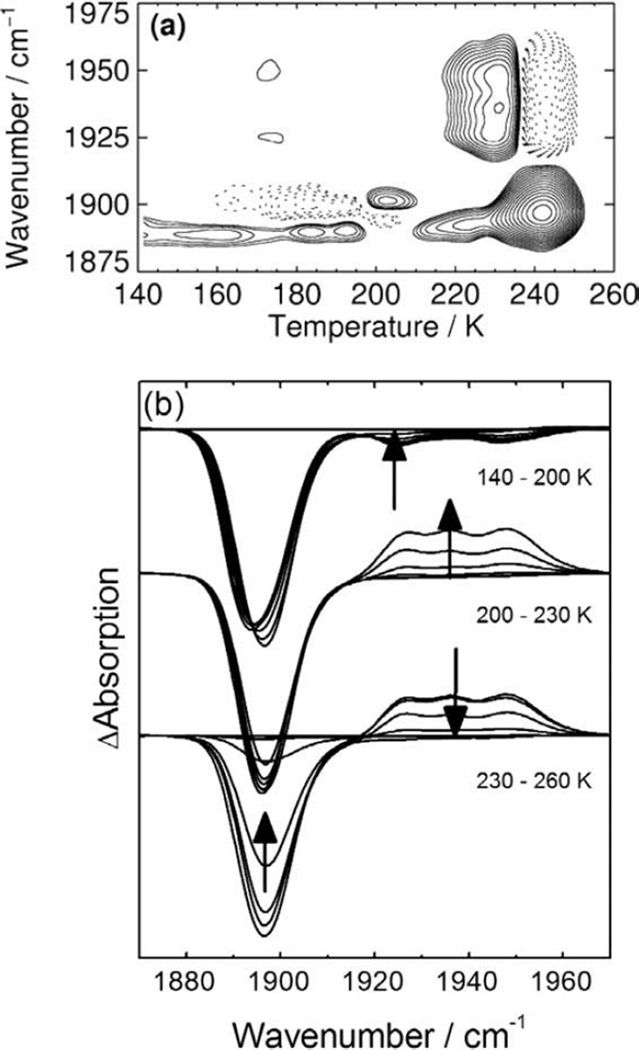
The A state TDS map obtained after slow cool illumination shows rebinding in AS from site B and, as the L-Trp concentration used was not sufficient to saturate the sample, in ASF from site D at ~170 K (Fig. 3d). The contours at ~1,890 cm−1 that stretch from ~70 to 200 K are not related to rebinding but rather to a slight band shift (Figs. 3d and 4a). At the highest temperature reached in the experiment (T = 180 K), ~60% of all CO ligands in AS are still trapped at the secondary docking site D. Apparently, the L-Trp bound at the active site prevents their recombination, presumably, by blocking the migration pathway from site D to site B, from where rebinding could occur.
Figure 4.
(a) A state TDS contour map of L-Trp-bound hIDO-CO obtained after slow cooling under continuous illumination from 160 to 140 K (rate 0.3 K/min). (b) FTIR spectra collected during the TDS experiment.
For complete relaxation of the sample, temperatures of ~250 K are required (Fig. 4). Starting at ~210 K, the cryosolvent (55% glycerol/45% buffer, vol/vol) turns liquid, which allows large-scale protein motions. As a consequence, L-Trp leaves the active site as, under anaerobic conditions, the ligand-free ferrous enzyme has a lower affinity for L-Trp as compared with the CO complex (29, 47). It is noted that in this context, “ligand-free” refers only to the actual binding site; the CO ligands are still inside the protein at the secondary docking site D. The removal of the L-Trp “road block” facilitates CO rebinding (29). As a consequence, “excess” substrate-free hIDO-CO is formed that was not present prior to photolysis, represented by the ASF bands with positive amplitude in Fig. 4b and the absorption increase in ASF at 210–230 K in Fig. 4a. Return of L-Trp to the active site of CO-ligated hIDO completes the relaxation process. The isosbestic point in Fig. 4b indicates a two-state transition from hIDO-CO to L-Trp bound hIDO-CO.
These data strongly suggest that, for the enzyme to be active, the ligand has to bind before the substrate because, otherwise, access to site B and thus to the binding site is significantly hindered. One may argue that site D is located on a migration side path and the substrate only blocks off the ligands that are trapped in site D, whereas ligands entering from the solvent are not affected and can easily access site B. Then, the docking site D would simply be a void space in the enzyme, without functional significance, that becomes occupied because of the special illumination conditions. This scenario would exclude the build-up of “excess” substrate-free hIDO-CO, however.
The Enzymatic Reaction
The molecular details of the mechanism by which O2 is activated and inserted into the substrate in the dioxygenase reactions are not yet known, presenting a major knowledge gap in heme oxygen chemistry. It is generally believed that the dioxygenase reaction of hIDO is initiated in the ferrous state. Recent data suggest that the ferrous enzyme first binds O2, followed by L-Trp, to generate the ternary complex. This binding sequence is supported by several observations. (i) The hIDO matrix provides at least two ligand docking sites, the primary site B and the more remote site D. L-Trp at the catalytic site acts like a road block for ligand dissociation (A → B) and association (D → A). (ii) Kinetic studies of CN− binding to hIDO at ambient temperature have shown that prebinding of L-Trp to hIDO retards cyanide association and dissociation, whereas prebinding of CN− facilitates L-Trp binding and retards L-Trp dissociation (47). (iii) Based on the Kd values of O2 and L-Trp binding to the ferrous, ligand-free enzyme, binding of O2 followed by L-Trp binding is also thermodynamically favored under physiological concentrations of O2 and L-Trp.
Once the ternary complex is formed, the two atoms of O2 are inserted into the C2=C3 bond of the indole moiety. On the basis of the ionic mechanism proposed early on (4), the first step of the dioxygenase reaction of hIDO involves the deprotonation of the indole NH group of L-Trp by an active base. The electron movement associated with the deprotonation reaction facilitates the electrophilic addition of the heme-bound O2 to the C2=C3 bond of the L-Trp indole, thereby leading to the formation of the heme iron-bound 3-indolenylperoxy intermediate, which is subsequently converted to the product, NFK, either via the dioxetane intermediate or via a Criegee type rearrangement (4, 16). In 2009, it was shown, however, that deprotonation of the indole N—H is not essential for catalysis (48). The most recent data support a consecutive two-step reaction in which the dioxygenase reaction is initiated by the insertion of the terminal atom of O2 into the C2=C3 of the indole ring, giving rise to a ferryl intermediate (49) and an indole 2,3-epoxide, which recombine to generate NFK (50).
The Inhibitory Site
At L-Trp concentrations below 50 µM, the steady-state activity of hIDO follows Michaelis–Menten behavior; yet a further increase in the L-Trp concentration leads to a decrease in the activity (51, 52). Substrate inhibition has been well known for rabbit IDO (53, 54). It was generally believed that, at high concentrations of L-Trp, the substrate binds directly to the ferric heme iron, thereby inhibiting its reduction to the active ferrous state (54). For hIDO, this scenario was discarded on the basis of two observations: (i) the dissociation constant of L-Trp from ferric hIDO (Kd = 0.9 mM) is significantly higher than the self-inhibition constant, Ksi (0.17 mM) (51), and (ii) the redox potential of L-Trp-bound ferric hIDO is ~46 mV higher than that of the substrate-free enzyme (55), indicating that L-Trp binding to the ferric enzyme does not prevent its reduction. On this basis, it was proposed that substrate inhibition of hIDO is a result of the binding of the second L-Trp molecule in an inhibitory substrate binding site of the enzyme (51). Computational docking experiments of 3-indole-ethanol (IDE) into the crystal structure of hIDO have suggested that a pocket occupied by a CHES molecule in the crystal structure (13) might at least partially coincide with the allosteric site (14). Unfortunately, such computational studies may yield ambiguous results because residues 360–380 are not resolved in the crystal structure of hIDO, but they may play an important role in substrate binding. FTIR and UV/Vis absorption data have provided the first, still indirect evidence for two distinct L-Trp binding sites (29). It was shown that CO escapes into the solvent upon photodissociation from the heme iron at T > 200 K. Concomitantly, L-Trp exits the active site and migrates to a secondary binding site or into the solvent. Although L-Trp is spectroscopically silent at this site, it is still noticeable due to its pronounced effect on the CO association kinetics, which is significantly slower than those of L-Trp-free hIDO.
Equilibrium titration studies of ferric hIDO-CN show explicitly that this species is capable of binding two L-Trp molecules, with Kd1 = 18 µM and Kd2 = 26 mM (47). In ligand-free hIDO, a second binding site was not observable. With O2 as a ligand, Kd2 has been determined as 0.17 mM (51). Binding of the effector IDE causes an elevation of Kd2, indicating that IDE competes with L-Trp for the second binding site and, once bound, does not retard the active site catalysis. Therefore, the apparent enzymatic activity is increased. In contrast, mitomycin C is an uncompetitive inhibitor that binds to the second site and inhibits the turnover at the active site (51).
CONCLUSIONS
The catalytic reaction of hIDO requires the concerted interaction between the protein, the ligand (O2), which binds to the heme iron, and the substrate (L-Trp). Hence, for optimal turnover of the enzyme, the protein scaffold has to provide easy access for both O2 and L-Trp to the heme active site. Binding of the voluminous L-Trp calls for a rather large “entry gate.” As both oxygen atoms are inserted in the reaction, efficient ligand escape pathways are most likely not required. But the reaction product NFK must be expelled from the active site to allow the uptake of new reagents. Binding of a molecule at the inhibitory site causes an allosteric structural transition that either advances or prevents the inhibition of the catalytic activity, depending on the nature of the molecule. In summary, the hIDO polypeptide chain is expected to be highly flexible to adjust to the actual situation.
ACKNOWLEDGEMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (Grants Ni291/3 and CFN) and the Fonds der Chemischen Industrie (to G.U.N.), as well as by the National Science Foundation (to SRY) under Grant No. 1026788.
REFERENCES
- 1.Anderson JL, Chapman SK. Ligand probes for heme proteins. Dalton Trans. 2005:13–24. doi: 10.1039/b413046d. [DOI] [PubMed] [Google Scholar]
- 2.Chapman SK, Daff S, Munro AW. Heme: the most versatile redox centre in biology? In: Sadler PJ, editor. Structure and Bonding. Berlin: Springer-Verlag; 1997. pp. 39–70. [Google Scholar]
- 3.King NJ, Thomas SR. Molecules in focus: indoleamine 2,3-dioxygenase. Int. J. Biochem. Cell Biol. 2007;39:2167–2172. doi: 10.1016/j.biocel.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Sono M, Roach MP, Coulter ED, Dawson JH. Heme-containing oxygenases. Chem. Rev. 1996;96:2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 5.Schutz G, Chow E, Feigelson P. Regulatory properties of hepatic tryptophan oxygenase. J. Biol. Chem. 1972;247:5333–5337. [PubMed] [Google Scholar]
- 6.Ren S, Correia MA. Heme: a regulator of rat hepatic tryptophan 2,3-dioxygenase? Arch. Biochem. Biophys. 2000;377:195–203. doi: 10.1006/abbi.2000.1755. [DOI] [PubMed] [Google Scholar]
- 7.Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, Brown C, Mellor AL. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 8.Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, Boon T, Van den Eynde BJ. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto A, Nikaido T, Ochiai K, Takakura S, Saito M, Aoki Y, Ishii N, Yanaihara N, Yamada K, Takikawa O, Kawaguchi R, Isonishi S, Tanaka T, Urashima M. Indoleamine 2,3-dioxygenase serves as a marker of poor prognosis in gene expression profiles of serous ovarian cancer cells. Clin. Cancer Res. 2005;11:6030–6039. doi: 10.1158/1078-0432.CCR-04-2671. [DOI] [PubMed] [Google Scholar]
- 10.Brandacher G, Perathoner A, Ladurner R, Schneeberger S, Obrist P, Winkler C, Werner ER, Werner-Felmayer G, Weiss HG, Gobel G, Margreiter R, Konigsrainer A, Fuchs D, Amberger A. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: effect on tumor-infiltrating T cells. Clin. Cancer Res. 2006;12:1144–1151. doi: 10.1158/1078-0432.CCR-05-1966. [DOI] [PubMed] [Google Scholar]
- 11.Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat. Med. 2005;11:312–319. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 12.Muller AJ, Scherle PA. Targeting the mechanisms of tumoral immune tolerance with small-molecule inhibitors. Nat. Rev. Cancer. 2006;6:613–625. doi: 10.1038/nrc1929. [DOI] [PubMed] [Google Scholar]
- 13.Sugimoto H, Oda S, Otsuki T, Hino T, Yoshida T, Shiro Y. Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase. Proc. Natl. Acad. Sci. USA. 2006;103:2611–2616. doi: 10.1073/pnas.0508996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macchiarulo A, Nuti R, Bellocchi D, Camaioni E, Pellicciari R. Molecular docking and spatial coarse graining simulations as tools to investigate substrate recognition, enhancer binding and conformational transitions in indoleamine-2,3-dioxygenase (IDO) Biochim. Biophys. Acta. 2007;1774:1058–1068. doi: 10.1016/j.bbapap.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Chauhan N, Basran J, Efimov I, Svistunenko DA, Seward HE, Moody PC, Raven EL. The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase. Biochemistry. 2008;47:4761–4769. doi: 10.1021/bi702405a. [DOI] [PubMed] [Google Scholar]
- 16.Macchiarulo A, Camaioni E, Nuti R, Pellicciari R. Highlights at the gate of tryptophan catabolism: a review on the mechanisms of activation and regulation of indoleamine 2,3-dioxygenase (IDO), a novel target in cancer disease. Amino acids. 2009;37:219–229. doi: 10.1007/s00726-008-0137-3. [DOI] [PubMed] [Google Scholar]
- 17.Nienhaus K, Nienhaus GU. Ligand dynamics in heme proteins observed by Fourier transform infrared-temperature derivative spectroscopy. Biochim. Biophys. Acta. 2010 doi: 10.1016/j.bbapap.2010.07.018. (in press). [DOI] [PubMed] [Google Scholar]
- 18.Li T, Quillin ML, Phillips GN, Jr, Olson JS. Structural determinants of the stretching frequency of CO bound to myoglobin. Biochemistry. 1994;33:1433–1446. doi: 10.1021/bi00172a021. [DOI] [PubMed] [Google Scholar]
- 19.Ray GB, Li X-Y, Ibers JA, Sessler JL, Spiro GS. How far can proteins bend the FeCO unit…. J. Am. Chem. Soc. 1994;116:162–176. [Google Scholar]
- 20.Vogel KM, Kozlowski PM, Zgierski MZ, Spiro TG. Determinants of the FeXO (X =C, N, O) vibrational frequencies in heme adducts from experiment and density functional theory. J. Am. Chem. Soc. 1999;121:9915–9921. [Google Scholar]
- 21.Braunstein DP, Chu K, Egeberg KD, Frauenfelder H, Mourant JR, Nienhaus GU, Ormos P, Sligar SG, Springer BA, Young RD. Ligand binding to heme proteins: III. FTIR studies of His-E7 and Val-E11 mutants of carbonmonoxymyoglobin. Biophys. J. 1993;65:2447–2454. doi: 10.1016/S0006-3495(93)81310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bredenbeck J, Helbing J, Nienhaus K, Nienhaus GU, Hamm P. Protein ligand migration mapped by nonequilibrium 2D-IR exchange spectroscopy. Proc. Natl. Acad. Sci. USA. 2007;104:14243–14248. doi: 10.1073/pnas.0607758104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nienhaus K, Deng P, Kriegl JM, Nienhaus GU. Structural dynamics of myoglobin: the effect of internal cavities on ligand migration and binding. Biochemistry. 2003;42:9647–9658. doi: 10.1021/bi034788k. [DOI] [PubMed] [Google Scholar]
- 24.Frauenfelder H, Nienhaus GU, Johnson JB. Rate Processes in Proteins. Ber. Bunsenges. Phys. Chem. 1991;95:272–278. [Google Scholar]
- 25.Alben JO, Beece D, Bowne SF, Doster W, Eisenstein L, Frauenfelder H, Good D, McDonald JD, Marden MC, Moh PP, Reinisch L, Reynolds AH, Shyamsunder E, Yue KT. Infrared spectroscopy of photodissociated carboxymyoglobin at low temperatures. Proc. Natl. Acad. Sci. U S A. 1982;79:3744–3748. doi: 10.1073/pnas.79.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vojtechovsky J, Chu K, Berendzen J, Sweet RM, Schlichting I. Crystal structures of myoglobin-ligand complexes at near-atomic resolution. Biophys. J. 1999;77:2153–2174. doi: 10.1016/S0006-3495(99)77056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang F, Phillips GN., Jr Crystal structures of CO-, deoxy- and met-myoglobins at various pH values. J. Mol. Biol. 1996;256:762–774. doi: 10.1006/jmbi.1996.0123. [DOI] [PubMed] [Google Scholar]
- 28.Johnson JB, Lamb DC, Frauenfelder H, Müller JD, McMahon B, Nienhaus GU, Young RD. Ligand binding to heme proteins. VI. Interconversion of taxonomic substates in carbonmonoxymyoglobin. Biophys. J. 1996;71:1563–1573. doi: 10.1016/S0006-3495(96)79359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickel E, Nienhaus K, Lu C, Yeh SR, Nienhaus GU. Ligand and substrate migration in human indoleamine 2,3-dioxygenase. J. Biol. Chem. 2009;284:31548–31554. doi: 10.1074/jbc.M109.039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terentis AC, Thomas SR, Takikawa O, Littlejohn TK, Truscott RJ, Armstrong RS, Yeh SR, Stocker R. The heme environment of recombinant human indoleamine 2,3-dioxygenase. Structural properties and substrate-ligand interactions. J. Biol. Chem. 2002;277:15788–15794. doi: 10.1074/jbc.M200457200. [DOI] [PubMed] [Google Scholar]
- 31.Capece L, Arrar M, Roitberg AE, Yeh SR, Marti MA, Estrin DA. Substrate stereo-specificity in tryptophan dioxygenase and indoleamine 2,3-dioxygenase. Proteins. 2010;78:2961–2972. doi: 10.1002/prot.22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nienhaus GU, Mourant JR, Chu K, Frauenfelder H. Ligand binding to heme proteins: the effect of light on ligand binding in myoglobin. Biochemistry. 1994;33:13413–13430. doi: 10.1021/bi00249a030. [DOI] [PubMed] [Google Scholar]
- 33.Berendzen J, Braunstein D. Temperature-derivative spectroscopy: a tool for protein dynamics. Proc. Natl. Acad. Sci. USA. 1990;87:1–5. doi: 10.1073/pnas.87.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mourant JR, Braunstein DP, Chu K, Frauenfelder H, Nienhaus GU, Ormos P, Young RD. Ligand binding to heme proteins: II. Transitions in the heme pocket of myoglobin. Biophys. J. 1993;65:1496–1507. doi: 10.1016/S0006-3495(93)81218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nienhaus K, Deng P, Olson JS, Warren JJ, Nienhaus GU. Structural dynamics of myoglobin: ligand migration and binding in valine 68 mutants. J. Biol. Chem. 2003;278:42532–42544. doi: 10.1074/jbc.M306888200. [DOI] [PubMed] [Google Scholar]
- 36.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 37.Parak F, Heidemeier J, Nienhaus GU. Protein structural dynamics as determined by Mössbauer spectroscopy. Hyperfine Interact. 1988;40:147–158. [Google Scholar]
- 38.Nienhaus GU, Heinzl J, Huenges E, Parak F. Protein crystal dynamics studied by time-resolved analysis of x-ray diffuse scattering. Nature. 1989;338:665–666. [Google Scholar]
- 39.Nienhaus K, Nienhaus GU. Ligand dynamics in heme proteins observed by Fourier transform infrared spectroscopy at cryogenic temperatures. Methods Enzymol. 2008;437:347–378. doi: 10.1016/S0076-6879(07)37018-3. [DOI] [PubMed] [Google Scholar]
- 40.Nienhaus K, Deng P, Kriegl JM, Nienhaus GU. Structural dynamics of myoglobin: spectroscopic and structural characterization of ligand docking sites in myoglobin mutant L29W. Biochemistry. 2003;42:9633–9646. doi: 10.1021/bi034787s. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt M, Nienhaus K, Pahl R, Krasselt A, Anderson S, Parak F, Nienhaus GU, Srajer V. Ligand migration pathway and protein dynamics in myoglobin: a time-resolved crystallographic study on L29W MbCO. Proc. Natl. Acad. Sci. USA. 2005;102:11704–11709. doi: 10.1073/pnas.0504932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nienhaus K, Ostermann A, Nienhaus GU, Parak FG, Schmidt M. Ligand migration and protein fluctuations in myoglobin mutant L29W. Biochemistry. 2005;44:5095–5105. doi: 10.1021/bi047513t. [DOI] [PubMed] [Google Scholar]
- 43.Scott EE, Gibson QH, Olson JS. Mapping the pathways for O2 entry into and exit from myoglobin. J. Biol. Chem. 2001;276:5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]
- 44.Nienhaus K, Maes EM, Weichsel A, Montfort WR, Nienhaus GU. Structural dynamics controls nitric oxide affinity in nitrophorin 4. J. Biol. Chem. 2004;279:39401–39407. doi: 10.1074/jbc.M406178200. [DOI] [PubMed] [Google Scholar]
- 45.Chu K, Ernst RM, Frauenfelder H, Mourant JR, Nienhaus GU, Philipp R. Light-induced and thermal relaxation in a protein. Phys. Rev. Lett. 1995;74:2607–2610. doi: 10.1103/PhysRevLett.74.2607. [DOI] [PubMed] [Google Scholar]
- 46.Chance MR, Courtney SH, Chavez MD, Ondrias MR, Friedman JM. O2 and CO reactions with heme proteins: quantum yields and geminate recombination on picosecond time scales. Biochemistry. 1990;29:5537–5545. doi: 10.1021/bi00475a018. [DOI] [PubMed] [Google Scholar]
- 47.Lu C, Lin Y, Yeh SR. Spectroscopic studies of ligand and substrate binding to human indoleamine 2,3-dioxygenase. Biochemistry. 2010;49:5028–5034. doi: 10.1021/bi1005078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chauhan N, Thackray SJ, Rafice SA, Eaton G, Lee M, Efimov I, Basran J, Jenkins PR, Mowat CG, Chapman SK, Raven EL. Reassessment of the reaction mechanism in the heme dioxygenases. J. Am. Chem. Soc. 2009;131:4186–4187. doi: 10.1021/ja808326g. [DOI] [PubMed] [Google Scholar]
- 49.Yanagisawa S, Yotsuya K, Horitani M, Hashiwaki Y, Horitani M, Sugimoto H, Shiro Y, Appleman EH, Ogura T. Identification of the FeO2 and the Fe5O heme species for indoleamine 2,3- dioxygenase during catalytic turnover. Chem. Lett. 2010;39:36–37. [Google Scholar]
- 50.Lewis-Ballester A, Batabyal D, Egawa T, Lu C, Lin Y, Marti MA, Capece L, Estrin DA, Yeh SR. Evidence for a ferryl intermediate in a heme-based dioxygenase. Proc. Natl. Acad. Sci. USA. 2009;106:17371–17376. doi: 10.1073/pnas.0906655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu C, Lin Y, Yeh SR. Inhibitory substrate binding site of human indoleamine 2,3-dioxygenase. J. Am. Chem. Soc. 2009;131:12866–12867. doi: 10.1021/ja9029768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sono M. Enzyme kinetic and spectroscopic studies of inhibitor and effector interactions with indoleamine 2,3-dioxygenase. 2. Evidence for the existence of another binding site in the enzyme for indole derivative effectors. Biochemistry. 1989;28:5400–5407. doi: 10.1021/bi00439a013. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto S, Hayaishi O. Tryptophan pyrrolase of rabbit intestine. D- and L-tryptophan-cleaving enzyme or enzymes. J. Biol. Chem. 1967;242:5260–5266. [PubMed] [Google Scholar]
- 54.Sono M, Taniguchi T, Watanabe Y, Hayaishi O. Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO-bound enzymes. J. Biol. Chem. 1980;255:1339–1345. [PubMed] [Google Scholar]
- 55.Papadopoulou ND, Mewies M, McLean KJ, Seward HE, Svistunenko DA, Munro AW, Raven EL. Redox and spectroscopic properties of human indoleamine 2,3-dioxygenase and a His303Ala variant: implications for catalysis. Biochemistry. 2005;44:14318–14328. doi: 10.1021/bi0513958. [DOI] [PubMed] [Google Scholar]