Abstract
The product of proto-oncogene Ron is a human receptor for the macrophage-stimulating protein (MSP). Upon activation, Ron is able to induce cell dissociation, migration and matrix invasion. Exon 11 skipping of Ron pre-mRNA produces Ron△165 protein that is constitutively active even in the absence of its ligand. Here we show that knockdown of SRSF2 promotes the decrease of exon 11 inclusion, whereas overexpression of SRSF2 promotes exon 11 inclusion. We demonstrate that SRSF2 promotes exon 11 inclusion through splicing and transcription procedure. We also present evidence that reduced expression of SRSF2 induce a decrease in the splicing of both intron 10 and 11, by contrast, overexpression of SRSF2 induce an increase in the splicing of intron 10 and 11. Through mutation analysis, we show that SRSF2 functionally target and physically interact with CGAG sequence on exon 11. In addition, we reveal that weak strength of splice sites of exon 11 is not required for the function of SRSF2 on the splicing of Ron exon 11. Our results indicate that SRSF2 promotes exon 11 inclusion of Ron proto-oncogene through targeting exon 11. Our study provides a novel mechanism by which Ron is expressed.
Keywords: Ron, proto-oncogene, pre-mRNA splicing, SRSF2, transcription, exon 11 inclusion
Introduction
RON receptor tyrosine kinase (Ron) along with c-Sea, c-Met and Stk are members of MET proto-oncogene family [1]. RON protein is an 180kDa-heterodimeric protein composed of a 40kDa-α chain and a 150kDa-β chain linked by disulfide bonds [2]. While the α chain contains the extracellular domain for ligand binding, the β chain includes the intracellular part that contains a kinase domain and a transmembrane domain [3]. Ron protein is a receptor for Macrophage stimulating protein (MSP), the binding stimulates the intrinsic kinase activity of Ron and subsequently leads to autophosphorylation on the tyrosine residues in its kinase domain and C-terminal docking sites for multiple transducers and adaptor proteins [4-6]. Whereas Ron overexpression and activation induce tumor progression and invasive growth of certain type of epithelial tumor cells, silencing of Ron expression promotes apoptosis [7-9]. Alternative splicing of Ron pre-mRNA produces various protein isoforms [10]. RONΔ165 protein, which was identified in gastric cancer cell line KATOIII, is generated by skipping of exon 11 [11]. RONΔ165 does not undergo proteolytic processing and is retained intracellularly, thus RONΔ165 is constitutively activated without the binding of MSP ligand [11]. Abnormal accumulation of RONΔ165 isoform was found in some types of breast and colon cancer cell lines [12]. Furthermore, overexpression of this splice variant can induce invasive growth and metastasis [11]. Whereas SRSF1 contact an enhancer on exon 12 to promote exon 11 skipping and furthermore control epithelial to mesenchymal transition, hnRNP A1 antagonizes the binding of SRSF1 and prevent exon skipping [12, 13]. In addition, hnRNP A2/B1 and hnRNP H are also known to regulate alternative splicing of Ron pre-mRNA [14, 15]. Alternative splicing generates proteomic diversity and is one of the critical gene regulatory mechanisms [16, 17]. Abnormal regulation of alternative splicing causes a variety of human diseases including cancer [18]. Alternative splicing is finely regulated by cis-acting elements and trans-acting elements [19, 20]. Cis-acting elements are RNA sequences on pre-mRNA which function as either enhancer or inhibitor to regulate exon inclusion or skipping. Trans-acting elements are proteins that regulate exon inclusion or skipping. The best identified trans-acting elements are SR proteins (serinearginine rich) and hnRNPs (heterogenous nuclear ribonucleoproteins) [21, 22]. SRSF2 is one of the SR proteins that are composed of RRM (RNA recognition motif) domain and RS (serine-arginine rich) domain [23]. SRSF2 participates in spliceosome assembly including U1 snRNP binding to 5’ splice site and U2 snRNP binding to branch point [24-26]. SRSF2 frequently binds to splicing enhancer sequences and regulates alternative splicing [27-29]. SRSF2 also play additional roles in transcription elongation, RNA stability, mRNA transport, and mRNA translation [30-34].
In this study, we demonstrate that SRSF2 promotes exon 11 inclusion of Ron pre-mRNA through activating splicing and transcription. We show that SRSF2 promotes splicing of both intron 10 and intron 11. Furthermore we identified that SRSF2 functionally targets and physically interacts with exon 11 to activate exon 11 inclusion. Our study added a new player in the splicing and transcription of Ron pre-mRNA.
Results
Knockdown of SRSF2 reduced exon 11 inclusion of Ron pre-mRNA
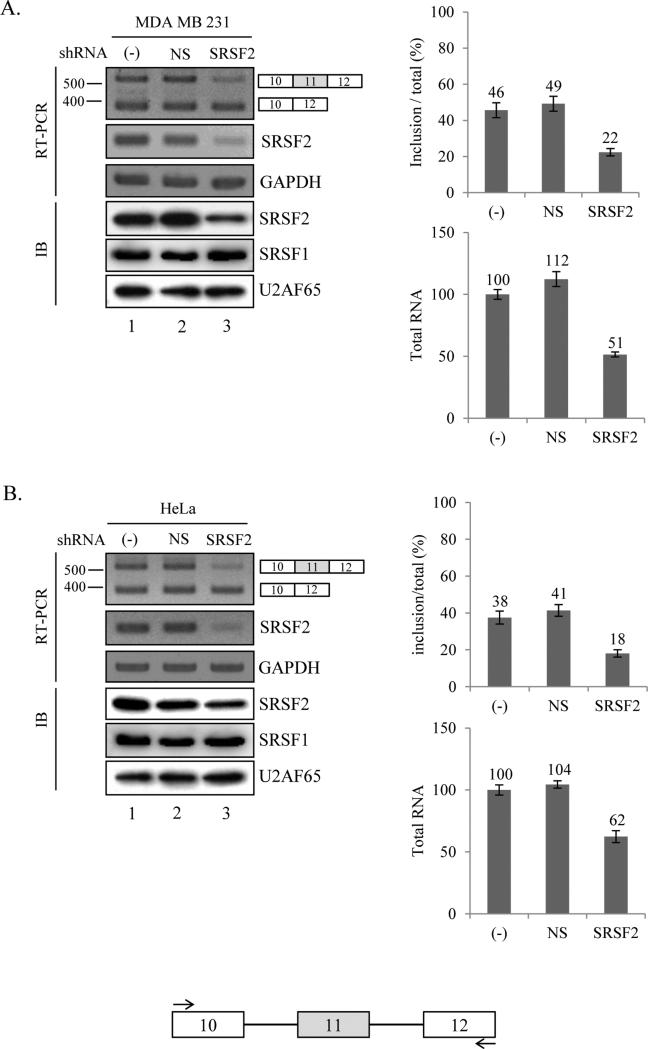
We initiated the project based on the fact that many potential SRSF2 binding sites on exon 11 of Ron pre-mRNA from a bioinformatics tool (ESE finder) [35] (supplementary figure 1). The bioinformatics searching results provide a potential that SRSF2 regulates alternative splicing of Ron exon 11. To test this idea, in the first set of experiment, we knocked down SRSF2 mRNA in cultured cells using lentivirus based shRNA. Three days after infection of SRSF2 shRNA virus, RNA was prepared for RT-PCR. The results in figure 1A show that in the MDA MB 231 cell, SRSF2 protein expression was decreased significantly (~60%) in SRSF2 knocked down cells as compared with that in untreated control. However non-silencing (NS) shRNA virus did not induce any decrease of SRSF2 expression. It has been shown that the effects of the knock down of one SR protein could be compensated by another SR protein [36]. To ask whether SRSF2 knock down increases another SR protein expression, we also analyzed SRSF1 protein expression level with immunoblotting analysis. As shown in figure 1A, SRSF1 expression was not altered in the SRSF2 knocked down cells. To determine whether knockdown of SRSF2 induces alteration of exon 11 splicing, we performed RT-PCR to analyze the alternative splicing of Ron exon 11. The results in figure 1A show that exon 11 included Ron mRNA is significantly decreased to ~24% of total Ron mRNA in the SRSF2 knockdown cells as compared with non-treated as well as non-silencing shRNA virus treated cells. Similar results were obtained from HeLa cell (~20% decrease) (figure 1B). Thus we conclude that reduced expression of SRSF2 decrease exon 11 inclusion of Ron pre-mRNA. In addition, as shown in the quantitation graph of total RNA in figure 1A and 1B, total RNA level that contains exon 11 skipped and included isoform is reduced significantly in SRSF2 knocked down cells (lane 3, ~49% and ~38% independently). The results in figure 1 demonstrate that SRSF2 knock down reduces exon 11 included isoform through regulating splicing and transcription events.
Figure 1.
Knockdown of SRSF2 with shRNA reduced exon 11 inclusion of Ron pre-mRNA. RT-PCR analysis of endogeneous Ron exon 11 alternative splicing with RNAs extracted from non-treated cells, non-silencing shRNA virus treated cells and SRSF2 shRNA virus treated cells are shown. Primers are located at exon 10 and exon 12 as shown with arrows. MDA MB 231 (A) and HeLa (B) cell lines are tested here. GAPDH is used as a control. Quantitative results are shown at right panels. Immunoblotting analysis in these cells using anti-SRSF2, anti-SRSF1 and anti-U2AF65 antibody are shown.
SRSF2 overexpression promotes exon 11 inclusion of Ron pre-mRNA
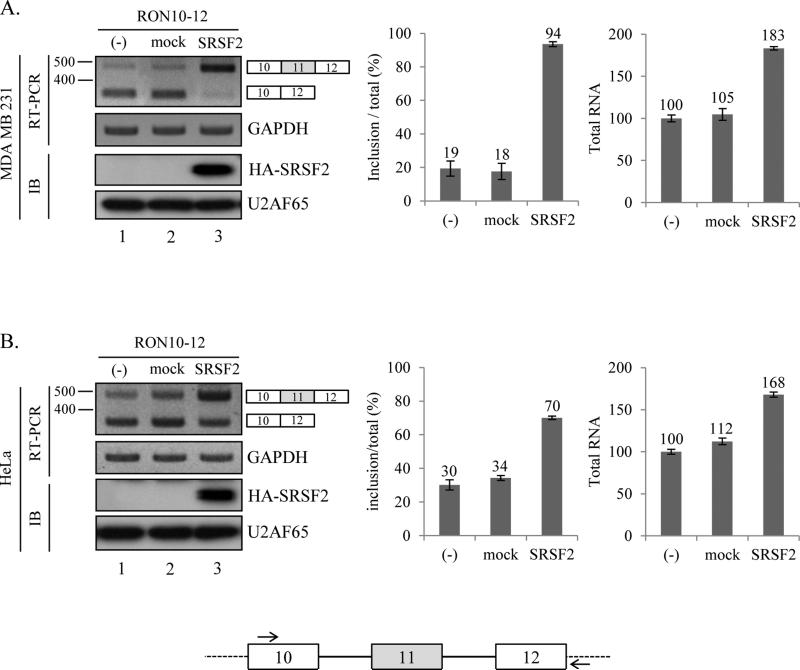
We next asked whether increased expression of SRSF2 has the opposite effects on the splicing and transcription of Ron proto-oncogene. We transfected cells with a SRSF2-expression plasmid and a Ron minigene that includes exon 10-12 part of Ron pre-mRNA (figure 2, lower panel). As shown in figure 2A, expression of SRSF2 induced a significant increase of exon 11 included form in MDA MB 231 and HeLa cells (lane 3, figure 2A and 2B). To compare the SRSF2 overexpression results with knockdown results, we performed similar analysis as in figure 1, in which both exon 11 inclusion rate and total RNA level were compared in SRSF2 knocked down cells. The results in figure 2 demonstrate that exon 11 inclusion rate was significantly increased (~75% and ~40% in MDA MB 231 cells and HeLa cells independently). In addition, we show that total Ron RNA was also significantly increased (~83% and ~68% in MDA MB 231 cells and HeLa cells independently). By contrast, overexpression of control plasmid did not induce a significant change in either exon 11 inclusion rate or transcription of Ron proto-oncogene (lane 2, figure 2). Thus the SRSF2 overexpression results in figure 2 are consistent with the SRSF2 knock down results in figure 1. To combinine the results in figure 1 and 2 together, we conclude that expression of SRSF2 promotes the both splicing and transcription of exon 11 included isoform in Ron proto-oncogene.
Figure 2.
SRSF2 overexpression promotes exon 11 inclusion of Ron pre-mRNA. (A) RT-PCR analysis of exogeneous Ron exon 11 alternative splicing with RNAs extracted from pcDNA-SRSF2 overexpressed, pcDNA overexpressed and non-treated MDA MB 231 (A) and HeLa (B) cells are shown. A minigene including exon 10, 11 and 12 is cloned into pcDNA3.1 (+) plasmid. The primer sets used for RT-PCR are shown. One primer is basepaired with pcDNA plasmid sequence (dot line); the other one is basepaired with exon 10. Quantitative results are shown at right panel.
SRSF2 promotes splicing of intron 10 as well as intron 11
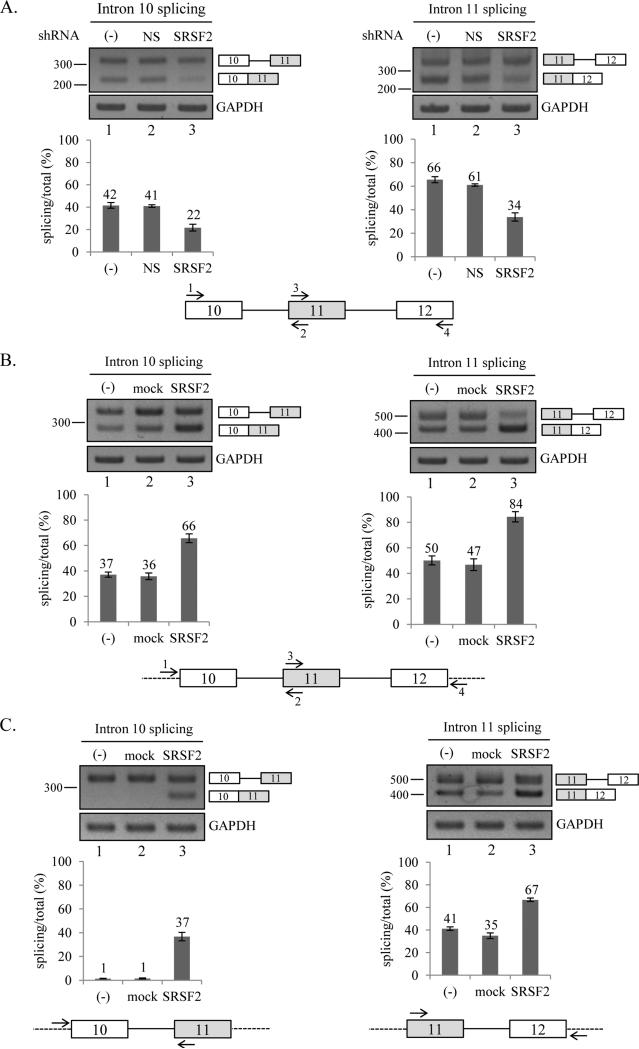
We next asked if SRSF2 promotes exon 11 inclusion by promoting splicing of both flanking introns or one of the flanking introns. To answer the question, we performed three lines of experiments. In the first line of experiment, we asked if the knockdown of SRSF2 affects splicing of intron 10 or 11. To determine the splicing of intron 10 and intron 11, we performed RT-PCR with primer sets that basepair with exon 10 and exon 11 to test the pre-mRNA splicing of intron 10, and another primer sets that basepair with exon 11 and exon 12 for the pre-mRNA splicing of intron 11 (figure 3A). As shown in figure 3A, splicing of intron 10 is significantly decreased (~20%) in the cells in which SRSF2 expression is reduced. In addition, splicing of intron 11 is also significantly reduced after knockdown of SRSF2 (~32%, Figure 3A). Thus we demonstrate that reduced expression of SRSF2 induced a reduced splicing of intron 10 and intron 11.
Figure 3.
SRSF2 promotes splicing of intron 10 as well as intron 11. (A) RT-PCR analysis of endogenous splicing of intron 10 (left panel) and 11 (right panel) with RNAs extracted from non-treated cells, non-silencing shRNA virus treated cells and SRSF2 shRNA virus treated cells are shown. The primer sets used to detect intron 10 (1 and 2) and intron 11 (3 and 4) RT-PCR are shown. Quantitation results are shown at lower panel. (B) RT-PCR analysis of exogenous intron 10 and intron 11 with RNAs from pcDNA-SRSF2 overexpressed, pcDNA overexpressed and non-treated MDA MB 231 cells are shown. The primer sets used to detect intron 10 (1 and 2) and intron 11 (3 and 4) RT-PCR are shown. (C) RT-PCR analysis of exogenous intron 10 and intron 11 with RNAs from SRSF2 overexpressed, pcDNA overexpressed and non-treated MDA MB 231 cells from minigenes containing exon 10-11 or exon 11-12 are shown.
In the second line of experiment, we asked if the overexpression of SRSF2 has the opposite effects on the splicing of intron 10 and 11. The results in figure 3B show that overexpression of SRSF2 caused a significant increase in the splicing of intron 10 and 11 (~29% and ~34% independently). By contrast, overexpression with control plasmid did not induce a significant alteration in the splicing of intron 10 and 11. Thus our results show that overexpression of SRSF2 promotes splicing of intron 10 and 11.
In the third line of experiment, we asked if the effects of SRSF2 on the splicing of intron 10 and 11 are dependent on the presence of distal exons or exon definition. To answer the question, we constructed minigenes that contain only exon 10-11 or exon 11-12 (figure 3C). We performed RT-PCR analysis with cells transfected with the minigenes along with SRSF2 expression plasmid. The results show that, in consistent with the results in figure 3B, SRSF2 promotes splicing of intron 10 and 11 significantly (~36% and ~26% independently), whereas control plasmid did not induce a significant alteration in the splicing of intron 10 and 11. Thus we conclude that SRSF2 splicing activation effects do not require exon 11 definition or distal exons. To combine together, SRSF2 facilitates splicing of intron 10 and 11.
SRSF2 interact with exon 11 to promotes exon 11 inclusion
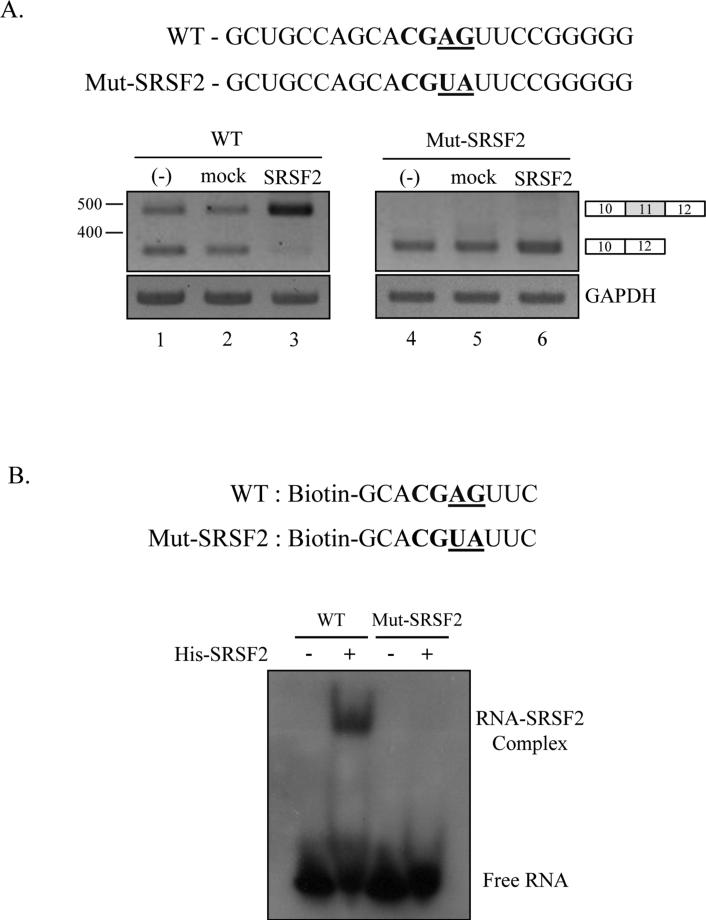
In order to locate the functional target of SRSF2, we performed various mutagenesis on the sequence that were predicted with ESE finder program. However, we were not able to locate the functional target through the mutation analysis, as shown in supplementary figure 1, showed none of the mutants disrupted the function of SRSF2 on Ron splicing and transcription. Later, we noticed that a Recent study identified new SRSF2 binding sequences-SSAG (S=C/G) [37]. We found that exon 11 includes a potential binding sequence (CGAG) for SRSF2 at the position 73 nt downstream from 3’ splice site of exon 11. In order to determine whether SRSF2 targets this sequence to promote exon 11 splicing, we mutated CGAG sequence into CGUA sequence (Mut-SRSF2) (figure 4A). If the sequence is the target of SRSF2, we expect that exon 11 inclusion is significantly reduced, and that the exon 11 promoting effects of SRSF2 is disrupted with the Mut-SRSF2 minigene. As we expected, figure 4A shows that exon 11 inclusion is hardly detectable for the Mut-SRSF2 minigene (lane 4). In addition, the effects of SRSF2 on promoting exon 11 are completely disrupted for the mutant minigene (lane 6). Thus we conclude that CGAG sequence is the functional target of SRSF2. We next asked if SRSF2 directly interacts with the functional targets sequence. We chemically synthesized 5’ biotin-labeled RNA that includes the potential target sequence and 3 nt flanking sequences. Correspondingly, we also synthesized Mut-SRSF2 RNA (figure 4B). The biotin-labeled RNAs were analyzed for binding with purified SRSF2 protein using Electrophoretic Mobility Shift Assay (EMSA). The results in figure 4B show that SRSF2 formed a complex with wild type RNA but not mutant RNA. Therefore we conclude that SRSF2 directly target CGAG sequence to promote exon 11 inclusion.
Figure 4.
SRSF2 interact with exon 11 to promote exon 11 inclusion. (A) The potential SRSF2 binding sites on exon 11 (bold) and the mutated nucleotide sequences (underlined) are highlighted. RT-PCR analysis of wild type Ron minigene and SRSF2 mutant construct with RNAs from non-treated, pcDNA overexpressed and SRSF2 overexpressed MDA MB 231 cells are shown. (B) 5’ biotinylated RNA sequences of wild type (WT) and mutant (Mut-SRSF2) are shown. Following incubation of biotinylated RNA with his-SRSF2 protein, we separated SRSF2-RNA complex from free RNA using native polyacrylamide gel electrophoresis. The biotin-labeled RNA was detected using streptavidin-HRP conjugates.
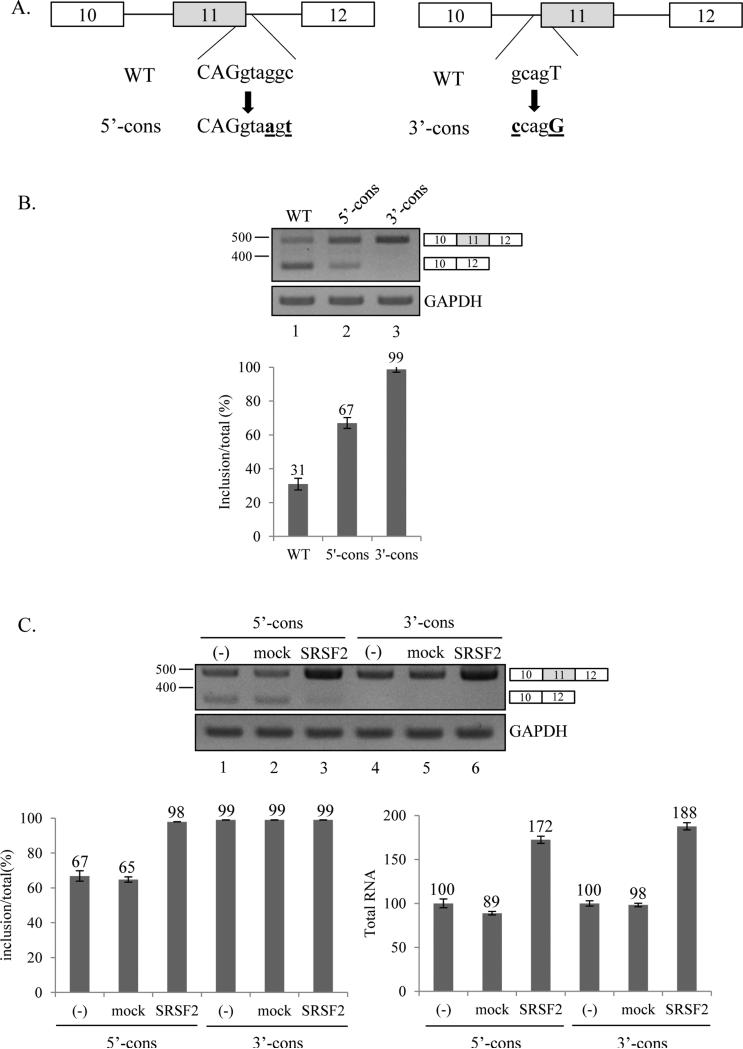
The strength of flanking splice sites on exon 11 is not required for the function of SRSF2 on exon 11 inclusion
To determine whether the strength of flanking splicing sites on exon 11 is required for function of SRSF2 on exon 11 inclusion, we performed mutagenesis on the splice site sequence. As shown in figure 5A, 5’ splicing site sequence (CAGgtaggc) of exon 11 is mutated into the conserved sequence (CAGgtaagt) (5’-cons). In the other mutant, 3’ splicing site sequence (gcagT) of exon 11 is mutated into the conserved sequence (ccagG) (3’-cons). We expect that the conserved sequences on the splice sites of exon 11 would facilitate splicing of intron 10 and 11, consequently exon 11 inclusion will be promoted. As we expected, the conserved sequence on both splice sites promotes exon 11 inclusion rate significantly in MDA MB 231 cells (~36% and ~68% independently, Figure 5B). If the strength of splice sites is required for exon 11 inclusion, we predict that SRSF2 would not promote exon 11 inclusion. However, we found that, as shown in figure 5C, SRSF2 can still promote exon 11 inclusion rate significantly in the 5’-cons construct (~31%). In addition, consistent with the results in figure 2B, the transcription of Ron is also promoted significantly (~72%). Therefore the 5’-cons construct did not disrupt the SRSF2 function in splicing and transcription of Ron exon 11. Therefore the strength of 5’ splice site on exon 11 is not required for the function of SRSF2 in splicing and transcription. For the 3’-cons construct, we noticed that exon 11 inclusion form is exclusively expressed, thus the increase of exon 11 inclusion rate is hard to detect with this construct. Not surprisingly, Ron transcription level is also significantly increased in SRSF2 overexpressed cells. Therefore we conclude that the strength of the flanking splice sites on exon 11 is not required for SRSF2 function in splicing and transcription of Ron exon 11.
Figure 5.
The strength of flanking splice sites on exon 11 is not required for the function of SRSF2 on exon 11 inclusion. (A) Mutated nucleotides of 5’ and 3’ splice sites are highlighted (bold). Sequence of wild type and mutants are shown. (B) RT-PCR analysis of exon 11 splicing in the wild type and two mutants. (C) RT-PCR analysis of minigene exon 11 splicing with RNAs from cells overexpressing pcDNA-SRSF2 overexpressed, pcDNA overexpressed and non-treated MDA MB 231 cells.
Discussions
In this study, we have studied the basis by which Ron proto-oncogene is spliced and transcribed. We have shown that SRSF2 promotes exon 11 inclusion by demonstrating that knockdown of SRSF2 induced a decrease of exon 11 included form, and that overexpression of SRSF2 promoted exon 11 included form of Ron pre-mRNA. Our results reveal that SRSF2 increases Ron exon 11 included form through promoting both exon 11 splicing and transcription. We further show that SRSF2 promotes splicing of both intron 10 and intron 11 by demonstrating that knockdown of SRSF2 induced a decrease in the splicing of intron 10 and 11, and that overexpression of SRSF2 induced an increase in the splicing of intron 10 and 11. In addition, we found that the strength of the flanking splice sites on exon 11 is not required for the function of SRSF2 on exon 11 splicing. Furthermore we demonstrate that a mutation of the potential binding site disrupted the function of SRSF2, and that SRSF2 directly interacts with exon 11. Our results strongly support the conclusion that SRSF2 is another regulator for Ron pro-oncogene expression.
Exon 11 splicing of Ron pre-mRNA was shown to be regulated by SRSF1 protein by contacting exon 12 [12]. It was also shown that hnRNP A1 antagonizes the binding of SRSF1 and prevents exon skipping [13]. Our results added SRSF2 as a new player in the regulation of Ron pre-mRNA splicing. SRSF1 and SRSF2 protein regulate Ron pre-mRNA splicing with antagonistic functions. The function of SR proteins on alternative splicing is partially determined by its binding locations. Alternative exon bound SRSF2 promotes exon inclusions; whereas flanking exon bound SRSF1 promotes alternative exon skipping. How these protein regulate splice site selection, spliceosome assembly, U2AF65 recruitment, U2snRNA-branchpoint basepairing and U1snRNA-5’ splice site basepairing need to be determined.
SRSF2 is a well-known alternative splicing regulatory protein. It has been shown that SRSF2 regulates alternative splicing of Tau, CD44, E-cadherin pre-mRNA either though promoting or inhibiting exon inclusion [38, 39]. Phosphorylation of SRSF2 plays a key role in the function of SRSF2 on Tau pre-mRNA splicing [27]. Our results indicate that SRSF2 promotes exon inclusion of Ron pre-mRNA. Our results also demonstrate that SRSF2 promotes splicing of both flanking introns, but not one of the flanking introns. In addition to protein-contact location on pre-mRNA, there are many possible modifications, for example phosphorylation, probably regulate the function of SRSF2 on Ron alternative splicing.
In addition to the role in pre-mRNA splicing, SRSF2 was also known to promote transcription elongation [31]. Consistent with the reports, our results demonstrated that SRSF2 significantly promotes Ron transcription procedure. In the previous kinetic model, it was shown that higher elongation rate promotes exon skipping, whereas lower elongation rate promotes exon inclusion [40]. As SRSF2 promotes transcription elongation, we can expect that SRSF2 promotes exon 11 skipping. However, our results clearly showed that SRSF2 promotes exon 11 inclusion. There is a possibility that SRSF2 plays other roles in Ron transcription instead of transcription elongation. We initiated the study because various potential binding sites of SRSF2 were predicted on Ron exon 11 from ESE finder program [35]. However, our results indicate that SRSF2 did not functionally target the potential high scores sequences. Surprisingly, a CGAG sequence, which was not shown in ESE finder program but found in the solution structure of SRSF2 with RNA, functionally targeted SRSF2 in Ron exon 11 splicing [37]. How the potential binding sites are functionally selected remains to be determined.
Whereas a group reported that SRSF2 expression is increased in ovarian cancer[41], downregulation of SRSF2 expression was also observed in clear cell renal cell carcinoma [42]. Unlike the fact that increased expression level of SRSF1 in tumors is directely related to delta Ron expression, the SRSF2 expression level is not matching quite well with Ron expression. A possibility is that mutations of the SRSF2 have been found in tumors as mutations of SRSF1 have not documented yet [43].
Materials and methods
Plasmid construction
The wild-type RON exon10-12 sequences were amplified from human genomic DNA using RON10-HindIII-for (5’-ATGTTAAGCTTCCTGAATATGTGGTCCGAGAC-3’) and RON12-XhoI-rev (5’-CTTACCTCGAGCTAGCTGCTTCCTCCGCCACC-3’) primers. RON exon10-11 construct was produced with RON10-HindIII-for and RON11-Xho-rev (5’-CTTAACTCGAGATGGGGCACCATCCTGGCCA-3’) primers. RON exon11-12 construct was produced with RON11-HindIII-for (5’-TAGATAAGCTTTATATTGGGCTGGGCGCTGTG-3’) and RON12-XhoI-rev primers. Mut-SRSF2 construct was produced by overlapping PCR using Mut-SRSF2-for (5’-CTGCCAGCACGTATTCCGGGGGGA-3’), Mut-SRSF2-rev (5’-TCCCCCCGGAATACGTGCTGGCAG-3’) primers. 5’-cons and 3’-cons mutants were produced by overlapping PCR using 5’-cons-for (5’-ATTGCAGGTAAGTAGCCCAGCTG-3’), 5’-cons-rev (5’-CAGCTGGGCTACTTACCTGCAAT-3’), 3’-cons-for (5’-ACCCTCTCTCCAGGGATATTGGGCT-3’) and 3’-cons-rev (5’-AGCCCAATATCCTGGAGAGAGGGT-3’) primers. All constructs were cloned into pcDNA3.1 (+) vector using HindIII and XhoI enzyme sites.
Cell culture and Transfection
MDA MB 231 cells were grown in RPMI-1640 medium and HeLa cells were grown in DMEM medium supplemented with 10% Fetal Bovine Serum (FBS) at 37°C in a humidified 5% CO2 condition. Ron mini-gene and mutant constructs were transfected into MDA MB 231 and HeLa cells using Polyethylenimine (PEI) reagent according to the manufacturer's protocol.
RT-PCR
Total RNA was extracted from MDA MB 231, HeLa and plasmid-DNA transfected cells using RiboEX regent (GeneAll) following manufacturer's protocol. 1ug of total RNA was reverse transcribed using oligo dT18 with ImProm-II™ reverse transcriptase (Promega) following manufacturer's protocol. 1uL of the cDNA was amplified by PCR reaction using G-Taq polymerase (Cosmo Genetech). Different Primers were used to detect splicing: primers for endogenous Ron exon 10-12 [Exon10-for (5’-CCGCTCGAGCGGACCATGTGTGAGAGGCAGCTTCCAG-3’), Exon12-rev (5’-CCGGAATTCCGGTCCTAGCTGCTTCCTCCGCC-3’)], Primers for Ron mini-gene [Exon10-for, pcDNA-rev (5’-CTAGAAGGCACAGTCGAGGCT-3’)], Primers for endogenous intron 11 splicing [Exon10-for and Exon11-rev (5’-ACAGCGCCCAGCCCAATAT-3’)], primers for endogenous intron 12 splicing [Exon11-for (5’-TATATTGGGCTGGGCGCTGTG-3’) and Exon12-rev], primers for detecting exogenous intron 11 splicing [T7-for (5’-TAATACGACTCACTATAGGG-3’) and Exon11-rev], primers for exogenous intron 12 splicing [Exon11-for and pcDNA-rev], primers for GAPDH [GADPH-for (5’-ACCACAGTCCATGCCATCA-3’), GAPDH-rev (5’-TCCACCACCCTGTTGCTGTA-3’)].
Immunoblotting
MDA MB 231 and HeLa cells were lysed with lysis buffer [1% NP-40, 50mM Tris-Cl (7.5), 150mM NaCl, 5mM EDTA, 1mM beta-mercaptoethanol, protease inhibitor cocktail] by incubating for 1 hour at 4°C, and then the supernatants were collected following centrifuge. The proteins were separated on SDS-PAGE gels and transferred to PVDF membrane. The membrane was blocked using 5% skim milk and incubated with following antibodies: anti-SRSF2 (AVIVA systems biology), anti-SRSF1 (Zymed), anti-HA (Santacruz biotechnology), anti-U2AF65 antibody. The signal was detected by Enhanced Chemiluminescence (ECL).
Knockdown of SRSF2 with shRNA
shRNA lentiviruses were produced by co-transfection of pLKO.1 shRNA plasmid coding the SRSF2 mRNA matched sequences or non-silencing sequences, PSPAX2 and PMD2G helper plasmids into 293T cells. After 24 hr transfection, supernatants containing lentiviruses were harvested with a 0.45 um filter. MDA MB231, HeLa cells were infected by the lentivirus with 10ug/ml polybrene. After 72 hr infection, media was removed then total RNA was extracted for RT-PCR.
Purification of His-tagged SRSF2
SRSF2 coding region was cloned into pcDNA6/myc-His A (Invitrogen) plasmid and transfected into HEK293 cells. After treatment with cell lysis buffer (50 mM NaH2PO4, 500 mM NaCl, 5 mM imidazole, 0.5% tween-20, 1 mM PMSF), Ni-NTA agarose beads (QIAGEN) were added and incubated overnight at 4°C in the binding buffer (50 mM NaH2PO4, 500 mM NaCl, 0.5% tween-20, 1 mM PMSF). After washing, Ni-NTA agarose beads were incubated with elution buffer (250 mM imidazole in binding buffer) for 20 min at room temperature.
Electrophoretic Mobility Shift Assay
One pmol of 5’ biotinylated RNA oligos (wild type and mutant) were incubated with purified his-SRSF2 protein in binding buffer [100 mM Tris (pH 7.5), 500 mM KCl, 10 mM DTT] for 30 min at 25°C. The mixtures were loaded to a 5% native polyacrylamide gel and transferred to the nylon membrane. After incubation with streptavidin-HRP conjugate (Sigma), the membrane was treated with ECL solution and detected with X-ray film.
Supplementary Material
Acknowledgements
This work was supported by the NRF-2013-R1A1A2062582 grants to Haihong Shen, the NRF-2013-R1A1A2061321 grant to Xuexiu Zheng funded by the National Research Foundation (NRF) of the Korean Ministry of Education, Science, and Technology (MEST) and a Systems Biology Infrastructure Establishment grant and integrative aging research grant at the Gwangju Institute of Science and Technology (GIST).
References
- 1.Ronsin C, Muscatelli F, Mattei MG, Breathnach R. A novel putative receptor protein tyrosine kinase of the met family. Oncogene. 1993;8:1195–1202. [PubMed] [Google Scholar]
- 2.Gaudino G, Follenzi A, Naldini L, Collesi C, Santoro M, Gallo KA, Godowski PJ, Comoglio PM. RON is a heterodimeric tyrosine kinase receptor activated by the HGF homologue MSP. The EMBO journal. 1994;13:3524–3532. doi: 10.1002/j.1460-2075.1994.tb06659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angeloni D, Danilkovitch-Miagkova A, Ivanov SV, Breathnach R, Johnson BE, Leonard EJ, Lerman MI. Gene structure of the human receptor tyrosine kinase RON and mutation analysis in lung cancer samples. Genes, chromosomes & cancer. 2000;29:147–156. [PubMed] [Google Scholar]
- 4.Danilkovitch A, Leonard EJ. Kinases involved in MSP/RON signaling. Journal of leukocyte biology. 1999;65:345–348. doi: 10.1002/jlb.65.3.345. [DOI] [PubMed] [Google Scholar]
- 5.Longati P, Bardelli A, Ponzetto C, Naldini L, Comoglio PM. Tyrosines1234-1235 are critical for activation of the tyrosine kinase encoded by the MET proto-oncogene (HGF receptor) Oncogene. 1994;9:49–57. [PubMed] [Google Scholar]
- 6.Iwama A, Yamaguchi N, Suda T. STK/RON receptor tyrosine kinase mediates both apoptotic and growth signals via the multifunctional docking site conserved among the HGF receptor family. The EMBO journal. 1996;15:5866–5875. [PMC free article] [PubMed] [Google Scholar]
- 7.Logan-Collins J, Thomas RM, Yu P, Jaquish D, Mose E, French R, Stuart W, McClaine R, Aronow B, Hoffman RM, Waltz SE, Lowy AM. Silencing of RON receptor signaling promotes apoptosis and gemcitabine sensitivity in pancreatic cancers. Cancer research. 2010;70:1130–1140. doi: 10.1158/0008-5472.CAN-09-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maggiora P, Marchio S, Stella MC, Giai M, Belfiore A, De Bortoli M, Di Renzo MF, Costantino A, Sismondi P, Comoglio PM. Overexpression of the RON gene in human breast carcinoma. Oncogene. 1998;16:2927–2933. doi: 10.1038/sj.onc.1201812. [DOI] [PubMed] [Google Scholar]
- 9.Wang MH, Yao HP, Zhou YQ. Oncogenesis of RON receptor tyrosine kinase: a molecular target for malignant epithelial cancers. Acta pharmacologica Sinica. 2006;27:641–650. doi: 10.1111/j.1745-7254.2006.00361.x. [DOI] [PubMed] [Google Scholar]
- 10.Lu Y, Yao HP, Wang MH. Multiple variants of the RON receptor tyrosine kinase: biochemical properties, tumorigenic activities, and potential drug targets. Cancer letters. 2007;257:157–164. doi: 10.1016/j.canlet.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Collesi C, Santoro MM, Gaudino G, Comoglio PM. A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Molecular and cellular biology. 1996;16:5518–5526. doi: 10.1128/mcb.16.10.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Molecular cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Bonomi S, di Matteo A, Buratti E, Cabianca DS, Baralle FE, Ghigna C, Biamonti G. HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic acids research. 2013;41:8665–8679. doi: 10.1093/nar/gkt579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golan-Gerstl R, Cohen M, Shilo A, Suh SS, Bakacs A, Coppola L, Karni R. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer research. 2011;71:4464–4472. doi: 10.1158/0008-5472.CAN-10-4410. [DOI] [PubMed] [Google Scholar]
- 15.Lefave CV, Squatrito M, Vorlova S, Rocco GL, Brennan CW, Holland EC, Pan YX, Cartegni L. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. The EMBO journal. 2011;30:4084–4097. doi: 10.1038/emboj.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nature reviews. Genetics. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 17.Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Progress in neurobiology. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 18.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes & development. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J, Zhou J, Zheng X, Cho S, Moon H, Loh TJ, Jo K, Shen H. Identification of a novel cis-element that regulates alternative splicing of Bcl-x pre-mRNA. Biochemical and biophysical research communications. 2012;420:467–472. doi: 10.1016/j.bbrc.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 20.Cho S, Moon H, Yang X, Zhou J, Kim HR, Shin MG, Loh TJ, Zheng X, Shen H. Validation of trans-acting elements that promote exon 7 skipping of SMN2 in SMN2-GFP stable cell line. Biochemical and biophysical research communications. 2012;423:531–535. doi: 10.1016/j.bbrc.2012.05.161. [DOI] [PubMed] [Google Scholar]
- 21.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors, Wiley interdisciplinary reviews. RNA. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. The Biochemical journal. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 23.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blencowe BJ, Bowman JA, McCracken S, Rosonina E. SR-related proteins and the processing of messenger RNA precursors. Biochemistry and cell biology = Biochimie et biologie cellulaire. 1999;77:277–291. [PubMed] [Google Scholar]
- 25.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. The Biochemical journal. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 26.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 27.Qian W, Liang H, Shi J, Jin N, Grundke-Iqbal I, Iqbal K, Gong CX, Liu F. Regulation of the alternative splicing of tau exon 10 by SC35 and Dyrk1A. Nucleic acids research. 2011;39:6161–6171. doi: 10.1093/nar/gkr195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apostolatos H, Apostolatos A, Vickers T, Watson JE, Song S, Vale F, Cooper DR, Sanchez-Ramos J, Patel NA. Vitamin A metabolite, all-trans-retinoic acid, mediates alternative splicing of protein kinase C deltaVIII (PKCdeltaVIII) isoform via splicing factor SC35. The Journal of biological chemistry. 2010;285:25987–25995. doi: 10.1074/jbc.M110.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meshorer E, Bryk B, Toiber D, Cohen J, Podoly E, Dori A, Soreq H. SC35 promotes sustainable stress-induced alternative splicing of neuronal acetylcholinesterase mRNA. Molecular psychiatry. 2005;10:985–997. doi: 10.1038/sj.mp.4001735. [DOI] [PubMed] [Google Scholar]
- 30.Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, Fu XD, Li X. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Molecular and cellular biology. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nature structural & molecular biology. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. The EMBO journal. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mo S, Ji X, Fu XD. Unique role of SRSF2 in transcription activation and diverse functions of the SR and hnRNP proteins in gene expression regulation. Transcription. 2013;4 doi: 10.4161/trns.26932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic acids research. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M, Jr., Fu XD. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Molecular cell. 2013;50:223–235. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daubner GM, Clery A, Jayne S, Stevenin J, Allain FH. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. The EMBO journal. 2012;31:162–174. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian W, Iqbal K, Grundke-Iqbal I, Gong CX, Liu F. Splicing factor SC35 promotes tau expression through stabilization of its mRNA. FEBS letters. 2011;585:875–880. doi: 10.1016/j.febslet.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma S, Liao W, Zhou X, Wong DT, Lichtenstein A. Exon 11 skipping of E-cadherin RNA downregulates its expression in head and neck cancer cells. Molecular cancer therapeutics. 2011;10:1751–1759. doi: 10.1158/1535-7163.MCT-11-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montes M, Cloutier A, Sanchez-Hernandez N, Michelle L, Lemieux B, Blanchette M, Hernandez-Munain C, Chabot B, Sune C. TCERG1 regulates alternative splicing of the Bcl-x gene by modulating the rate of RNA polymerase II transcription. Molecular and cellular biology. 2012;32:751–762. doi: 10.1128/MCB.06255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischer DC, Noack K, Runnebaum IB, Watermann DO, Kieback DG, Stamm S, Stickeler E. Expression of splicing factors in human ovarian cancer. Oncology reports. 2004;11:1085–1090. [PubMed] [Google Scholar]
- 42.Piekielko-Witkowska A, Wiszomirska H, Wojcicka A, Poplawski P, Boguslawska J, Tanski Z, Nauman A. Disturbed expression of splicing factors in renal cancer affects alternative splicing of apoptosis regulators, oncogenes, and tumor suppressors. PloS one. 2010;5:e13690. doi: 10.1371/journal.pone.0013690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida K, Ogawa S. Splicing factor mutations and cancer, Wiley interdisciplinary reviews. RNA. 2014;5:445–459. doi: 10.1002/wrna.1222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.