Abstract
Background
Perception and transduction of temperature changes result in altered growth enabling plants to adapt to increased ambient temperature. While PHYTOCHROME-INTERACTING FACTOR4 (PIF4) has been identified as a major ambient temperature signaling hub, its upstream regulation seems complex and is poorly understood. Here, we exploited natural variation for thermo-responsive growth in Arabidopsis thaliana using quantitative trait locus (QTL) analysis.
Results
We identified GIRAFFE2.1, a major QTL explaining ~18 % of the phenotypic variation for temperature-induced hypocotyl elongation in the Bay-0 x Sha recombinant inbred line population. Transgenic complementation demonstrated that allelic variation in the circadian clock regulator EARLY FLOWERING3 (ELF3) is underlying this QTL. The source of variation could be allocated to a single nucleotide polymorphism in the ELF3 coding region, resulting in differential expression of PIF4 and its target genes, likely causing the observed natural variation in thermo-responsive growth.
Conclusions
In combination with other recent studies, this work establishes the role of ELF3 in the ambient temperature signaling network. Natural variation of ELF3-mediated gating of PIF4 expression during nightly growing periods seems to be affected by a coding sequence quantitative trait nucleotide that confers a selective advantage in certain environments. In addition, natural ELF3 alleles seem to differentially integrate temperature and photoperiod information to induce architectural changes. Thus, ELF3 emerges as an essential coordinator of growth and development in response to diverse environmental cues and implicates ELF3 as an important target of adaptation.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0566-6) contains supplementary material, which is available to authorized users.
Keywords: Ambient temperature signaling, Auxin, ELF3, PIF4, Thermomorphogenesis
Background
In analogy to photomorphogenesis, the term thermomorphogenesis describes the effect of temperature on morphogenesis [1]. Hypocotyl elongation [2] and leaf hyponasty [3] belong to the most sensitive thermomorphogenic changes in plant development. Physiologically, these coordinated responses likely enhance evaporative leaf cooling [4, 5] and thus enable plants to adapt to warmth. Within the context of globally increasing ambient temperatures, it is imperative to improve our understanding of the basic processes plants employ to react to such environmental perturbations.
A major hub in the ambient temperature signaling network is the basic helix-loop-helix (bHLH) transcription factor PHYTOCHROME-INTERACTING FACTOR4 (PIF4). PIF4 protein binds to the promoters of auxin biosynthesis and response genes [6–9]. It thereby transcriptionally activates auxin responses, resulting in elongation growth. PIF4 itself seems to be transcriptionally regulated in a temperature-dependent manner by the bZIP transcription factor ELONGATED HYPOCOTYL5 (HY5) [10]. Accumulating data on PIF4 regulation from light signaling, photomorphogenesis and the circadian clock [11–13] indicate a more complex regulation of PIF4 activity on several levels.
The objective of this study was to exploit natural variation within the gene pool of Arabidopsis thaliana to identify additional components of the complex signaling network that plants use to adapt growth to changes in ambient temperature. Based on a quantitative genetic approach, we here show that two naturally occurring alleles of EARLY FLOWERING3 (ELF3) cause a differential response in thermomorphogenesis.
Results
We previously observed extensive natural variation for the thermomorphogenic signature phenotype we termed temperature-induced hypocotyl elongation (TIHE; [10, 14]). To identify the underlying genetic variants, we performed quantitative trait locus (QTL) analyses based on two natural accessions from geographically distant locations. When comparing growth at 20 and 28 °C, the selected accessions Bay-0 (Germany) and Sha (Tajikistan) differed significantly in several thermomorphogenic responses (Fig. 1a, b; Additional file 1). Identification of the underlying genetic variants would help to improve our understanding of how ambient temperature changes are translated into growth responses.
Fig. 1.
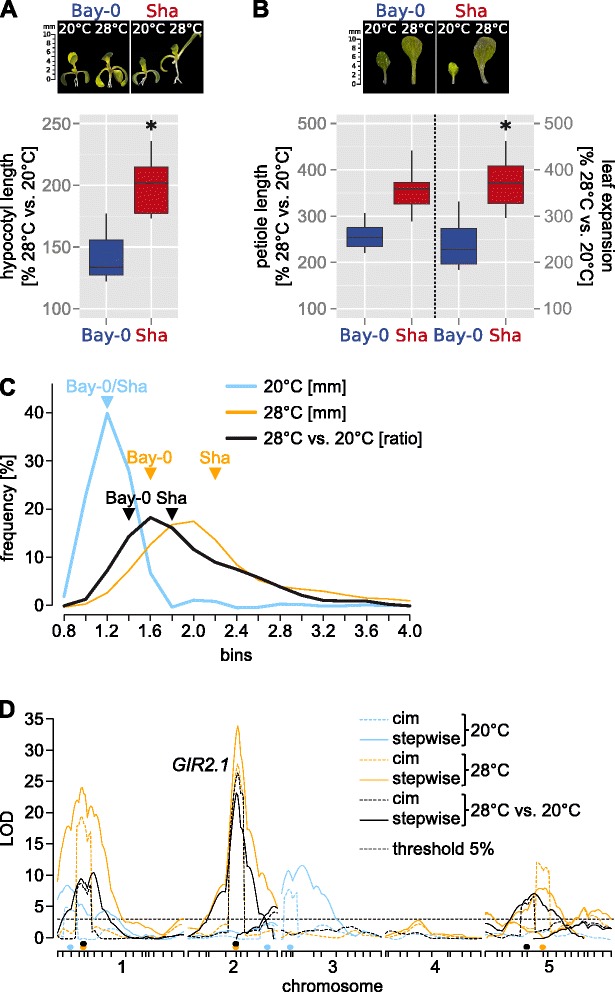
Quantitative trait locus analysis of temperature-induced growth responses in Arabidopsis thaliana. a Relative hypocotyl length (28 °C/20 °C in %, n = 15) of 10 days-old A. thaliana accessions Bay-0 and Sha. b Relative petiole length and leaf expansion of 12 days-old seedlings. a,b Box plots show medians (horizontal bar), interquartile ranges (IQR, boxes), and data ranges (whiskers). Black dots mark outliers (defined as > 1.5 × IQR). Asterisks mark significant differences in temperature responses (P < 0.05) as assessed by two-way ANOVA (i.e. genotype x treatment effect) of the absolute data presented in Additional file 1. c Frequency plot of phenotypic classes observed in a Bay-0 x Sha-derived recombinant inbred line population for hypocotyl length of 10 days-old seedlings grown at 20 °C (n = 400) or 28 °C (n = 395), and for the ratio of 28 vs. 20 °C (n = 387) means. Parental phenotype classes are indicated to illustrate the transgression effects within the population. d LOD scores (y axis) from composite interval mapping (cim) and multiple QTL mapping (stepwiseqtl) are plotted against all chromosomes (x axis). Tick marks on the x axis correspond to molecular markers in the genetic map. Colored dots on the x axis show co-variates set for cim. Thresholds are based on 1000 permutations and an alpha of 0.05. The 28 °C vs. 20 °C GIR2.1 QTL, which is the subject of this study, is highlighted
QTL analysis of temperature-induced hypocotyl elongation
We phenotyped a Bay-0 x Sha recombinant inbred line population [15] for the TIHE response. We grew seedlings in different ambient temperatures (10 days 20 °C vs. 10 days 28 °C) under a long-day diurnal cycle. QTL analysis based on composite interval and multiple QTL mapping using R/qtl was performed for hypocotyl length at either temperature alone or the ratio between hypocotyl length at 28 and 20 °C (28 °C/20 °C). In total, we identified 14 different QTLs (Fig. 1c, d, Additional files 2 and 3). Focusing on growth differences between high and low temperature (28 °C/20 °C ratio) identified five QTLs, which were named GIRAFFE1/2.1/2.2/5.1/5.2 (GIR1, GIR2.1, GIR2.2, GIR5.1, GIR5.2), according to their respective chromosomal location. Together the five QTLs explained ~43 % of the phenotypic variation within the mapping population. The strongest QTL, GIR2.1 (LOD score of 23, stepwiseqtl procedure), explained ~18 % of this variation (Additional files 2 and 3), suggesting that a sizeable part of the natural variation between Bay-0 and Sha can be attributed to this locus.
To facilitate map-based cloning of GIR2.1, we first validated it using heterogeneous inbred families (HIFs; [16]; Fig. 2a, Additional file 4). Phenotypic differences between two HIF lines, carrying either parental allele in the target region, while being otherwise genetically identical, can be attributed to genetic variation in the target QTL interval. In addition to GIR2.1, we also included GIR1 and GIR5.1 in this analysis. We were unable to validate GIR5.1, but observed significant differences in TIHE for the HIF lines separating the two parental alleles for GIR1 (194-B and 194-S) and GIR2.1 (84-B and 84-S) under long-day photoperiod (Fig. 2b, Additional files 4 and 5). Due to the high impact on the phenotypic variation further analyses focused on GIR2.1. Here, the Sha allele conferred long hypocotyls and the Bay-0 allele conferred short hypocotyls (Fig. 2b; interestingly, this situation is reversed for GIR1 [Additional file 4]). We found that the differences in TIHE between the GIR2.1 HIF lines did not persist under continuous light, darkness or short-day conditions (Fig. 2b, Additional file 5). Hence, diurnal cycling with an extensive light phase seems to be necessary for natural variation in TIHE caused by GIR2.1. Furthermore, parental differences under monochromatic lights seem to be independent of GIR2.1 (Additional file 5).
Fig. 2.
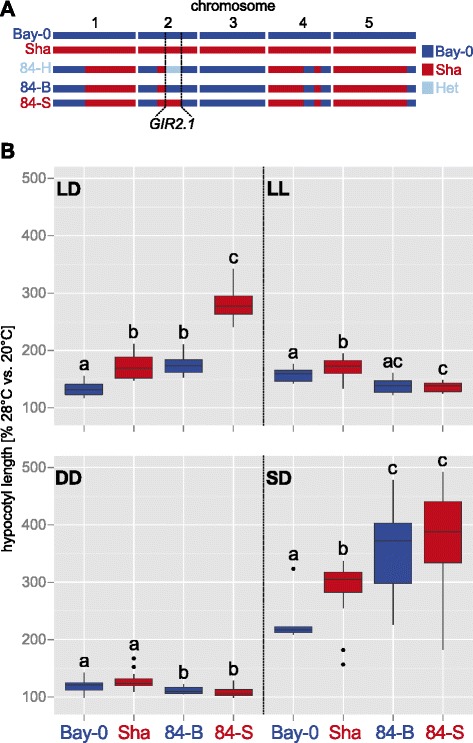
GIR2.1 validation and photoperiod specificity. a Haplotype overview of the heterogeneous inbred family (HIF) 84 that segregates for Bay-0 and Sha alleles within the GIR2.1 interval. b Box plots show relative hypocotyl length (28 °C/20 °C in %, n > 15) of seedlings grown for 8 days under long-day (LD), continuous light (LL), in darkness (DD) or short-day (SD) photoperiods. Horizontal bars, boxes, and whiskers show medians, interquartile ranges (IQR), and data ranges, respectively. Black dots mark outliers (defined as > 1.5 × IQR). Different letters denote statistical differences in temperature responses (within one photoperiod) as assessed by two-way ANOVA (i.e. genotype x treatment effect, P < 0.05) of the absolute data presented in Additional file 5
GIR2.1 possibly constitutes a ghost QTL
An F1 derived by crossing 84-B with 84-S showed that the long-hypocotyl phenotype inherited by the Sha allele is dominant over the Bay-0 short-hypocotyl phenotype (Additional file 5). In the process of fine-mapping the GIR2.1 interval, genotyping of F2 and F3 recombinants with long hypocotyls (longer than 84-B) at 28 °C revealed several plants for which the long-hypocotyl phenotype could be attributed to different, non-overlapping Sha intervals within the GIR2.1 region. This indicated that the exact localization of the GIR2.1 LOD score peak was possibly caused by two or more contributing loci. This phenomenon is frequently observed in QTL analysis and has been called ghost QTL [17]. Recomputing the QTL analysis with additional co-variates separated this peak into two neighbors, supporting this scenario (Additional file 6). Interestingly, Jiménez-Gómez et al. [18] reported a similar phenomenon for this region in the same Bay-0 × Sha population for the regulation of shade avoidance responses, which phenocopies the high temperature response. However, as we only reproducibly observed long hypocotyls for one of the two Sha intervals (chr. 2: 9,199,751-10,426,485 bp), we focused subsequent analyses on this robust interval.
A single nucleotide polymorphism in EARLY FLOWERING3 is underlying the GIR2.1 QTL
Knowing that a diurnal photoperiod was a prerequisite for TIHE differences between Bay-0 and Sha (Fig. 2b), we identified EARLY FLOWERING3 (ELF3) as a candidate gene located in the GIR2.1 target interval. ELF3 is a component of the circadian clock [19] that functions in the evening complex to repress growth [20] and had previously been shown to regulate hypocotyl elongation in response to shade avoidance [18]. Using a transgenic approach, Anwer et al. [11] recently showed that the ELF3Bay-0 and ELF3Sha alleles differentially regulate period length of the circadian clock.
Consistent with a role of ELF3 in thermomorphogenesis, we found that elf3-4 null mutants conferred long hypocotyls in comparison to their Ws-2 wild-type (Fig. 3). To investigate whether TIHE differences between the Bay-0 and Sha HIF lines can indeed be attributed to ELF3, we followed a transgenic complementation approach. We took advantage of the same transgenic lines generated by Anwer et al. [11] that contain either the ELF3Bay-0 or the ELF3Sha allele in the elf3-4 mutant genome (Fig. 3a), which enabled us to study allele-specific TIHE effects in an independent elf3 loss-of-function background. Figure 3b shows that transgenic lines carrying either parental allele complemented the elf3-4 phenotype at 20 °C. At 28 °C, however, transgenics carrying ELF3Bay-0 (elf3-4 [ProBay-0:ELF3Bay-0]) repressed hypocotyl elongation significantly more than those carrying the ELF3Sha allele (elf3-4 [ProSha:ELF3Sha]). This demonstrated that allelic variation in ELF3 affects TIHE in the Bay × Sha population.
Fig. 3.
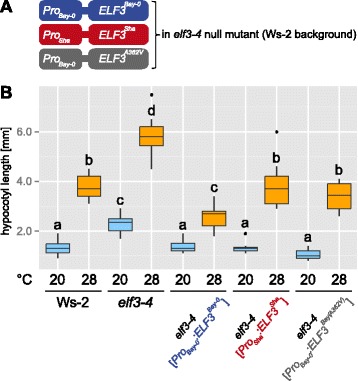
Transgenic complementation of ELF3-mediated TIHE variation. a Overview of transgenic constructs used for complementation of the elf3-4 null mutation (Ws-2 background). b Box plot shows absolute hypocotyl length of 8 days-old seedlings grown in LD at 20 or 28 °C, respectively. Horizontal bars, boxes, and whiskers show medians, interquartile ranges (IQR), and data ranges, respectively. Black dots mark outliers (defined as > 1.5 × IQR). Different letters denote statistical differences as assessed by one-way ANOVA and Tukey HSD (P < 0.05)
Bay-0 and Sha ELF3 variants display several promoter polymorphisms [11] and differ (i) in a nonsynonymous SNP causing an amino acid change at position 362, encoding an alanine-to-valine transition (A362V), and (ii) in the length of a C-terminal glutamine stretch [21, 22]. Although this might depend on the genetic background [23], Tajima et al. [22] suggested that there was no apparent correlation between the length of the polyglutamine region and hypocotyl elongation. We therefore focused on the A362V polymorphism and investigated its potential role in conferring allelic differences in thermomorphogenesis. Again, we made use of transgenic lines in the elf3-4 mutant background generated by Anwer et al. [11]. We inspected TIHE in transgenic elf3-4 lines carrying either the ProBay-0:ELF3Bay-0 allele or the ProBay-0:ELF3Bay(A362V) allele, differing only in the A362V SNP [11]. We found that temperature-induced hypocotyls in transgenic lines carrying a valine at position 362 (as in Sha) were similar to the ELF3Sha allele and, importantly, significantly longer than those with its alanine counterpart at the same position (Fig. 3b). Theoretically, differences in thermomorphogenesis among elf3-4 [ProSha:ELF3Sha] and elf3-4 [ProBay:ELF3Bay] might also be affected by circadian or other differences in ELF3 expression. The significant differences observed between elf3-4 plants expressing the ProBay:ELF3Bay and ProBay-0:ELF3Bay(A362V), however, demonstrate that the SNP underlying the A362V change in ELF3 causes phenotypic variation in TIHE and establishes ELF3 as a negative regulator of thermomorphogenesis.
Differential transcriptional responses caused by natural ELF3 variants
It has recently been shown that the evening complex of the circadian clock consisting of ELF3, ELF4, and LUX ARRHYTHMO (LUX) underlies the molecular basis for circadian gating of hypocotyl growth by directly down-regulating the expression of PIF4 in the early evening [20]. As a result, hypocotyl elongation peaks at dawn under diurnal cycles. We, therefore, tested the hypothesis that adopts this model for ambient temperature signaling and investigated whether PIF4 expression and possibly also the transcript levels of PIF4-regulated genes that mediate cell elongation are affected in response to elevated temperature.
To assess temperature responsiveness, we grew seedlings for 7 days at 20 °C under long-day photoperiods, kept control plates at 20 °C, and shifted the remaining seedlings to 28 °C at lights off (t = 16). Control and 28 °C seedlings were subsequently harvested 4 h after the shift. We found that loss of ELF3 in elf3-4 results in up-regulation of PIF4 transcript levels at both temperatures (Fig. 4a). At 20 °C, we observe that complementation of elf3-4 with either transgenic allele (ProBay-0:ELF3Bay-0; ProSha:ELF3Sha; ProBay-0:ELF3Bay(A362V)) restores wild-type PIF4 levels (Fig. 4a), demonstrating functionality of the constructs, but suggesting that allelic differences are absent at 20 °C. This observation is consistent with similar hypocotyl length of the three lines at 20 °C (Fig. 3b). At 28 °C, however, PIF4 expression levels in elf3-4 [ProSha:ELF3Sha] and elf3-4 [ProBay-0:ELF3Bay(A362V)] are significantly higher than in elf3-4 [ProBay-0:ELF3Bay-0] (Fig. 4a), again reflecting the hypocotyl phenotype (Fig. 3b). Importantly, this expression behavior at 20 vs. 28 °C explains the detection of the GIR2.1 QTL (=28 °C/20 °C ratio), at 28 °C and the absence of a QTL at 20 °C (Fig. 1d, Additional file 3). Together, this suggests that the natural variation observed for TIHE is attributable to temperature-dependent differences in PIF4 expression levels caused by the A362V SNP in ELF3Sha.
Fig. 4.
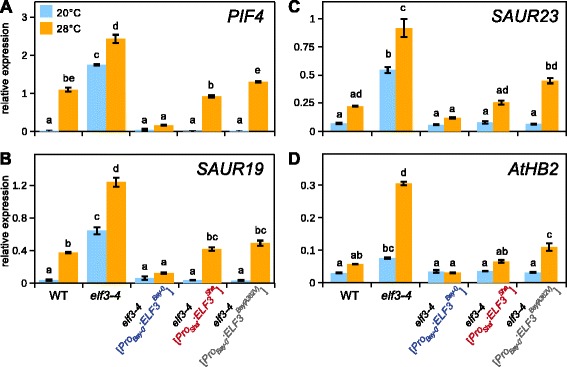
Effects of ELF3 allelic variation on the expression of PIF4 and auxin-responsive genes. qRT-PCR analysis of (a) PIF4, (b) SAUR19, (c) SAUR23, and (d) AtHB2 expression in WT (Ws-2) and transgenic complementation lines (see Fig. 3a). Seedlings were grown for 7 days at 20 °C and transferred to 28 °C or kept at 20 °C (control). Seedlings were harvested after 4 h in the middle of the 8 h dark period. Relative expression levels of three biological replicates per treatment were assessed using At1g13320 as control gene. Bar plots show means and SEM. Different letters denote statistical differences among samples as analyzed by one-way ANOVA and Tukey HSD test (P < 0.05)
We then connected this scenario to the level of PIF4 target genes responsible for cell elongation. Genes involved in cell elongation such as the SMALL AUXIN UPREGULATED RNA (SAUR) family or ARABIDOPSIS THALIANA HOMEOBOX PROTEIN2 (AtHB2) have previously been shown to be activated by PIF4 in a temperature-dependent manner [6, 24, 25]. As Fig. 4b–d shows, we found that temperature-induced expression levels of the tested genes in elf3-4 [ProBay-0:ELF3Bay(A362V)] were significantly higher than those in elf3-4 [ProBay-0:ELF3Bay-0]. This strongly suggested that the A362V SNP causes natural variation of temperature-induced PIF4 expression levels directly resulting in alterations of auxin-mediated cell elongation.
Discussion
Our findings shown here illustrate the power of natural variation approaches and support ELF3 as a negative regulator of ambient temperature signaling. Physiological and gene expression data indicate that ELF3 protein might be involved in down-regulating transcript levels of the major ambient temperature signaling hub PIF4, and thereby affect thermo-responsive growth. Transgenic complementation assays furthermore demonstrated that a nonsynonymous SNP between the natural accessions Bay-0 and Sha significantly affects the ability of ELF3 to regulate temperature-induced PIF4 transcript levels, its target genes, and hypocotyl elongation.
In general, different types of polymorphisms, such as nonsynonymous SNPs or expression level polymorphisms, can contribute to the expression of a particular trait [26]. In line with this phenomenon, distinct types of naturally occurring ELF3 polymorphisms seem to contribute to hypocotyl elongation in response to different temperatures. Box et al. [27] recently used a different quantitative genetic approach based on the MAGIC lines [28], and elegantly showed that both protein-coding and expression level polymorphisms in ELF3 are likely responsible for TIHE differences in natural accessions. The authors presented convincing evidence that warmth relieves the gating of growth by ELF3 at night. Specifically, ELF3 gating of transcriptional targets responds rapidly to changes in temperature by temperature-dependent binding of ELF3 to target promoters including PIF4. Together with Box et al.’s [27] non-transgenic quantitative complementation assays, our transgenic complementations unequivocally establish the role of ELF3 in thermomorphogenesis signaling.
Intriguingly, the ELF3 QTLs in both studies were identified in different photoperiods. ELF3 polymorphisms causal for variation within the MAGIC population were identified under short-day conditions. In contrast, our study identified the ELF3 polymorphism under a long-day photoperiod and subsequent analysis of HIF lines showed long-day specificity (Fig. 2b). Furthermore, a direct comparison of Bay-0 and Sha with two of the parental lines used in the study of Box et al. [27] revealed the short-day-specificity of the Sf-2 and Zu-0 alleles in promoting hypocotyl elongation (Additional file 7). This photoperiod specificity of the different natural alleles represents an interesting observation in itself requesting further investigations. Another unexpected difference between the two studies relates to the growth temperature at which the ELF3 QTL was detected. Whereas we identified the ELF3 QTL peak for hypocotyl growth at 28 °C, but not at 20 °C (Fig. 1d), Box et al. [27] did not detect ELF3 at high temperature but rather at 22 °C standard growth conditions. This difference could be attributed to the differential integration of temperature and photoperiod by natural ELF3 alleles. Alternatively, the genetic backgrounds and interactions with other contributing loci might be involved. In support of this, it is known that the capacity of ELF3 to mediate growth depends on the context of the genome [23]. Hence, Box et al. [27] and this study confer complementary evidence for a central role of ELF3 as a major signaling hub acting upstream of PIF4 in the ambient temperature signaling network and add yet another layer to its complex regulation (Fig. 5).
Fig. 5.
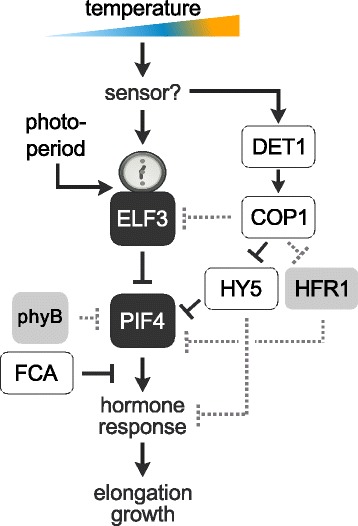
Simplified model of ambient temperature signaling. ELF3 functions as a transcriptional repressor of PIF4 and integrates temperature and photoperiod information in the regulation of thermomorphogenesis. In addition, PIF4 regulation in response to temperature involves the regulatory components of the photomorphogenesis pathway DET1, COP1, and HY5 [10]. PIF4-mediated transcriptional regulation of target genes can be terminated by the RNA binding protein FCA, causing dissociation of PIF4 from target gene promoters [33]. Additional mechanisms previously shown to contribute to PIF4 regulation e.g., transcriptional regulation of target genes via the BAP/HLH/bHLH module of transcription factors [34]. might contribute also to the regulation of temperature responses. Other putatively involved modulators of PIF4 activity known from other biological contexts are depicted by gray dashed lines. These involve direct binding of DET1 to PIF4 [12], competition of PIF4 and other transcription factors for similar binding sites [35], PIF4 protein sequestration by HFR1 [36] and phyB-mediated phosphorylation and degradation of PIF4 [37]
Anwer et al. [11] recently identified ELF3 as a QTL for the regulation of the circadian clock in the same Bay-0 × Sha population. In fact, they showed that the ELF3Sha protein variant failed to properly localize to the nucleus and its ability to accumulate in the dark was compromised. If the same scenario holds for ambient temperature responses, then ELF3Sha might fail to accumulate in the nucleus during nighttime when hypocotyl growth peaks. As a consequence of a weaker potential of ELF3Sha to repress PIF4 expression, a temperature increase could result in a much earlier activation of PIF4-mediated signaling processes during the night. Indeed, Box et al. [27] demonstrated that hypocotyl growth dynamics at elevated temperatures are considerably different from those at standard conditions and show a prolonged growth throughout the first night and an additional growth peak in the beginning of the dark period in subsequent nights. However, it is currently unknown in which manner temperature might affect ELF3 protein localization.
Conclusions
In summary, remarkable progress has been made in understanding the functions of ELF3. In combination with recent studies on the role of ELF3 in the shade avoidance response [18, 21] and the circadian clock [11, 13, 29], this work contributes to understanding its role in the fine-tuned integration of a variety of environmental stimuli that in concert regulate plant growth and development (Fig. 5). Natural variation in thermomorphogenesis caused by ELF3 variants could be mediated at several levels. First, transcriptional regulation of ELF3 itself caused by expression level polymorphisms can result in varying amounts of PIF4-repressing ELF3 protein [27]. In addition, coding sequence polymorphisms might affect the ability of ELF3 protein to interact with PIF4 protein and thereby inhibit its transcriptional activity, as shown by Nieto et al. [13]. However, it is unknown whether this protein-protein interaction is temperature-dependent and the existence of natural variation for this mechanism has yet to be reported. Lastly, nonsynonymous SNPs may affect nuclear accumulation of ELF3, which - like expression level polymorphisms described above - would result in variation of the amount of nuclear ELF3 [11] capable of transcriptionally repressing PIF4, and other targets. Possibly, the latter mechanism is responsible for natural variation between Bay-0 and Sha thermomorphogenesis reported here.
Our work in combination with the study of Box et al. [27] adds further insight into the essential role of ELF3 in integrating multiple signals to promote architectural changes. The photoperiod-specific function of natural ELF3 alleles could provide new avenues to elucidate the clock-mediated growth regulation in general and the ELF3 mode of action specifically.
Methods
Plant material
Plant material used for QTL analyses has been obtained from the Versailles Arabidopsis Stock Center: Bay-0 (accession number 41AV), Sha (236AV), heterogeneous inbred families (HIFs, 33HV84, 33HV194), Bay-0 × Sha population (33RV). Lines used for complementation assays have been described in Anwer et al. [11]. Natural accessions Sf-2 (N6857) and (Zu-0 N6902) were obtained from the Nottingham Arabidopsis Stock Centre.
Thermo-responsive growth assays
Seeds were surface-sterilized and kept in deionized H2O for 3 days at 4 °C before sowing. Seedlings were germinated and grown under sterile conditions and the indicated temperatures on Arabidopsis thaliana solution (ATS) medium [30]. Hypocotyl growth was quantified in seedlings cultivated for 8–10 days under 250 μmol m−2 s−1 white light and a long-day photoperiod (16/8) unless stated otherwise. Hypocotyl length was measured using the RootDetection software package (http://www.labutils.de/). Petiole length and leaf expansion were measured on 12 days-old seedlings using ImageJ. All growth assays including phenotyping of the Bay-0 × Sha population have been repeated at least three times with similar results of which one representative data set is shown.
QTL mapping
Described QTL mapping was applied using Haley-Knott Regression [31] at 1 cM steps with the R/qtl package [32]. Logarithm of odds (LOD) score thresholds were based on 1000 permutations and an alpha error rate of 0.05. Detailed instructions on the QTL mapping procedure are found in Additional file 8. QTL mapping has been performed on all three repetitions of phenotyping of the Bay-0 × Sha mapping population independently with similar results. QTL mapping data of one representative data set are shown. Phenotypic data used for QTL mapping is provided as Additional file 9.
Light response assays
Seeds were surface-sterilized, stratified at 4 °C for 2 days, and dispersed on 0.8 % agar (w/v) Murashige and Skoog medium. Monochromatic red (670 nm peak wavelength, 0.89 μmol m−2 s−1), far-red (735 nm peak wavelength, 0.024 μmol m−2 s−1), or blue light (470 nm peak wavelength, 4.93 μmol m−2 s−1) was generated with an LED SNAP-LITE (Quantum Devices, Barnereld, WI). Green light (540 nm peak wavelength, 0.32 μmol m−2 s−1) was generated from a filtered fluorescent light bulb. Peak wavelength verification and light intensity measurements were done with a SPEC-UV/PAR spectroradiometer (Apogee Instruments, Logan, UT). Plant images were taken after plants were incubated in darkness or under light for the indicated time. Hypocotyl length was measured using ImageJ software.
qRT-PCR
Surface-sterilized seeds were placed on ATS medium and grown for 7 days under long-day photoperiod (16/8) and 100 μmol m−2 s−1 white light at 20 °C. Temperature-induced samples were shifted to 28 °C at dusk, while control plants remained at 20 °C. Samples for qRT-PCR analyses were harvested in the middle of the night 4 h before subjective dawn. Sample preparation and qRT-PCR (including primer sequences) were performed as previously described [10].
Statistical analyses
Statistical differences among absolute length or expression values were analyzed by one-way ANOVA using default parameters in R (www.r-project.org). To assess differences in the treatment response among different genotypes, we used the “genotype x treatment” interaction results of a two-way ANOVA analysis performed on the absolute values in R. Statistically significant differences were highlighted in box plots showing the relative length data.
Acknowledgements
We thank Christine Camilleri and INRA Versailles for providing HIF lines. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to MQ (Qu 141/3-1) and by the Leibniz Association.
Abbreviations
- ATS
Arabidopsis thaliana solution
- AtHB2
ARABIDOPSIS THALIANA HOMEOBOX PROTEIN2
- BAP
BZR-ARF-PIF
- bHLH
Basic helix-loop-helix
- COP1
CONSTITUTIVE PHOTOMORPHOGENIC1
- DD
Darkness
- DET1
DE-ETIOLATED1
- ELF3
EARLY FLOWERING3
- FCA
FLOWERING TIME CONTROL PROTEIN
- GIR
GIRAFFE
- HFR1
LONG HYPOCOTYL IN FAR-RED1
- HIF
Heterogeneous inbred family
- HLH
Helix-loop-helix
- HY5
ELONGATED HYPOCOTYL5
- IQR
Interquartile ranges
- LD
Long day
- LL
Continuous light
- LOD
Logarithm of odds
- LUX
LUX ARRHYTHMO
- PIF4
PHYTOCHROME-INTERACTING FACTOR4
- QTL
Quantitative trait locus
- SAUR
SMALL AUXIN UPREGULATED RNA
- SD
Short day
- SNP
Single nucleotide polymorphism
- TIHE
Temperature-induced hypocotyl elongation
Additional files
Temperature-induced growth responses in Bay-0 and Sha. Absolute values for (A) temperature-induced hypocotyl elongation (TIHE), (B) temperature-induced petiole elongation (TIPE), and (C) temperature-induced leaf expansion (TILE). Data correspond to the relative data presented in Fig. 1. Box plots show medians (horizontal bar), interquartile ranges (IQR, boxes), and data ranges (whiskers). Black dots mark outliers (defined as > 1.5 × IQR). Statistical differences were assessed by one-way ANOVA and Tukey HSD (P < 0.05) and are denoted by different lower- and uppercase letters for 20 °C and 28 °C values, respectively.
Descriptive statistics of phenotypes analyzed in the Bay-0 × Sha population.
QTL summary statistics.
Validation of the GIR1 QTL. (A) Haplotype overview of the heterogeneous inbred family (HIF) 194 that segregates for Bay-0 and Sha within the GIR1 interval and was used for validation and mapping of the GIR1 QTL. (B) Box plots show relative (28 °C/20 °C in %) hypocotyl length of 10 days-old seedlings derived from the absolute hypocotyl length data presented in (C). (D) Haplotype overview of the heterogenous inbred family (HIF) 214 that segregates for Bay-0 and Sha within the GIR5.1 interval and served for the attempted validation of this QTL. (E) Box plots show relative (28 °C/20 °C in %) hypocotyl length of 10 days-old seedlings derived from the absolute hypocotyl length data presented in (F). (B,C,E,F) Horizontal bars, boxes, and whiskers show medians, interquartile ranges (IQR), and data ranges, respectively. Black dots mark outliers (defined as > 1.5 × IQR). (B+E) Different letters denote statistical differences in temperature responses as assessed by two-way ANOVA (P < 0.05) of the absolute hypocotyl length data. (C+F) Statistical differences were assessed by one-way ANOVA and Tukey HSD (P < 0.05) and are denoted by different lower- and uppercase letters for 20 °C and 28 °C values, respectively. The significant differences in TIHE observed for the parental lines Bay-0 and Sha was not reflected by the two HIF lines 214-B and 214-S that carried a Bay-0 or Sha allele within the GIR5.1 interval, respectively. As such, the GIR5.1 QTL could not be validated with the available genetic material.
Effect of altered light conditions on GIR2.1-mediated hypocotyl elongation. (A) Box plots show relative (28 °C/20 °C in %) and absolute hypocotyl length of 10 days-old seedlings of Bay-0, Sha, and HIF lines homozygous for either Bay-0 (84-B) or Sha (84-S) in the GIR2.1 interval. Horizontal bars, boxes, and whiskers show medians, interquartile ranges (IQR), and data ranges, respectively. Black dots mark outliers (defined as > 1.5 × IQR). F1 plants derived from a cross of 84-B and 84-S correspond to the haplotype 84-H in Fig. 2a and illustrate the dominance of the Sha over the Bay-0 allele. Different letters denote statistical differences in temperature responses as assessed by two-way ANOVA (i.e. genotype x treatment effect, P < 0.05) of the absolute hypocotyl length data. (B) Absolute hypocotyl length corresponding to the relative data presented in Fig. 2b. Statistical differences were assessed by one-way ANOVA and Tukey HSD (P < 0.05) and are denoted by different lower- and uppercase letters for 20 °C and 28 °C values, respectively. (C) Hypocotyl length in monochromatic light conditions. Significant differences among Bay-0 and Sha are observed in 4 days-old seedlings grown at 20 °C in constant blue (4.93 μmol m−2 sec−1), green (0.32 μmol m−2 sec−1), or red (0.89 μmol m−2 sec−1) light. These differences seem to be regulated independent of GIR2.1 as 84-B and 84-S did not differ in their growth response. No differences among genotypes were detected in seedlings grown in far-red (0.024 μmol m−2 sec−1) light. Statistical differences were assessed by one-way ANOVA and Tukey HSD (P < 0.05) and are denoted by different lower- and uppercase letters for 20 °C and 28 °C values, respectively.
GIR2 constitutes a ghost QTL. Setting additional covariates in the GIR2.1 target region separates the single peak into two linked peaks (compare with Fig. 1d), indicating the potential existence of two linked loci. Tick marks on the x axis correspond to molecular markers in the genetic map of the Bay-0 and Sha mapping population. Circles on the x axis show co-variates set for composite interval mapping.
Photoperiod and allele effects on ELF3-mediated TIHE. TIHE comparison of 7 days-old seedlings grown either in short day (SD) or long day (LD) photoperiods. Box plots show (A) relative (28/20 °C in %) and (B) absolute hypocotyl length for Bay-0, Sha and MAGIC population parental lines Sf-2 and Zu-0 that also carry polymorphisms in ELF3. While Sf-2 and Zu-0 show a strong TIHE response in SD, the response for Sf-2 and Zu-0 is much weaker under LD. Horizontal bars, boxes, and whiskers show medians, interquartile ranges (IQR), and data ranges, respectively. Black dots mark outliers (defined as > 1.5 × IQR). (A) Different letters in denote statistical differences in temperature responses as assessed by two-way ANOVA (i.e. genotype x treatment effect, P < 0.05) of the absolute hypocotyl length data. (B) Statistical differences were assessed by one-way ANOVA and Tukey HSD (P < 0.05) and are denoted by different lower- and uppercase letters for 20 °C and 28 °C values, respectively.
Methods S1. Estimation of heritability and QTL analysis procedure.
Supplementary Dataset 1. Phenotypic data used for QTL mapping. This dataset has been uploaded to figshare and can be accessed via http://dx.doi.org/10.6084/m9.figshare.1339892.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MQ designed the study. AR, CI, SB, AG, JT, KD, and CD performed the thermo-responsive growth assays. KKU and BS conducted the QTL analysis. AR and CI fine-mapped the GIR2.1 QTL. CI and CD conducted the qRT-PCR analysis. XS and MN performed the light response assays. MUA and SJD contributed materials and edited the manuscript. AR, CI, CD and MQ wrote the manuscript. All authors read and approved the manuscript.
Authors’ information
Anja Raschke and Carla Ibañez Co-first authors.
Contributor Information
Anja Raschke, Email: anja.raschke@landw.uni-halle.de.
Carla Ibañez, Email: cibanez@ipb-halle.de.
Kristian Karsten Ullrich, Email: Kristian.Ullrich@biologie.uni-marburg.de.
Muhammad Usman Anwer, Email: anwer@mpipz.mpg.de.
Sebastian Becker, Email: sebastian.becker@student.uni-halle.de.
Annemarie Glöckner, Email: annemarie.gloeckner@uni-hamburg.de.
Jana Trenner, Email: Jana.Trenner@ipb-halle.de.
Kathrin Denk, Email: kdenk@ipb-halle.de.
Bernhard Saal, Email: saal@baypmuc.de.
Xiaodong Sun, Email: xdsun@plantsciences.ucdavis.edu.
Min Ni, Email: nixxx008@umn.edu.
Seth Jon Davis, Email: seth.davis@york.ac.uk.
Carolin Delker, Email: cdelker@ipb-halle.de.
Marcel Quint, Email: mquint@ipb-halle.de.
References
- 1.Erwin JE, Heins RD, Karlsson MG. Thermomorphogenesis in Lilium longiflorum. Am J Bot. 1989;76:47–52. doi: 10.2307/2444772. [DOI] [Google Scholar]
- 2.Gray WM, Östin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci U S A. 1998;95:7197–202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Zanten M, Voesenek LACJ, Peeters AJM, Millenaar FF. Hormone- and light-mediated regulation of heat-induced differential petiole growth in arabidopsis. Plant Physiol. 2009;151:1446–58. doi: 10.1104/pp.109.144386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford AJ, McLachlan DH, Hetherington AM, Franklin KA. High temperature exposure increases plant cooling capacity. Curr Biol. 2012;22:R396–7. doi: 10.1016/j.cub.2012.03.044. [DOI] [PubMed] [Google Scholar]
- 5.Bridge LJ, Franklin KA, Homer ME. Impact of plant shoot architecture on leaf cooling: a coupled heat and mass transfer model. J R Soc Interface. 2013;10:20130326. doi: 10.1098/rsif.2013.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci U S A. 2011;108:20231–35. [DOI] [PMC free article] [PubMed]
- 7.Sun J, Qi L, Li Y, Chu J, Li C. PIF4–mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Proveniers MCG, van Zanten M. High temperature acclimation through PIF4 signaling. Trends Plant Sci. 2013;18:59–64. doi: 10.1016/j.tplants.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 9.De Wit M, Lorrain S, Fankhauser C. Auxin-mediated plant architectural changes in response to shade and high temperature. Physiol Plant. 2014;151:13–24. doi: 10.1111/ppl.12099. [DOI] [PubMed] [Google Scholar]
- 10.Delker C, Sonntag L, James GV, Janitza P, Ibañez C, Ziermann H, et al. The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 2014;9:1983–89. [DOI] [PubMed]
- 11.Anwer MU, Boikoglou E, Herrero E, Hallstein M, Davis AM, James GV, et al. Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife. 2014;3:e02206. [DOI] [PMC free article] [PubMed]
- 12.Dong J, Tang D, Gao Z, Yu R, Li K, He H, et al. Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell. 2014;26:3630–45. [DOI] [PMC free article] [PubMed]
- 13.Nieto C, López-Salmerón V, Davière J-M. Prat pp. ELF3–PIF4 interaction regulates plant growth independently of the Evening Complex. Curr Biol. 2015;25:187–193. doi: 10.1016/j.cub.2014.10.070. [DOI] [PubMed] [Google Scholar]
- 14.Delker C, Pöschl Y, Raschke A, Ullrich K, Ettingshausen S, Hauptmann V, et al. Natural variation of transcriptional auxin response networks in Arabidopsis thaliana. Plant Cell. 2010;22:2184–200. [DOI] [PMC free article] [PubMed]
- 15.Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet. 2002;104:1173––84. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- 16.Tuinstra MR, Ejeta G, Goldsbrough PB. Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet. 1997;95:1005–11. doi: 10.1007/s001220050654. [DOI] [Google Scholar]
- 17.Martínez O, Curnow RN. Estimating the locations and the sizes of the effects of quantitative trait loci using flanking markers. Theor Appl Genet. 1992;85:480–8. doi: 10.1007/BF00222330. [DOI] [PubMed] [Google Scholar]
- 18.Jiménez-Gómez JM, Wallace AD, Maloof JN. Network analysis identifies ELF3 as a QTL for the shade avoidance response in arabidopsis. PLoS Genet. 2010;6:e1001100. doi: 10.1371/journal.pgen.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, et al. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell. 2012;24:428–43. [DOI] [PMC free article] [PubMed]
- 20.Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. [DOI] [PMC free article] [PubMed]
- 21.Coluccio MP, Sanchez SE, Kasulin L, Yanovsky MJ, Botto JF. Genetic mapping of natural variation in a shade avoidance response: ELF3 is the candidate gene for a QTL in hypocotyl growth regulation. J Exp Bot. 2011;62:167–76. doi: 10.1093/jxb/erq253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tajima T, Oda A, Nakagawa M, Kamada H, Mizoguchi T. Natural variation of polyglutamine repeats of a circadian clock gene ELF3 in Arabidopsis. Plant Biotechnol. 2007;24:237–40. doi: 10.5511/plantbiotechnology.24.237. [DOI] [Google Scholar]
- 23.Undurraga SF, Press MO, Legendre M, Bujdoso N, Bale J, Wang H, et al. Background-dependent effects of polyglutamine variation in the Arabidopsis thaliana gene ELF3. Proc Natl Acad Sci U S A. 2012;109:19363–7. [DOI] [PMC free article] [PubMed]
- 24.Kunihiro A, Yamashino T, Nakamichi N, Niwa Y, Nakanishi H, Mizuno T. PHYTOCHROME-INTERACTING FACTOR 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol. 2011;52:1315–29. doi: 10.1093/pcp/pcr076. [DOI] [PubMed] [Google Scholar]
- 25.Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inze D, et al. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012;70:978–90. [DOI] [PMC free article] [PubMed]
- 26.Delker C, Quint M. Expression level polymorphisms: heritable traits shaping natural variation. Trends Plant Sci. 2011;16:481–8. doi: 10.1016/j.tplants.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, et al. ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol. 2015;25:194–9. [DOI] [PubMed]
- 28.Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, et al. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000551. [DOI] [PMC free article] [PubMed]
- 29.Mizuno T, Nomoto Y, Oka H, Kitayama M, Takeuchi A, Tsubouchi M, et al. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC nighttime repressor in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:958–76. [DOI] [PubMed]
- 30.Lincoln C, Britton J, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–80. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–24. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- 32.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–90. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 33.Lee H-J, Jung J-H, Cortés Llorca L, Kim S-G, Lee S, Baldwin IT, et al. FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat Commun. 2014;5:5473. [DOI] [PubMed]
- 34.Oh E, Zhu J-Y, Bai M-Y, Arenhart RA, Sun Y, Wang Z-Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife. 2014;3:e03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, et al. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 2014;10:e1004416. [DOI] [PMC free article] [PubMed]
- 36.Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28:3893–902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–23. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]


