Abstract
Protein interaction surface mapping using MS is widely applied but comparatively resource intensive. Here a workflow adaptation for use of isotope coded tandem mass tags for the purpose is reported. The key benefit of improved throughput derived from sample acquisition multiplexing and automated analysis is shown to be maintained in the new application. Mapping of the epitopes of two monoclonal antibodies on their respective targets serves to illustrate the novel approach. We conclude that the approach enables mapping of interactions by MS at significantly larger scales than hereto possible.
Most proteins do not function in isolation in living organisms, which is why mapping of protein interactions with other proteins and cellular components has become instrumental to our understanding of biologic systems. Knowledge of the detailed nature of the interfaces of such interactions is key to understanding the molecular mechanisms at work and is often crucial to therapeutic intervention.
High-resolution approaches such as X-ray crystallography and NMR are able to define the atomic contacts but suffer from limited throughput and high cost. Various alternative technologies including peptide arrays1, cell-surface display2–4, and mass spectrometry (MS) based methods have been used to elucidate protein interfaces, however there are significant tradeoffs in terms of definition, throughput, and cost associated with each of these strategies.
Mass spectrometry based methods have become widespread due to their adaptability to different kind of interactions and their ability to elucidate even complex interfaces with moderate material requirements and throughput in potentially complex environments. MS approaches generally rely on protein surface labeling to deduce interaction sites by comparing labeling and/or enzymatic cleavage patterns between apo-protein and the respective holo complexes5, or the identification of cross-linked peptides6–11. Surface labeling methods used to characterize protein interaction sites and epitopes include hydrogen deuterium exchange (HDx)12–21, hydroxyl radical labeling17,22–33, and amino acid labeling34–39 approaches. The complexity of the labeling product mixtures depend on reaction chemistry and site-specificity of the reagents and has an immediate bearing on the time required for data analysis, which in a lot of cases is the throughput limiting factor.
In this work, we show that commercially available isotope coded reagents originally designed for proteome quantitation can be used for rapid characterization of protein interactions. Improved throughput is obtained using the multiplexing capabilities of the isotope coded reagents and the availability of automated analysis software. The new application is demonstrated by profiling the epitopes of two monoclonal antibodies on their respective antigens.
EXPERIMENTAL
Materials
Formic acid, acetonitrile (ACN), and M2 anti-FLAG monoclonal antibody were purchased from Sigma Chemical Company (St. Louis, MO). Tandem Mass Tag (TMT) reagents were obtained from Thermo Scientific (Waltham, MA). Green Fluorescent Protein (GFP) with a N-terminal 6xHis-TEV-3xFlag tandem purification tag sequence of MGSDKIHHHHHHENLYFQGDYKDHDGDYKDHDIDYKDDDDK), human ErbB2 D3-4 (residues R340-R647), anti-ErbB2 monoclonal antibody (Trastuzumab variable domains and specificity), and anti-Lysozyme monoclonal antibody were expressed using standard methods. The GFP construct is referred to as 3xFlag-GFP from hereon.
Epitope Mapping
All samples were buffer exchanged and concentrated into 100mM triethylamine bicarbonate (TEAB), pH8.0, 150mM NaCl using 10kDa molecular weight cutoff micro-concentrators (Milipore Amicon Ultra). 3xFlag-GFP/anti-Lysozyme mAb, 3xFlag-GFP/M2 mAb and ErbB2/anti-ErbB2 mAb complexes were prepared by mixing an equimolar amount of each antigen with the matching mAb. Complexes were equilibrated for 30 min. at room temp before labeling.
For mapping of the M2 anti-Flag epitope on the 3xFlag-GFP protein, duplicate samples of the antigen (0.5µM in 95µL TEAB buffer) were labeled with 5µL TMT reagents 126 and 127 in anhydrous acetonitrile at four time points (10, 30, 60, 300s). Isotope coding was used to facilitate technical repeats. Duplicates of the 3xFlag-GFP/anti-Lysozyme mAb and 3xFlag-GFP/M2 mAb complexes were then labeled with the 128 and 129, and 130 and 131 reagents at the same time points respectively.
The anti-ErbB2 mAb epitope on ErbB2 D3-4 was mapped by labeling of the antigen (0.5 µM in 95µL TEAB buffer) in triplicate using 5µL TMT reagents 126, 127, and 128 in anhydrous acetonitrile. A single 60s time point was evaluated. Similarly, triplicate samples of the ErbB2 D3-4/anti-ErbB2 mAb complex were labeled with the 129, 130 and 131 reagents.
Each labeling reaction was quenched with 25uL 5% hydroxylamine and equal volumes of the six labeling reactions were mixed together. Samples were deglycosylated with PNGaseF (according to the manufacturer’s directions), and run on an SDS-PAGE gel to separate components. The antigen bands (3xFlag-GFP, and ErbB2) were cut out and in-gel digested with elastase. Peptide digests were analyzed by LC-MS/MS on a QExactive mass spectrometer (Thermo Scientific, Waltham, MA).
LC-MS/MS
HPLC was performed using a vented column setup40, with a 2cm Poros 10 R2 (Life Technologies, Carlsbad, CA) self-packed pre-column, and a PepMap EASY-Spray C18 analytical column (Thermo Scientific, Waltham, MA). Proteolytic fragments were eluted using a gradient from 2 to 40% acetonitrile / 0.1% formic acid in 120min at a flow rate of 300ηL/min. The chromatography system was coupled with a QExactive mass spectrometer (Thermo Scientific, Waltham, MA). Data-dependent MS/MS experiments were performed for peptide identification.
Data Analysis
Proteome Discoverer 1.4 was used for peptide identification by Mascot and Sequest-HT database searching. Search parameters included fixed carbamidomethyl modification of cysteine, and variable oxidation of methionine, deamidation of N, pyroGlu of n-terminal Q, and TMT(6-plex) modification of K residues. Scaffold 4 Q+ was used for filtering search results and quantification of reporter ion intensities. The Scaffold quantitative settings used included normalization between samples based on an average protein reference. For each spectrum, average ratios were determined from the replicate channels. Relative sample abundances were compared by dividing the average reporter ion ratio of the complex samples by the average ratio of the control samples. Then, the ratios were averaged for each spectrum derived from the same peptide and charge-state. To determine the ratio of a particular lysine residue, the average ratios from peptides that contained the same modified lysine residue were averaged. A macro written in Microsoft Excel VBA was used to compile quantification reports, and map peptide data to individual lysine residues.
Intact LC-MS
Intact proteins were analyzed by LC-MS using an Agilent 1200 series HPLC system and an Agilent 6520 QTOF mass spectrometer. The system was configured with a PLRP-S, 1000Å, 2.1 × 50mm, 8µm particle column (Agilent, Santa Clara, CA) and was operated at a flow rate of 500µl/min. A linear gradient of acetonitrile containing 0.1% formic acid was delivered over 1min after sample loading. Average spectra were de-convoluted using the MassHunter Qualitative Analysis software.
RESULTS AND DISCUSSION
Protein surface labeling is controlled by structure, local pH, temperature and a number of other factors whose interplay in controlling labeling rates at each specific site are poorly understood. A detailed analysis of TMT labeling and the observed reactivity difference as well as the difficulties involved in correlating rates with structure derived parameters such as solvent-accessible surface area was recently published41. Therefore, protein surface labeling experiments typically employ a time course as signals for various sites maximize at different labeling times. As labeling experiments integrate over the full folding equilibrium, which is potentially disturbed by the labeling process, interpretations based on a single structure or protein conformer might be flawed. Nevertheless, a wealth of data is now available in the literature showing that in most cases useful protein interaction site information can be obtained by a simple differential experiment that compares the labeling observed between the constituents of a complex labeled independently with the degree of labeling observed when the complex is labeled under identical conditions. The differential labeling observed in such an experiment is interpreted on the basis of the simplistic model that surfaces participating in a protein interaction are protected from labeling due to shielding in the complex by the binding partner, whereas they are “freely” accessible if the constituents are labeled alone. This model does not provide a means to explain potentially increased labeling rates in the complex and can’t account for differences resulting from conformational change in the complex induced by the binding event. The simple assumption is that sequences participating in a protein-protein interface are those that show the largest protection signals in the difference experiment. If no dominant protection signals are observed inference of an interfacial region is not possible.
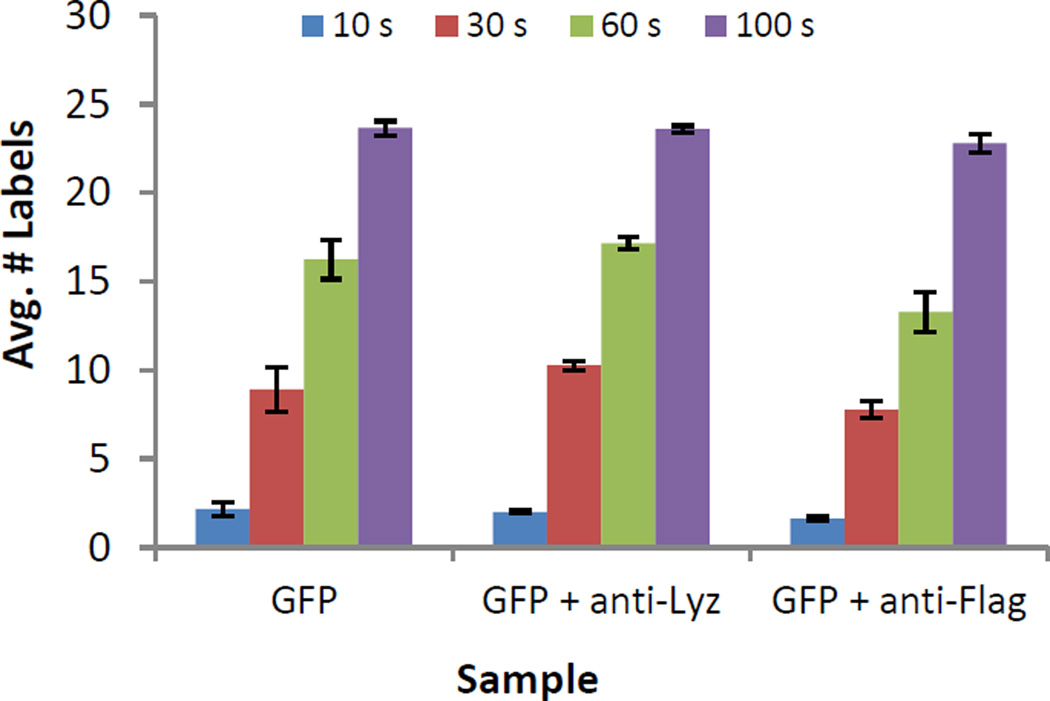
Isotope coded reagents that derivatize amino groups are commonly used for the MS based quantification of complex peptide mixtures42,43, and have been previously employed to label intact proteins44–48. We evaluated both iTRAQ and TMT reagents for intact protein labeling and found TMT reagents to label more efficiently in the low percentage organic conditions we employed. Fig 1. shows an example of TMT labeling of an intact 3xFlag-GFP construct in the absence and presence of an anti-Lysozyme mAb and an anti-Flag mAb. Labeling was determined as being close to quantitative after 5 min and resulted in the addition of 23.6 TMT labels on average to 3xFlag-GFP (Fig. 1, 3xFlag-GFP contains 26 free amines). This shows that the lysines of GFP are rapidly derivatized.
Figure 1.
Average number of TMT labels on 3xFlag-GFP measured by intact mass spectrometry at four time points (10, 30, 60, and 300s). 3xFlag-GFP has 25 lysine residues (error bars correspond to 1 standard deviation).
The presence of the control anti-Lysozyme mAb did not reduce TMT labeling as compared to the labeling of GFP alone. However, the average number of TMT labels was reduced in the 3xFlag-GFP/M2 anti-Flag mAb complex in comparison to the controls (GFP and GFP+ anti-Lyz, Figure 1). The difference in the number of TMT labels was greatest at the 60s time point, at which labeling was reduced by ~4 TMT in comparison to the controls. This result is consistent with protection of several lysine residues in the 3xFlag-GFP construct by the M2 mAb, whose epitope is known to comprise the DYKD motif49.
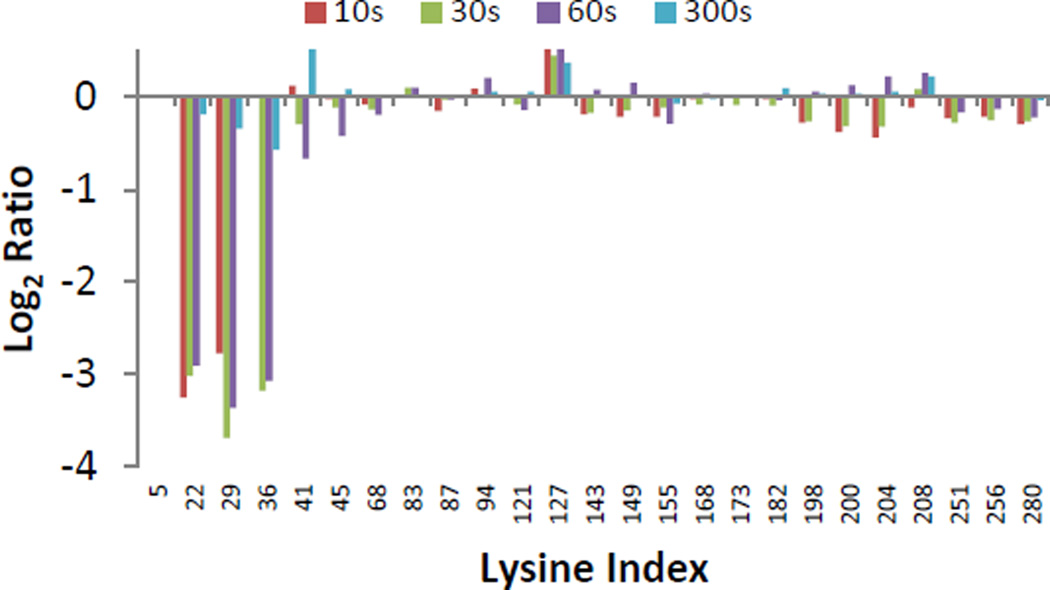
To elucidate precisely which lysine residues were protected, we subjected the derivatized 3xFlag-GFP protein to proteolytic digestion and quantified the resulting peptide mixture. It should be noted that trypsin does not cleave after modified lysine residues which necessitates the use of proteases with alternative specificity like pepsin, elastase or chymotrypsin. Peptide reporter ion abundances, derived from in-gel elastase digests, were compared between the 3xFlag-GFP, 3xFlag-GFP/anti-Lyz, and the 3xFlag-GFP/anti-Flag mAb complexes. Ratios of reporter ion intensities of the 3xFlag-GFP/anti-Flag complex compared to the controls were determined for peptides containing all but one of the lysine residues in the 3xFlag-GFP construct (Fig. 2, K5 and the n-terminus were not covered by the data). The ratios for 3xFlag-GFP and 3xFlag-GFP/anti-Lyz mAb complex were identical within experimental error and only data for 3xFlag-GFP is shown in Figure 2. In this strategy, ratios below 1 (log2 < 0) represent a decrease in the extent of TMT derivatization. The M2 anti-Flag mAb clearly protects several lysine residues including K22, K29, and K36, all of which are found within the DYKD motif of the known epitope of this mAb. No major differences in the extent of labeling could be discerned between the three aforementioned lysines, suggesting that all three Flag repeats are similarly blocked by the M2 anti-Flag mAb. This result is in agreement with data obtained on the complex using hydrogen deuterium exchange mass spectrometry50. The disappearance of the difference signal at 300s is consistent with competitive elution of FLAG tagged proteins with FLAG peptide in protein purification applications and reported off-rates51,52.
Figure 2.
Log2 of the ratio of average reporter ion intensities for labeled lysine side chains in the 3xFlag-GFP + anti-Flag M2 mAb complex compared to the 3xFlag-GFP control.
A second test was performed in the ErbB2/anti-ErbB2 system. We chose this target for a proof of concept study because of the availability of high quality structures.
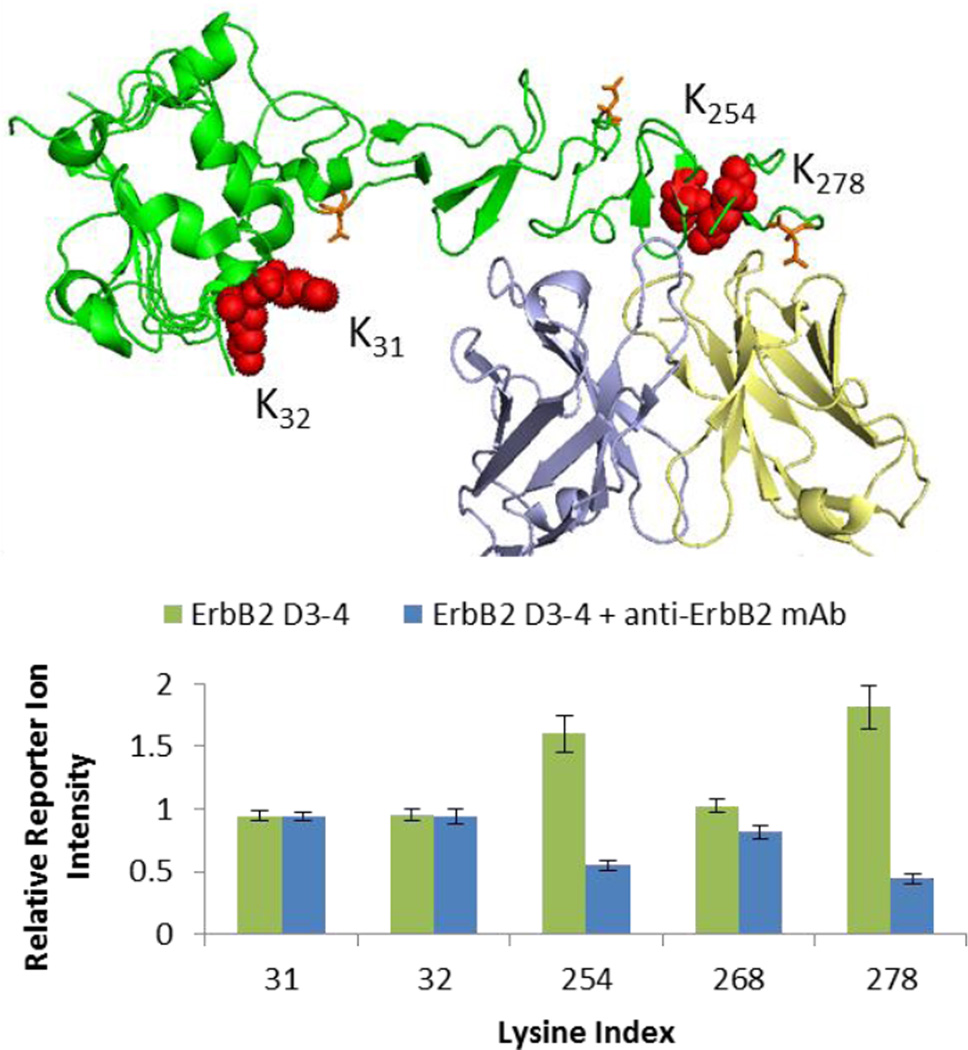
Reporter ion ratios of modified peptides covering five of the six lysines of ErbB2 D3-4 could be mapped to specific lysine residues as shown in Figure 3. Panel (b) of Figure 3 shows strong, protection of K254 and K278 as indicated by the dramatically lower relative reporter ion intensities observed for the complex (blue columns). The protected residues are buried in the protein interface of the complex as seen in the structure shown in Figure 3 (a). In contrast, the two remotely located lysine residues K31 and K32 do not show any protection within experimental error in good agreement with expectations. The only other quantifiable lysine residue K268 is not observed in the available structure data but shows minor protection suggesting it is only indirectly affected by antibody binding.
Figure 3.
(a) Structure (PDB: 1N8Z) of ErbB2 D3-4 (green) in complex with anti-ErbB2 Fab (light chain yellow, heavy chain blue, N-linked glycans orange sticks, lysines red balls). (b) Average relative reporter ion intensities for control (green columns) and complex (blue columns) of ErbB2 lysines (error bars correspond to 1 standard deviation).
The two mapping studies shown above are proof of concept examples of protein interaction mapping with TMT reagents. The major advantages of using isotope coded labeling reagents for proteome quantitation are multiplexing and the availability of automated analysis tools, which allows greatly improved throughput. The examples show that those advantages can be translated to protein interaction studies, which opens up the potential for mapping of tenth to hundredth of interactions within a few days. The stability of the TMT modification does not put any limitations on the post-labeling processing of samples, facilitating the analysis of even heavily postranslationally modified targets using standard proteomics sample processing strategies (i.e. reduction, deglycosylation, etc.). A target’s inherent lysine distribution thus remains the major limitation for comprehensive interaction profiling. However, because of the relatively large number of lysine residues in most proteins, and the propensity of lysine residues to be at the surface, this method should still be widely applicable.
CONCLUSION
We have demonstrated that tandem mass tags can be used to identify protein interaction sites by quantifying the relative protection observed toward labeling of lysine residues. The strategy was successfully demonstrated in an epitope mapping setting. The method translates the design features of isotope encoded mass tags into the new application and ameliorates throughput issues of surface labeling methods at both sample acquisition and data analysis stages for a compounding of benefits. As it was shown that all the benefits of isotope coded labels that enable large scale proteomic quantitation studies are also applicable in a protein interaction setting, it is expected that the demonstrated workflow facilitates protein interaction mapping at considerably larger scales, too.
ACKNOWLEDGMENT
This work was supported by the NIH, National Institute of General Medical Sciences, Protein Structure Initiative Grant U54 GM094586. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Kazim AL, Atassi MZ. The Biochemical journal. 1981;197:507–510. doi: 10.1042/bj1970507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larman HB, Zhao Z, Laserson U, Li MZ, Ciccia A, Gakidis MA, Church GM, Kesari S, Leproust EM, Solimini NL, Elledge SJ. Nature biotechnology. 2011;29:535–541. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christmann A, Wentzel A, Meyer C, Meyers G, Kolmar H. Journal of immunological methods. 2001;257:163–173. doi: 10.1016/s0022-1759(01)00461-6. [DOI] [PubMed] [Google Scholar]
- 4.Chao G, Cochran JR, Wittrup KD. Journal of molecular biology. 2004;342:539–550. doi: 10.1016/j.jmb.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 5.Papac DI, Hoyes J, Tomer KB. Protein science : a publication of the Protein Society. 1994;3:1485–1492. doi: 10.1002/pro.5560030914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Back JW, de Jong L, Muijsers AO, de Koster CG. Journal of molecular biology. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 7.Petrotchenko EV, Borchers CH. Mass spectrometry reviews. 2010;29:862–876. doi: 10.1002/mas.20293. [DOI] [PubMed] [Google Scholar]
- 8.Rappsilber J. Expert review of proteomics. 2012;9:485–487. doi: 10.1586/epr.12.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh P, Panchaud A, Goodlett DR. Analytical chemistry. 2010;82:2636–2642. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- 10.Sinz A. Mass spectrometry reviews. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 11.Sinz A. Analytical and bioanalytical chemistry. 2010;397:3433–3440. doi: 10.1007/s00216-009-3405-5. [DOI] [PubMed] [Google Scholar]
- 12.Chalmers MJ, Busby SA, Pascal BD, West GM, Griffin PR. Expert review of proteomics. 2011;8:43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engen JR. Analytical chemistry. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Englander SW. Journal of the American Society for Mass Spectrometry. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoofnagle AN, Resing KA, Ahn NG. Annual review of biophysics and biomolecular structure. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 16.Konermann L, Pan J, Liu YH. Chemical Society reviews. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 17.Konermann L, Tong X, Pan Y. Journal of mass spectrometry : JMS. 2008;43:1021–1036. doi: 10.1002/jms.1435. [DOI] [PubMed] [Google Scholar]
- 18.Marcsisin SR, Engen JR. Analytical and bioanalytical chemistry. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Percy AJ, Rey M, Burns KM, Schriemer DC. Analytica chimica acta. 2012;721:7–21. doi: 10.1016/j.aca.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 20.Tsutsui Y, Wintrode PL. Current medicinal chemistry. 2007;14:2344–2358. doi: 10.2174/092986707781745596. [DOI] [PubMed] [Google Scholar]
- 21.Wales TE, Engen JR. Mass spectrometry reviews. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Rempel DL, Gau BC, Gross ML. Journal of the American Chemical Society. 2012;134:18724–18731. doi: 10.1021/ja307606f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Rempel DL, Gross ML. Journal of the American Chemical Society. 2010;132:15502–15504. doi: 10.1021/ja106518d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gau BC, Sharp JS, Rempel DL, Gross ML. Analytical chemistry. 2009;81:6563–6571. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan JQ, Vorobiev S, Almo SC, Chance MR. Biochemistry. 2002;41:5765–5775. doi: 10.1021/bi0121104. [DOI] [PubMed] [Google Scholar]
- 26.Hambly DM, Gross ML. Journal of the American Society for Mass Spectrometry. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Konermann L, Pan Y. Expert review of proteomics. 2012;9:497–504. doi: 10.1586/epr.12.42. [DOI] [PubMed] [Google Scholar]
- 28.Konermann L, Stocks BB, Pan Y, Tong X. Mass spectrometry reviews. 2010;29:651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 29.Maleknia SD, Downard K. Mass spectrometry reviews. 2001;20:388–401. doi: 10.1002/mas.10013. [DOI] [PubMed] [Google Scholar]
- 30.Takamoto K, Chance MR. Annual review of biophysics and biomolecular structure. 2006;35:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 31.Wong JW, Maleknia SD, Downard KM. Analytical chemistry. 2003;75:1557–1563. doi: 10.1021/ac026400h. [DOI] [PubMed] [Google Scholar]
- 32.Wong JW, Maleknia SD, Downard KM. Journal of the American Society for Mass Spectrometry. 2005;16:225–233. doi: 10.1016/j.jasms.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Xu G, Chance MR. Chemical reviews. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 34.Mendoza VL, Antwi K, Baron-Rodriguez MA, Blanco C, Vachet RW. Biochemistry. 2010;49:1522–1532. doi: 10.1021/bi901748h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendoza VL, Baron-Rodriguez MA, Blanco C, Vachet RW. Biochemistry. 2011;50:6711–6722. doi: 10.1021/bi2004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendoza VL, Vachet RW. Analytical chemistry. 2008;80:2895–2904. doi: 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendoza VL, Vachet RW. Mass spectrometry reviews. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Vachet RW. Journal of the American Society for Mass Spectrometry. 2012;23:899–907. doi: 10.1007/s13361-012-0349-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Vachet RW. Journal of the American Society for Mass Spectrometry. 2012;23:708–717. doi: 10.1007/s13361-011-0332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Licklider LJ, Thoreen CC, Peng J, Gygi SP. Analytical chemistry. 2002;74:3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- 41.Gautier V, Boumeester AJ, Lossl P, Heck AJ. Proteomics. 2015 doi: 10.1002/pmic.201400462. [DOI] [PubMed] [Google Scholar]
- 42.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Analytical chemistry. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 43.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Analytical chemistry. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 44.Hung CW, Tholey A. Analytical chemistry. 2012;84:161–170. doi: 10.1021/ac202243r. [DOI] [PubMed] [Google Scholar]
- 45.Sinclair J, Timms JF. Methods (San Diego, Calif.) 2011;54:361–369. doi: 10.1016/j.ymeth.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y, Vachet RW. Analytical chemistry. 2013;85:9664–9670. doi: 10.1021/ac401978w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prudova A, auf dem Keller U, Butler GS, Overall CM. Molecular & cellular proteomics : MCP. 2010;9:894–911. doi: 10.1074/mcp.M000050-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Proteomics. 2007;7:340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 49.Roosild TP, Castronovo S, Choe S. Acta crystallographica. Section F, Structural biology and crystallization communications. 2006;62:835–839. doi: 10.1107/S1744309106029125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venable JD, Okach L, Agarwalla S, Brock A. Analytical chemistry. 2012;84:9601–9608. doi: 10.1021/ac302488h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiang CM, Roeder RG. Peptide research. 1993;6:62–64. [PubMed] [Google Scholar]
- 52.Fujii Y, Kaneko M, Neyazaki M, Nogi T, Kato Y, Takagi J. Protein expression and purification. 2014;95:240–247. doi: 10.1016/j.pep.2014.01.009. [DOI] [PubMed] [Google Scholar]