Amphioxus, also called lancelet or cephalochordate, is a promising model organism owning to its particularly evolutionary position, simple genome content and comparable body plan to that of vertebrates (Holland et al., 2004; Bertrand and Escriva, 2011). However, use of amphioxus as a model organism has been limited for many years because of lack of an efficient genomic modification method. Recently, several revolutionary gene targeting methods that could induce directed mutations, insertions and deletions at intended target sites, have been developed (Gaj et al., 2013). Among these methods, the transcription activator-like effector nuclease (TALEN) has drawn much interest because of its efficiency in generating target gene alterations, simplicity in vector design and nearly limitless in targeting range (Huang et al., 2011; Miller et al., 2011; Bedell et al., 2012; Lei et al., 2012). Up to date, this method has been shown to be effective in inducing mutations in a broad range of organisms including zebrafish, frog, rat, mouse and human (Tong et al., 2012; Gaj et al., 2013; Liu et al., 2013a), suggesting a great potential use for adopting it in amphioxus genome engineering. Here, we presented the first report of an effective TALEN-mediated genome editing method in Chinese amphioxus, Branchiostoma belcheri. We chose B. belcheri because it is the only amphioxus species which could spawn consecutively all year round (Li et al., 2012, 2013) and be raised through generations in captivity (Zhang et al., 2007). Besides, the species is one of the four amphioxus frequently used in evolutionary/developmental studies and its genome sequence (http://mosas.sysu.edu.cn/genome/gbrowser_wel.php) and embryo microinjection are available in our lab (Liu et al., 2013b).
To establish an efficient TALEN system suitable for genome editing in amphioxus, we examined the effectiveness of three TALEN backbone vector systems (namely, Goldy, HZ and BZ systems) in inducing mutations in amphioxus embryonic cells. These three systems have been optimized and demonstrated to be highly active in lots of model organisms (Huang et al., 2011; Bedell et al., 2012; Lei et al., 2012; Ma et al., 2013; Qiu et al., 2013; Xiao et al., 2013). Six TALEN pairs targeting the first coding exon of amphioxus Pax1/9 gene were constructed using these three systems (two TALEN pairs for each system). Unexpectedly, among these six pairs, only the two pairs generated using the Goldy backbone could mutagenize the targeted loci with mutation ratio at 34.3% and 21.9%, respectively (Fig. 1A and Fig. S1A; Table 1). Goldy TALEN-induced mutations included small insertions or deletions (indels), which were the characteristics of non-homologous end joining (NHEJ) mediated repairs (Fig. 1A and Fig. S1A). Two additional TALEN pairs constructed using the HZ system targeting amphioxus Pax3/7 and Pax4/6 failed to induce indel mutations in amphioxus embryos. In contrast, 60% and 27.8% mutation frequencies were respectively obtained when the same loci were targeted using the Goldy TALEN system. The apparent failure of the HZ and BZ systems appeared unrelated to their translation based on the detection of immunoreactive TALEN protein on Western blots comparable to the levels translated from the positive control DrAmmecr1 TALEN mRNAs (Fig. S1B). Mislocalization of TALEN protein was also unlikely since the backbone vectors of the two systems contained exactly the same nuclear localization signal peptide (PKKKRKV) in their N-termini as that of the Goldy vector (Fig. S2).
Fig. 1. Goldy TALEN-mediated genome editing in amphioxus embryos.
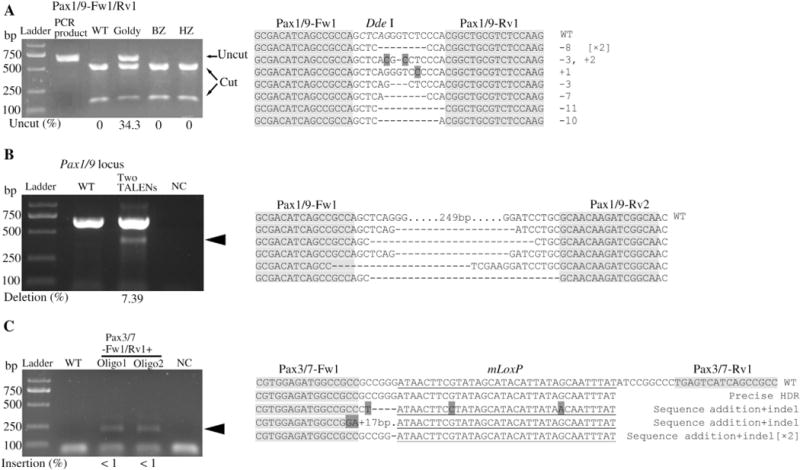
A: Activities of three TALEN systems at amphioxus Pax1/9 locus targeted by TALEN pair Pax1/9-Fw1/Rv1. The induced mutation ratios (estimated as percentages of uncut PCR products) are labeled under the gel image. B: Goldy TALEN-induced large DNA fragment deletions at amphioxus Pax1/9 locus by injecting two TALENs. PCR products amplified from the genomic DNA extracted from injected and un-injected (WT) embryos, and their sequences are shown. No DNA template was added in the negative control (NC). PCR products marked by black arrowheads would be obtained when the targeted deletion occured. The left half sequence of the first TALEN and the right half sequence of the second TALEN are shown in the first line (WT) in each alignment, with TALEN site shaded in grey and spacer unmarked. C: Goldy TALEN-mediated homology-directed repair (HDR) at amphioxus Pax3/7 locus. PCR products amplified from the genomic DNA extracted from injected (Pax3/7-Fw1/Rv1 mRNAs + ssDNA oligonucleotides) and un-injected (WT) embryos, and their sequences are shown. TALEN binding sites are shaded in grey and the mLoxP site is underlined. PCR was conducted using a forward gene-specific primer and a reverse mLoxP primer. PCR amplicons marked by black arrowheads would be obtained when HDR and mLoxP insertion occurred, while no PCR products would be obtained when no mLoxP insertion happened. The amplicons amplified from genomic DNA of the embryos injected with TALEN mRNAs and oligo1 were cloned into pGEM-T vector, and five or six selected clones were sequenced for each locus. No DNA template was added in NC (negative control). Abbreviations: WT, wild type; Goldy, Goldy TALEN system; HZ, Hui Zhao’s TALEN system; BZ, Bo Zhang’s TALEN system; NC, negative control.
Table 1.
Assembled TALENs, TALEN-targeted genes, their binding sequences, and TALEN-induced mutation ratios estimated by direct DNA sequencing or restriction enzyme (RE) analyzing in B. belcheri
| TALEN name | Target gene | Target site sequence | Lengths of left arm/spacer/right arm (bp) | TALEN system used | Mutation rate via sequencing (%)a | Mutation rate via RE reaction (%) | RE used for efficacy evaluation |
|---|---|---|---|---|---|---|---|
| Pax1/9-Fw1/Rv1b | Pax1/9 | TGCGACATCAGCCGCCAGCTCAGGGTCTCCCACGGCTGCGTCTCCAAGA | 16/15/16 | Goldy, HZ and BZ | 40.0 | 34.3 | Dde I |
| Pax1/9-Fw2/Rv2 | Pax1/9 | TGCCCTCGGTCTCGTCCATCTCGCGGATCCTGCGCAACAAGATCGGCAACA | 16/17/16 | Goldy, HZ and BZ | 25.0 | 21.9 | BamH I |
| Pax2/5/8-Fw0/Rv0 | Pax2/5/8 | TCGGCGGCGTCTACGTCAACGGCCGCCCGCTCCCTGATGTGGTTCGGCA | 16/15/16 | Goldy | 22.2 | 21.9 | Eag I |
| Pax2/5/8-Fw1/Rv1 | Pax2/5/8 | TACAAGCGACAGAACCCsACCATGTTCGCATGGGAGATCAGAGACAGGCTA | 16/17/16 | Goldy | 60.0 | NA | NA |
| Pax3/7-Fw1/Rv1 | Pax3/7 | TCGTGGAGATGGCCGCCGCCGGGATCCGGCCCTGCGTCATCAGCCGCCA | 16/15/16 | Goldy and HZ | 60.0 | 48.5 | BamH I |
| Pax4/6-Fw0/Rv0 | Pax4/6 | TCGGGGGCGTGTTCGTGAACGGCCGCCCGCTCCCCGATTCCACGCGGCAGAA | 17/17/16 | Goldy and HZ | 27.8 | NA | Eag I |
| Bra-Fw1/Rv1 | Bra1 & Bra2 | TTCCCCGTGCTGAAGGTGAACGTGTCCGGACTGGACCCTAACGCCATGTA | 16/16/16 | Goldy | 35.0 & NA | 52.6 & 50.3 | BspE I |
| Bra-Fw2/Rv2 | Bra1 & Bra2 | TACTCCTTCCTGCTGGACTTCACGGCCGCCGACAACCACCGCTGGAAGTA | 16/16/16 | Goldy | NA & 21.1 | 55.0 & 42.0 | Eag I |
| Fgf8/17/18-Fw1/Rv1 | Fgf8/17/18 | TGCGATGCTTGCCTTTGTTACGGCTCGTGCCTCTGTAAGGACAAGTTTGTA | 16/17/16 | Goldy | 70.0 | 64.1 | BssS I |
| En-Fw1/Rv1 | En | TTCGGCGCTCGCAAGAGAGACAGTAAAAGTTGTGACTCCTCTCCCGTGTCCA | 17/16/17 | Goldy | 29.4 | NA | NA |
20 individual colonies were sequenced for each target site.
Pax1/9-Fw1 was the name of an assembled TALEN vector bound to the left side targeted sequence of Pax1/9 gene in red color of target site sequence, whereas Pax1/9-Rv1 was the name of an assembled TALEN vector bound to the right side targeted sequence in blue color. Restriction enzyme recognition sequences are underlined. NA, no available.
After choosing the Goldy TALEN system, we conducted a series of experiments to establish optimum conditions for mutagenizing amphioxus embryos by testing different concentrations of TALEN transcripts and sampling times using Pax3/7-Fw1/Rv1 TALEN vectors. Injection solutions containing 50 ng/μL, 260 ng/μL, 600 ng/μL and 1.3 μg/μL of the TALEN mRNAs were tested. We observed the ratios of normally developing embryos were between 39.7% and 43.5% after the microinjections with TALEN transcripts over a concentration range of 50–600 ng/μL. This is comparable to the proportion of normally developing embryos (47%) observed in the control microinjections with 200 mmol/L KCl solution (Fig. S3A). A high concentration of TALEN transcripts (1.3 μg/μL) resulted in a relatively high ratio of death and deformation (67.4%), indicating a modest deleterious effect of the increasing TALEN mRNAs on amphioxus development (Fig. S3A). Importantly, a much higher somatic mutation ratio (48.5%) was obtained in the embryos microinjected with 1.3 μg/μL of the TALEN mRNAs than those injected with 600 ng/μL (29.9%), 260 ng/μL (11.2%) or 50 ng/μL (1.6%) of the TALEN mRNAs, which were estimated by loss of BamH I recognition site in the spacer region (Fig. S3B). Since the DNA sequencing revealed a high mutation ratio (12 of 20 alleles) in 1.3 μg/μL injected embryos (Fig. S3D), we adopted 1.3 μg/μL as an optimal mutagenizing concentration of injection solution for all TALEN transcripts in subsequent experiments unless indicated otherwise. We also examined the temporal mutagenic activity of Pax3/7-Fw1/Rv1 TALEN vector in amphioxus embryos collected at 40 min post-fertilization (just before the first cleavage), blastula, gastrula, early neurula and late neurula stages. The mutations, which inactivated the restriction site, were detectable at the one cell stage and increased throughout the development process, reaching an apparent plateau by the early neurula stage (Fig. S3C).
To examine the variation of gene-specific activity of Goldy TALEN, we assembled seven additional pairs of TALENs targeting six endogenous amphioxus genes (Pax2/5/8, Pax4/6, Bra1, Bra2, Fgf8/17/18 and En). It should be noted that Bra-Fw1/Rv1 and Bra-Fw2/Rv2 were designed to target conserved sequences found both in Bra1 and Bra2 genes. Thus each of the TALEN pairs could induce mutations at both Bra1 and Bra2 gene loci concurrently. We found efficient genome modifications in the embryos injected with in vitro synthesized mRNAs transcribed from these seven pairs of TALENs. The somatic mutation frequencies ranged from 22.2% to 70% (Table 1 and Fig. S4). As expected, the two TALEN pairs (Bra-Fw1/Rv1 and Bra-Fw2/Rv2) targeting Bra1 and Bra2 genes could mutagenize the two genes simultaneously. It should be noted that the mutation frequencies determined by direct DNA sequencing were lower than those estimated by the analysis of restriction enzyme digestion for some target sites (e.g., Bra-Fw1/Rv1 and Bra-Fw2/Rv2) (Table 1). It was caused by sequence polymorphisms existing in the cleavage site of restriction enzyme.
Co-injection of mRNAs encoding two pairs of TALENs that target adjacent regions within the same chromosome would generate two tandem double-stranded breaks (DSBs). These two DSBs could then be fused via NHEJ-mediated repair concomitant with the deletion of the intervening region (Carlson et al., 2012; Ma et al., 2012; Gupta et al., 2013; Xiao et al., 2013). We used this strategy to determine the utility of TALEN-medicated segment deletion in amphioxus. Transcripts of six TALEN pairs targeting the amphioxus Pax1/9 and Pax2/5/8 loci and the tail-to-tail amphioxus Bra1 and Bra2 gene loci, which were expected to generate deletions of 249 bp, 5.6 kb, 16.3 kb and 16.4 kb, were used in this analysis (Fig. S5A). We detected the expected lengths of segmental deletions in the embryos treated with all four TALEN injections (Fig. 1B and Fig. S5B). Based on the intensity of bands amplified from the modified region and the control region (full length amplicon or unmodified genomic DNA), we estimated deletion ratios at the Pax1/9, Pax2/5/8 and two Bra loci were 7.39%, 5.34%, 7.33% and 5.83%, respectively. Sequence analysis of the PCR amplicons further confirmed the deletion of the genomic fragments between the TALEN target sites and revealed a set of distinct junction sequences (Fig. 1B and Fig. S5B). Notably, the sequence outside TALEN binding sites was also deleted from four of five sequenced alleles from embryos injected with the Bra-Fw2/Rv2 mRNAs (Fig. S5B).
Both indel mutations and segmental deletions induced by TALEN were generally produced via NHEJ of DSBs. Nevertheless, the TALEN-induced DSBs could also be repaired via homology-directed repair (HDR) if a donor template was provided. Using this principle, Bedell et al. (2012) and Zu et al. (2013) successfully inserted exogenous DNA sequences including an EcoR V restriction site, a LoxP site and a GFP gene into zebrafish genome. To determine whether TALEN would induce HDR in amphioxus genome, we designed and synthesized two distinct ssDNA oligonucleotides (oligo 1 and oligo 2) that included a modified LoxP (mLoxP) site in the middle and two 25-bp homology arms of TALEN-targeted gene at both 3′- and 5′-ends (Fig. S6A). Two highly active TALENs (Pax3/7-Fw1/Rv1 and Pax2/5/8-Fw1/Rv1) were applied in the study. After co-injection of mRNAs of each TALEN pair and their corresponding ssDNAs (Fig. S6B), somatic insertions of the mLoxP sequence could be detected in the injected embryos using PCR detection (Fig. 1C and Fig. S6C). However, when we selected about 100 positive colonies containing the target region from injected embryos, which were generated by PCR plus TA-cloning, for further detection, no clone including the mLoxP sequence was found, indicating that the insertion ratio was less than 1%. Therefore, to determine whether the mLoxP sequence was correctly inserted into the targeted sites, we directly amplified the target segment from the DNA template of the injected embryos using gene-specific forward and mLoxP reverse primers. The PCR products were then cloned into the pGEM-T vector. Among six identified clones, we observed three precise homologous recombinations that resulted in mLoxP insertion at the 3′-end of the Pax2/5/8 locus (Fig. S6C). The remains included small indels at the modification site. We also observed a similar result in the embryos co-injected with Pax3/7-Fw1/Rv1 mRNA and its oligo 1 (Fig. 1C). Based on these results, we concluded that the Goldy TALEN system could induce precise fragment insertions at 5′-end of a targeted site. Taken together, our results suggest that Goldy TALEN system could induce precise genome insertions of a small DNA sequence in amphioxus via co-injection of TALEN mRNAs and ssDNA donor templates.
In summary, we successfully established a Goldy TALEN-mediated genome editing method for amphioxus. The method could efficiently induce somatic mutations, segmental deletions and insertions in amphioxus genome. So far, the germline transmission of TALEN-mediated genome modifications was successfully carried out in all examined model animals (Bedell et al., 2012; Carlson et al., 2012; Lei et al., 2012; Qiu et al., 2013; Xiao et al., 2013; Zu et al., 2013). We undoubtedly expected to create genome modified amphioxus immediately by germline transmission after this successful application of TALEN technique. However, several factors hinder us to obtain this amphioxus at the moment. The first restricting factor is the long generation time of amphioxus B. belcheri. The neonatal amphioxus needs about one year for growing and developing before its sexual maturity (Zhang et al., 2007). Secondly, the reproduction of amphioxus follows a strict r-selection strategy: a single mature female releases several thousands of eggs in one spawning, but over 90% of them die before metamorphosis (Zhang et al., 2007). Therefore, optimizing animal rearing method for shortening amphioxus generation time and increasing their surviving ratio is under urgent requirement.
Supplementary Material
Acknowledgments
We wish to express our great appreciation to Prof. Bo Zhang at Peking University for her kindly providing the plasmids and useful suggestions, and to Dr. Edmund J Stellwag at East Carolina University for his editing the manuscript. We also thank Dr. Anlong Xu for his kindly providing access to the unpublished genome sequence of B. belcheri. This work was supported by the National Natural Science Foundation of China (Nos. 31071110, 30830023 and 31101631) and the Scientific and Technical Innovation Committee of Shenzhen, China (No. CXZZ20120614164555920).
Footnotes
SUPPLEMENTARY DATA
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgg.2014.02.003.
References
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug2nd RG, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S, Escriva H. Evolutionary crossroads in developmental biology: amphioxus. Development. 2011;138:4819–4830. doi: 10.1242/dev.066720. [DOI] [PubMed] [Google Scholar]
- Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Hall VL, Kok FO, Shin M, McNulty JC, Lawson ND, Wolfe SA. Targeted chromosomal deletions and inversions in zebrafish. Genome Res. 2013;23:1008–1017. doi: 10.1101/gr.154070.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ, Laudet V, Schubert M. The chordate amphioxus: an emerging model organism for developmental biology. Cell Mol Life Sci. 2004;61:2290–2308. doi: 10.1007/s00018-004-4075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Cheng CH, Dawid IB, Chen Y, Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) Proc Natl Acad Sci USA. 2012;109:17484–17489. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Yang X, Shu Z, Chen X, Wang Y. Consecutive spawnings of Chinese amphioxus, Branchiostoma belcheri, in captivity. PLoS ONE. 2012;7:e50838. doi: 10.1371/journal.pone.0050838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Shu ZH, Wang YQ. Year-round reproduction and induced spawning of Chinese amphioxus, Branchiostoma belcheri, in laboratory. PLoS ONE. 2013;8:e75461. doi: 10.1371/journal.pone.0075461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li C, Yu Z, Huang P, Wu H, Wei C, Zhu N, Shen Y, Chen Y, Zhang B, Deng WM, Jiao R. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J Genet Genomics. 2013a;39:209–215. doi: 10.1016/j.jgg.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Liu X, Li G, Feng J, Yang X, Wang YQ. An efficient microinjection method for unfertilized eggs of Asian amphioxus Branchiostoma belcheri. Dev Genes Evol. 2013b;223:269–278. doi: 10.1007/s00427-013-0441-0. [DOI] [PubMed] [Google Scholar]
- Ma S, Zhang S, Wang F, Liu Y, Liu Y, Xu H, Liu C, Lin Y, Zhao P, Xia Q. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS ONE. 2012;7:e45035. doi: 10.1371/journal.pone.0045035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AC, Lee HB, Clark KJ, Ekker SC. High efficiency in vivo genome engineering with a simplified 15-RVD GoldyTALEN design. PLoS ONE. 2013;8:e65259. doi: 10.1371/journal.pone.0065259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Qiu Z, Liu M, Chen Z, Shao Y, Pan H, Wei G, Yu C, Zhang L, Li X, Wang P, Fan HY, Du B, Liu B, Liu M, Li D. High-efficiency and heritable gene targeting in mouse by transcription activator-like effector nucleases. Nucleic Acids Res. 2013;41:e120. doi: 10.1093/nar/gkt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C, Huang G, Ashton C, Wu H, Yan H, Ying QL. Rapid and cost-effective gene targeting in rat embryonic stem cells by TALENs. J Genet Genomics. 2012;39:275–280. doi: 10.1016/j.jgg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, Zu Y, Li W, Huang P, Tong X, Zhu Z, Lin S, Zhang B. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013;41:e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QJ, Sun Y, Zhong J, Li G, Lu XM, Wang YQ. Continuous culture of two lancelets and production of the second filial generations in the laboratory. J Exp Zool B Mol Dev Evol. 2007;308:464–472. doi: 10.1002/jez.b.21172. [DOI] [PubMed] [Google Scholar]
- Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Hu Y, Luo Z, Huang P, Wu Q, Zhu Z, Zhang B, Lin S. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat Methods. 2013;10:329–331. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


