Abstract
We recently identified the long non-coding RNA (ncRNA) TRPM2-AS as a key regulator of survival in prostate cancer [1]. This essential role, coupled to the TRPM2-AS low-expression levels in healthy tissues, makes this ncRNA a suitable therapeutic target for further clinical studies. To get insights into the survival mechanism controlled by this molecule, we ablated its expression in prostate cancer cells and performed a genome-wide analysis of the transcripts differentially regulated in dying cells. Here, we describe in detail the experimental system, methods and quality control for the generation of the microarray data associated with our recent publication [1]. The data are related to [1]. Data have been deposited to the Gene Expression Omnibus (GEO) database repository with the dataset identifier GSE40687.
Keywords: Prostate cancer, ncRNA, Apoptosis, Microarray, Ion channel
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens |
| Sex | Male |
| Sequencer or array type | Illumina HumanHT-12 V3.0 expression beadchip |
| Data format | Microarray raw and normalized data: TXT files |
| Experimental factors | PC3 cells without or with TRPM2-AS expression |
| Experimental features | Microarray gene expression profiling to identify transcripts that are regulated by TRPM2-AS |
| Consent | N/A |
| Sample source location | N/A |
Direct link to the deposited data
Microarray data is deposited at the NCBI Gene Expression Omnibus (GEO) database under GEO Series and is available at the following link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40687.
Experimental design, materials and methods
Cell line
The human, androgen-independent prostate cancer cell line PC3 was obtained from ATCC (CRL-1435, ATCC).
RNA interference
During an in-silico search for antisense transcripts [2], [3], [4], we have identified TRPM2-AS as antisense transcript in respect to TRPM2 gene, which encodes an oxidative stress-activated ion channel [5]. This long ncRNA is overexpressed in several tumor types [1], [4] and its knock-out leads to massive apoptosis of PC3 and DU145 prostate cancer cells [1]. To gain a molecular understanding of the mechanisms by which TRPM2-AS maintains cell survival in prostate cancer cells, PC3 cells were transfected for 48 h either with a non-specific siRNA or with a siRNA ablating TRPM2-AS RNA.
Microarray and quality control
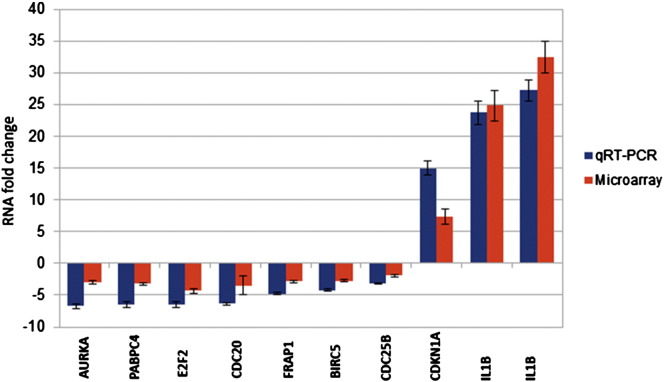
To identify the transcripts that are regulated by TRPM2-AS, we isolated total RNA from four samples (two technical replicates) of PC3 cell lines that had been transfected with the control siRNA or with the experimental siRNA targeting TRPM2-AS expression. The quantity and quality of the RNA samples was measured and assessed by a NanoDrop spectrophotometer and Agilent 2100 Bioanalyzer (Agilent Technologies). Illumina microarray analysis was performed using the Illumina HumanHT-12 V3.0 expression beadchip. Quantitative real-time PCR (qRT-PCR) was used to check the microarray data, analyzing transcript levels that changed in both directions. In particular segments corresponding to the following transcripts were amplified: AURKA, PABPC4, E2F2, CDC20, FRAP1, BIRC5, CDC25B, CDKN1A, IL1B, IL8. Results, shown in Fig. 1, indicated that while, due to the large dynamic range of qRT-PCR [6], microarray data were mostly slightly compressed, there was an excellent qualitative correlation among the two techniques (correlation coefficient: 0.97), thereby validating the microarray experiment.
Fig. 1.
Validation of microarray data by qRT-PCR. Ten transcripts were quantified by qRT-PCR both in control and in TRPM2-AS KO PC3 cells. The resulting expression fold change is plotted against the expression fold change obtained from the Illumina HumanHT-12 V3.0 microarray data. A correlation coefficient of 0.97 was found between the two datasets.
Discussion
We report here a dataset composed of microarray gene expression profiling of TRPM2-AS-regulated transcripts. With this experiment, we were able to show that TRPM2-AS coordinates the expression of a large number of genes involved in controlling the survival, the unfolded protein response (UPR) response and the cell-cycle progression in prostate cancer cells. Moreover, targets of existing drugs and treatments were found to be consistently regulated by TRPM2-AS KO, underlying the clinical relevance of this long ncRNA as a novel therapeutic target. According to the dual role of the UPR in promoting cell survival or commitment to programmed cell death [7], several pro-survival molecules and known oncological targets, including the chaperonin HSP90B1 and the oncogenes FYN and AKT1, were also induced by TRPM2-AS KO, suggesting that the dataset as a whole would be particularly valuable for identifying new molecular targets and underlying mechanisms.
References
- 1.Orfanelli U., Jachetti E., Chiacchiera F., Grioni M., Brambilla P. Antisense transcription at the TRPM2 locus as a novel prognostic marker and therapeutic target in prostate cancer. Oncogene. 2014 doi: 10.1038/onc.2014.144. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 2.Lavorgna G., Dahary D., Lehner B., Sorek R., Sanderson C.M., Casari G. In search of antisense. Trends Biochem. Sci. 2004;29:88–94. doi: 10.1016/j.tibs.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Lavorgna G., Triunfo R., Santoni F., Orfanelli U., Noci S. AntiHunter 2.0: increased speed and sensitivity in searching BLAST output for EST antisense transcripts. Nucleic Acids Res. 2005;33:W665–W668. doi: 10.1093/nar/gki448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orfanelli U., Wenke A.K., Doglioni C., Russo V., Bosserhoff A.K., Lavorgna G. Identification of novel sense and antisense transcription at the TRPM2 locus in cancer. Cell Res. 2008;18:1128–1140. doi: 10.1038/cr.2008.296. [DOI] [PubMed] [Google Scholar]
- 5.Knowles H., Li Y., Perraud A.L. The TRPM2 ion channel, an oxidative stress and metabolic sensor regulating innate immunity and inflammation. Immunol. Res. 2013;55:241–248. doi: 10.1007/s12026-012-8373-8. [DOI] [PubMed] [Google Scholar]
- 6.Wong M.L., Medrano J.F. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 7.Wang G., Yang Z.Q., Zhang K. Endoplasmic reticulum stress response in cancer: molecular mechanism and therapeutic potential. Am. J. Transl. Res. 2010;2:65–74. [PMC free article] [PubMed] [Google Scholar]