Abstract
The mulberry white caterpillar, Rondotia menciana Moore (Lepidoptera: Bombycidae) is a species with closest relationship with Bombyx mori and Bombyx mandarina, and the genetic information of R. menciana is important for understanding the diversity of the Bombycidae. In this study, the mitochondrial genome (mitogenome) of R. menciana was amplified by polymerase chain reaction and sequenced. The mitogenome of R. menciana was determined to be 15,301 bp, including 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, 22 transfer RNA genes, and an AT-rich region. The A+T content (78.87%) was lower than that observed for other Bombycidae insects. All PCGs were initiated by ATN codons and terminated with the canonical stop codons, except for coxII, which was terminated by a single T. All the tRNA genes displayed a typical clover-leaf structure of mitochondrial tRNA. The length of AT-rich region (360 bp) of R. menciana mitogenome is shorter than that of other Bombycidae species. Phylogenetic analysis showed that the R. menciana was clustered on one branch with B. mori and B. mandarina from Bombycidae.
Keywords: mitogenome, Bombycidae, diversity, phylogeny
Insect mitochondrial genomes (mitogenomes) are typically circular molecules 14–19 kb in length that contain 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes (Wolstenholme 1992, Boore 1999), and an A+T-rich region, which contains initiation sites for transcription and replication (Zhang et al. 1995, Zhang and Hewitt 1997, Taanman 1999).
Mitogenome sequences, which exhibit very low levels of recombination, are widely used in population genetics, comparative and evolutionary genomics, reconstruction of phylogenetic relationships, and evolutionary biology (Avise 1987, Ballard 2000, Ballard and Rand 2005, Cameron and Whiting 2008, Hao et al. 2012). The silk-producing insects in the lepidoptera with economic value belong to two families of moth, Bombycidae and Saturniidae (Mahendran et al. 2006). The complete mitogenomes of Bombyx mori and Bombyx mandarina of Bombycidae (Yukuhiro et al. 2002; Hu et al. 2010; Li et al. 2010a; Liu et al. 2013), and Antheraea pernyi (Liu et al. 2012b), Antheraea yamamai (Kim et al. 2009), Eriogyna pyretorum (Jiang et al. 2009), Samia cynthia ricini (Kim et al. 2012), Actias selene (Liu et al. 2012a), and Caligula boisduvalii (Hong et al. 2008) of Saturniidae have been sequenced. The origin of bombycidae insects had been studied more according to the mitogenomes (Hu et al. 2010; Li et al. 2010a).
The mulberry white caterpillar, Rondotia menciana Moore (Lepidoptera: Bombycidae) is a silk-producing insects from Bombycidae and has been exploited since the Yangshao culture period (approximately 5,500–6,000 years ago). As all the other insects from lepidoptera, R. menciana is a bivoltine insect that exhibits four molts and a dormant period after the formation of resting eggs, too (Xu et al. 1994). The number of chromosomes (22) in R. menciana differs from that of B. mori, (28) or B. mandarina (27 or 28) (Deng and Xiang 1993), and, thus, the genetic information of R. menciana is important for understanding the diversity of the Bombycidae. R. menciana larvae feed on mulberry leaves and can, in serious cases, defoliate trees. So, the natural R. menciana populations have been decreasing, due to effective control of the insect by the Chinese government to prevent destruction of mulberry trees in recent years. At the same time, the research on genetic or the other aspects about R. menciana was rare. In this study, the complete mitogenome sequence of R. menciana was obtained (GenBank accession number: KC881286), and the phylogenetic analyses based on the mitogenome of the selected insects from lepidoptera were performed using the maximum-likelihood (ML) method.
Materials and Methods
Specimen Sampling and DNA Extraction
Adult specimens of R. menciana were collected from the Tsinling Mountains (106° 55′19″ E, 34° 14′29″ N), Shaanxi Province, China, in September 2011, preserved in 100% ethanol, and stored at −80°C until DNA extraction. Total genomic DNA was extracted from heads excised from frozen insects using the MagSi Tissue DNA Kit (Omega, GA).
Polymerase Chain Reaction Amplification and Sequencing
To amplify the entire mitogenome of R. menciana, 10 primer sets (Table 1) were designed according to known mitochondrial DNA sequences from Bombycidea insects. Purified genomic DNA was amplified using the polymerase chain reaction (PCR) technique and the Taq PCR Kit (NEB, MA), under the following cycling parameters: 94°C for 3 min; 35 cycles of 30 s at 94°C, 40 s at 55–60°C, 1–3 min at 72°C; and 72°C for 10 min. The PCR products were detected by 1.0% agarose-gel electrophoresis and purified using a DNA gel extraction kit (TaKaRa, Japan). The purified PCR products were ligated into the T-vector (TaKaRa) and sequenced at least three times at Sangon.
Table 1.
Primers used in this study
| Primers | Location | Sequence(5′–3′) | Mismatch |
|---|---|---|---|
| nad2-coxIF | 335 –354 | TGATTTGGDTGTTGAATTGGHYTAGAA | 1 |
| nad2-coxIR | 1,952 –1,927 | GCTCCTAAGATTGAWGAAATACCWGC | 2 |
| coxI-coxIIF | 1,830 –1,850 | TGGTGCAGGAACAGGATGAAC | 3 |
| coxI-coxIIR | 3,791 –3,771 | GAGACCADTACTTGCTTTCAG | 1 |
| coxII-coxIIIF | 3,673 –3,692 | ATTTGTGGRGCTAATCWTAG | 2 |
| coxII-coxIIIR | 4,788 –4,769 | GGTCAAGGWCTATAATCYAC | 1 |
| coxIII-nad3F | 4,511 –4,528 | TCGACCTGGAACTTTAGC | 1 |
| coxIII-nad3R | 5,727 –5,709 | TGGATCAAATCCACATTCA | 1 |
| nad3-nad5F | 5,444 –5,466 | GAAGCAGCAGCTTGATATTGACA | 2 |
| nad3-nad5R | 7,487 –7,462 | GCAGCTATAGCMGCTCCTACTCCWGT | 1 |
| nad5-nad4F | 7,421 –7,444 | CCCCTGCTGTTACTAAAGTTGAWG | 0 |
| nad5-nad4R | 9,079 –9,055 | GGCTCTTTACCTTTATTAATRGGAA | 1 |
| nad4-cytbF | 8,892 –8,914 | GGAGCTTCTACATGAGCTTTTGG | 3 |
| nad4-cytbR | 10,906 –10,885 | CCCCTCAAAAWGATATTTGACC | 0 |
| cytb-rrnLF | 10,717 –10,739 | CGTACTCTTCATGCWAATGGRGC | 4 |
| cytb-rrnLR | 12,974 –12,941 | CTAATCAAYAGAAAAGWTTGCGACCTCGATGTTG | 4 |
| rrnLF | 12,858 –12,881 | CGGTTTGAACTCAGATCATGTAAG | 0 |
| rrnLR | 13,920 –13,895 | TATTGTATCTTGTGTATCAGAGTTTA | 1 |
| rrnL-nad2F | 13,304 –13,335 | ATGCTACCTTTGCACRGTCAAAATACYGCRGC | 1 |
| rrnL-nad2R | 588 –563 | TCAAAAATGAAATGGKGYTGAWCCTAT | 3 |
Sequence Analysis and Gene Annotation
The BLASTN (http://www.ncbi.nlm.nih.gov/blast) was used to determine sequence similarity. Sequence assembly was performed using DNAStar software. The location of tRNA genes and potential stem-loop secondary structures were identified using tRNAscan-SE software version 1.21 (Lowe and Eddy 1997), specifying mito and chloroplast DNA as the source and using invertebrate mitochondrial genetic code predictors. Thirteen PCGs were identified using an open reading frame finder, using the invertebrate mitochondrial genetic code. The nucleotide composition and codon usage were calculated using MEGA 5.05 (Tamura et al. 2011) and the composition skewness to the formulas: AT skew = [A–T]/[A + T]; GC skew = [G–C]/[G + C] (Perna and Kocher 1995). The putative control region was identified by alignment with sequences from the closely related species B. mori and B. mandarina, and the tandem repeats in the control region were predicted using the Tandem Repeats Finder program (Benson 1999).
Phylogenetic Analysis
The complete mitogenomes of 29 lepidopteran species (Table 2) were used to reconstruct the phylogenetic relationship. The mitogenomes of Drosophila melanogaster (Diptera: Drosophilidae) and Tribolium castaneum (Coleoptera: Tenebrionidae) were used as outgroups (Clary et al. 1982, Friedrich and Muqim 2003). The amino acid sequences of each of the 13 mitochondrial PCGs were aligned by Clustal X 1.83 using default settings (Thompson et al. 1997) and then backtranslated into nucleotide sequences after alignment. The concatenated set of nucleotide sequences were performed in phylogenetic analysis, using ML method with the MEGA version 5.05 program.
Table 2.
List of taxa used in this study
| Superfamily | Family | Insect species | Accession number | References |
|---|---|---|---|---|
| Bombycoidea | Bombycidae | R. mencianaa | KC881286 | This study |
| R. menciana | KJ647172 | Kim et al. (2014) | ||
| B. mori Xiafang | AY048187 | Li et al. (2010b) | ||
| B. mori C108 | AB070264 | Yukuhiro et al. (2002) | ||
| B. mandarina Qingzhou | FJ384796 | Hu et al. (2010) | ||
| B. mandarina Japanese | GU966593 | Li et al. (2010a) | ||
| Saturniidae | A. pernyi | AY242996 | Liu et al. (2012b) | |
| A. yamamai | EU726630 | Kim et al. (2009) | ||
| Eriogyna pyretorum | FJ685653 | Jiang et al. (2009) | ||
| Samia cynthia ricini | JN215366 | Kim et al. (2012) | ||
| Ac. selene | JX186589 | Liu et al. (2012a) | ||
| Saturnia boisduvalii | EF622227 | Hong et al. (2008) | ||
| Sphingidae | M. sexta | NC_010266 | Cameron and Whiting (2008) | |
| Geometridae | Geometridae | Phthonandria atrilineata | EU569764 | Yang et al. (2009) |
| Biston panterinaria | JX406146 | Yang et al. (2013) | ||
| Noctuoidea | Noctuidae | Helicoverpa armigera | NC_014668 | Yin et al. (2010) |
| Spodoptera exigua | JX316220 | Wu et al. (2013) | ||
| Sesamia inferens | JN039362 | Chai and Du (2012) | ||
| Ochrogaster lunifer | AM946601 | Salvato et al. (2008) | ||
| Arctiidae | Hyphantria cunea | NC_014058 | Liao et al. (2010) | |
| Pyraloidea | Crambidae | Chilo suppressalis | HQ860290 | Yin et al. (2011) |
| Diatraea saccharalis | FJ240227 | Li et al. (2011) | ||
| Ostrinia nubilalis | NC_003367 | Coates et al. (2005) | ||
| Cnaphalocrocis medinalis | JQ305693 | Yin et al. (2014) | ||
| Tortricoidea | Tortricidae | Adoxophyes honmai | DQ073916 | Lee et al. (2006) |
| Grapholita molesta | HQ116416 | Gong et al. (2012) | ||
| Spilonota lechriaspis | HM204705 | Zhao et al. (2011) | ||
| Choristoneura longicellana | HQ452340 | Unpublished | ||
| Acleris fimbriana | HQ662522 | Unpublished | ||
| Diptera | Drosophilidae | D. melanogaster | DMU35741 | Clary et al. (1982) |
| Coleoptera | Tenebrionidae | Tribolium castaneum | AJ312413 | Friedrich and Muqim (2003) |
a This study.
Results
Genome Organization and Base Composition
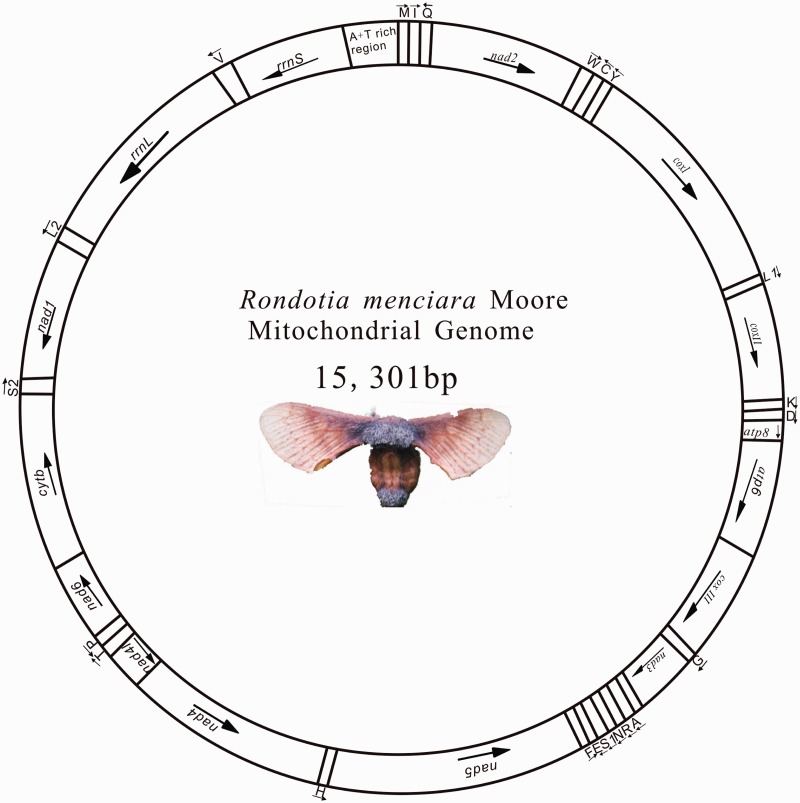
In this study, the organization of R. menciana mitogenome was shown in Fig. 1. The complete mitogenome is a closed circular molecule of 15,301 bp in length, containing 13 PCGs (coxІ-III, nad1-6, nad4L, cyt B, atp6, atp8), 22 tRNA genes, 2 rRNAs (rrnL and rrnS), and an A + T-rich region (Table 3). The order and orientation of R. menciana mitochondrial genes are identical to those found in available lepidopteran mitogenomes (Cameron and Whiting 2008, Liu et al. 2013, Yang et al. 2013). The gene order trnM, trnI, and trnQ of the lepidopteran mitogenomes differs from the most common type trnI, trnQ, and trnM, which was found in a variety of insect orders and inferred to be ancestral for insects (Boore et al. 1998). The nucleotide composition of the R. menciana mitogenome was as follows: A (41.42%), T (37.45%), G (7.82%), and C (13.31%), and the A + T content (78.87%) was the lowest in the Bombycidae (Table 4). The AT skewness and GC skewness of R. menciana mitogenome was 0.050 and −0.26, respectively, as observed in other Bombycidae, more biased toward A (the value of AT skewness is above zero) and C (Table 4).
Fig. 1.
Map of the mitogenome of R. menciana. The direction of all PCGs is designed by the underlined arrows. The transfer RNA genes are designated by single-letter amino acids codes. L1, L2, S1, and S2 denote trnL(UUR), trnL(CUN), trnS(AGN), and trnS(UCN), respectively.
Table 3.
Summary of the mitogenome of R. menciana
| Genes | Location | Size (bp) | Intergenic nucleotides | Direction | Anticodon | Start codon/stop codon | A + T(%) |
|---|---|---|---|---|---|---|---|
| trnM(CAU) | 1 –68 | 68 | F | cat | 77 .94 | ||
| trnI(GAU) | 69 –132 | 64 | 0 | F | gat | 78 .13 | |
| trnQ(UUG) | 130 –198 | 69 | –3 | R | ttg | 84 .06 | |
| nad2 | 251 –1,264 | 1,014 | 52 | F | att/taa | 84 .12 | |
| trnW(UCA) | 1,276 –1,345 | 70 | 11 | F | tca | 82 .86 | |
| trnC(GCA) | 1,338 –1,404 | 67 | –8 | R | gca | 76 .12 | |
| trnY(GUA) | 1,405 –1,473 | 69 | 0 | R | gta | 78 .26 | |
| coxI | 1,471 –3,015 | 1,545 | –3 | F | att/taa | 69 .9 | |
| trnL(UUR) | 3,011 –3,078 | 68 | –5 | F | taa | 73 .53 | |
| cox II | 3,079 –3,760 | 682 | 0 | F | atg/t | 75 .07 | |
| trnK(CUU) | 3,761 –3,831 | 71 | 0 | F | ctt | 74 .65 | |
| trnD(GUC) | 3,831 –3,897 | 67 | –1 | F | gtc | 88 .06 | |
| atp8 | 3,898 –4,059 | 162 | 0 | F | att/taa | 89 .51 | |
| atp6 | 4,053 –4,730 | 678 | –7 | F | atg/taa | 76 .84 | |
| cox III | 4,736 –5,524 | 789 | 5 | F | atg/taa | 71 .99 | |
| trnG(UCC) | 5,527 –5,592 | 66 | 2 | F | tcc | 86 .36 | |
| nad3 | 5,590 –5,946 | 357 | –3 | F | att/tag | 79 .55 | |
| trnA(UGC) | 5,969 –6,034 | 66 | 22 | F | tgc | 80 .30 | |
| trnR(UCG) | 6,037 –6,101 | 65 | 2 | F | tcg | 78 .46 | |
| trnN(GUU) | 6,103 –6,167 | 65 | 1 | F | gtt | 80 .00 | |
| trnS(AGN) | 6,170 –6,238 | 69 | 2 | F | gct | 81 .16 | |
| trnE(UUC) | 6,240 –6,306 | 67 | 1 | F | ttc | 92 .54 | |
| trnF(GAA) | 6,306 –6,372 | 67 | –1 | R | gaa | 85 .07 | |
| nad5 | 6,372 –8,102 | 1,731 | –1 | R | ata/taa | 80 .59 | |
| trnH(GUG) | 8,107 –8,173 | 67 | 4 | R | gtg | 83 .58 | |
| nad4 | 8,186 –9,526 | 1,341 | 12 | R | atg/taa | 78 .23 | |
| nad4L | 9,526 –9,816 | 291 | –1 | R | atg/taa | 83 .51 | |
| trnT(UGU) | 9,821 –9,888 | 68 | 4 | F | tgt | 80 .88 | |
| trnP(UGG) | 9,889 –9,953 | 65 | 0 | R | tgg | 78 .46 | |
| nad6 | 9,956 –10,468 | 513 | 2 | F | ata/taa | 81 .68 | |
| cytb | 10,471 –11,622 | 1,152 | 2 | F | ata/taa | 74 .39 | |
| trnS(UCN) | 11,632 –11,699 | 68 | 9 | F | tga | 80 .88 | |
| nad1 | 11,717 –12,655 | 939 | 17 | R | atg/tag | 75 .08 | |
| trnL(CUN) | 12,657 –12,726 | 70 | 1 | R | tag | 78 .57 | |
| rrnL | 12,726 –14,090 | 1,365 | –1 | R | 83 .37 | ||
| trnV(UAC) | 14,091 –14,159 | 69 | 0 | R | TAC | 82 .61 | |
| rrnS | 14,160 –14,941 | 782 | 0 | R | 84 .4 | ||
| A + T rich region | 14,942 –15,301 | 360 | 0 | 91 .11 |
Table 4.
Comparison of the nucleotides composition and skewness of Bombycoidea insects
| Insect species | Whole genome |
PCGs codona |
rrnL |
rrnS |
A + T rich |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Size(bp) | A + T% | AT skewness/GC skewness | Size | A + T% | Size | A + T% | Size | A + T% | Size | A + T% | |
| R. mencianab | 15,301 | 78.87 | 0.050/–0.260 | 11,157 | 77.05 | 1,365 | 83.37 | 782 | 84.4 | 360 | 91.11 |
| R. menciana | 15,364 | 82.14 | 0.021/–0.195 | 11,178 | 80.96 | 1,398 | 85.77 | 775 | 85.03 | 360 | 91.11 |
| B. mori Xiafang | 15,664 | 81.35 | 0.058/–0.215 | 11,142 | 79.51 | 1,376 | 84.38 | 783 | 85.44 | 498 | 95.38 |
| B. mori C108 | 15,656 | 81.36 | 0.059/–0.216 | 11,160 | 79.53 | 1,378 | 84.4 | 783 | 85.57 | 494 | 94.55 |
| B. mandarina Qingzhou | 15,717 | 81.42 | 0.057/–0.211 | 11,142 | 79.5 | 1,380 | 84.64 | 788 | 85.66 | 495 | 95.56 |
| B. mandarina Japanese | 15,928 | 81.73 | 0.054/–0.212 | 11,166 | 79.64 | 1,377 | 84.68 | 783 | 85.95 | 747 | 95.18 |
| A. pernyi | 15,566 | 80.16 | –0.021/–0.216 | 11,181 | 78.46 | 1,369 | 83.86 | 775 | 84.13 | 552 | 90.4 |
| A. yamamai | 15,338 | 80.3 | –0.022/–0.22 | 11,187 | 78.89 | 1,380 | 83.99 | 776 | 84.41 | 334 | 89.52 |
| Eriogyna pyretorum | 15,327 | 80.82 | –0.031/–0.204 | 11,193 | 79.35 | 1,338 | 84.6 | 778 | 84.45 | 358 | 92.18 |
| Samia cynthia ricini | 15,384 | 79.78 | –0.006/–0.228 | 11,196 | 78.26 | 1,358 | 84.02 | 779 | 83.83 | 361 | 90.86 |
| Ac. selene | 15,236 | 78.91 | –0.023/–0.236 | 11,184 | 77.3 | 1,364 | 83.58 | 762 | 83.99 | 339 | 87.91 |
| Saturnia boisduvalii | 15,360 | 80.62 | –0.024/–0.217 | 11,199 | 79.11 | 1,391 | 84.76 | 774 | 84.11 | 330 | 91.52 |
| M. sexta | 15,516 | 81.78 | –0.005/–0.181 | 11,157 | 80.24 | 1,391 | 85.26 | 777 | 85.71 | 324 | 95.37 |
a Termination codons excluded.
b This study.
PCGs and Codon Usage
The total length of 13 PCGs is 11,194 bp, and the A + T content of them is between 69.9% (coxI) and 89.51% (atp8) (Table 3). All PCGs in the R. menciana mitogenome were initiated by typical ATN codons: ATT for nad2, coxI, atp8, and nad3 genes, ATA for nad5, nad6, and cytb genes, and ATG for the other six genes. Twelve of the PCGs were terminated with the canonical stop codons TAA or TAG, and coxII gene was terminated with a single T (Table 3). The presence of an incomplete stop codon seems a common phenomenon and had been found in several invertebrate mitochondrial genes (Jiang et al. 2009, Liu et al. 2013, Yang et al. 2013).
Codon usage of the PCGs exhibited a notable AT bias with an A + T composition of 77.05% (Table 4), which plays a major role in the A + T bias of the entire mitogenome. The six most frequently used codons in the R. menciana mitogenome (TTA for Leu, ATT for Ile, TTT for Phe, ATA for Met, AAT for Asn, and TAT for Tyr) are composed of T or a combination of A and T, and the least frequent codons (CCG for Pro, TCG, AGG, and AGC for Ser, CGC for Arg, CTG for Leu, and CGG for Arg) have a high CG content (Table 5).
Table 5.
Codon usage of PCGs in R. menciana mitogenome
| Codon | No. of codons | RSCUa | Codon | No. of codons | RSCUa |
|---|---|---|---|---|---|
| AAA(Lys) | 92 | 1.69 | TAAb | 10 | 1.67 |
| AAG(Lys) | 17 | 0.31 | TAGb | 2 | 0.33 |
| AAC(Asn) | 39 | 0.31 | TAC(Tyr) | 27 | 0.28 |
| AAT(Asn) | 211 | 1.69 | TAT(Tyr) | 167 | 1.72 |
| ACA(Thr) | 78 | 1.88 | TGA(Trp) | 83 | 1.77 |
| ACG(Thr) | 5 | 0.12 | TGG(Trp) | 11 | 0.23 |
| ACC(Thr) | 22 | 0.53 | TGC(Cys) | 6 | 0.38 |
| ACT(Thr) | 61 | 1.47 | TGT(Cys) | 26 | 1.63 |
| AGA(Ser) | 86 | 2.21 | TCA(Ser) | 76 | 1.95 |
| AGG(ser) | 2 | 0.05 | TCG(Ser) | 2 | 0.05 |
| AGC(Ser) | 3 | 0.08 | TCC(Ser) | 23 | 0.59 |
| AGT(Ser) | 31 | 0.8 | TCT(Ser) | 88 | 2.26 |
| ATA(Met) | 248 | 1.78 | TTC(Phe) | 37 | 0.2 |
| ATG(Met) | 30 | 0.22 | TTT(Phe) | 338 | 1.8 |
| ATC(Ile) | 43 | 0.2 | TTA(Leu) | 442 | 4.74 |
| ATT(Ile) | 394 | 1.8 | TTG(Leu) | 32 | 0.34 |
| GTA(Val) | 51 | 1.32 | CTA(Leu) | 51 | 0.55 |
| GTG(Val) | 10 | 0.26 | CTG(Leu) | 2 | 0.02 |
| GTC(Val) | 8 | 0.21 | CTC(Leu) | 6 | 0.06 |
| GTT(Val) | 86 | 2.22 | CTT(Leu) | 27 | 0.29 |
| GAA(Glu) | 53 | 1.49 | CAA(Gln) | 54 | 1.71 |
| GAG(Glu) | 18 | 0.51 | CAG(Gln) | 9 | 0.29 |
| GAC(Asp) | 7 | 0.21 | CAC(His) | 17 | 0.49 |
| GAT(Asp) | 60 | 1.79 | CAT(His) | 53 | 1.51 |
| GCA(Ala) | 38 | 1.32 | CCA(Pro) | 51 | 1.62 |
| GCG(Ala) | 4 | 0.14 | CCG(Pro) | 1 | 0.03 |
| GCC(Ala) | 14 | 0.49 | CCC(Pro) | 24 | 0.76 |
| GCT(Ala) | 59 | 2.05 | CCT(Pro) | 50 | 1.59 |
| GGA(Gly) | 86 | 1.78 | CGA(Arg) | 31 | 2.34 |
| GGG(Gly) | 41 | 0.85 | CGG(Arg) | 3 | 0.23 |
| GGC(Gly) | 7 | 0.15 | CGC(Arg) | 2 | 0.15 |
| GGT(Gly) | 59 | 1.22 | CGT(Arg) | 17 | 1.28 |
a Relative synonymous codon usage.
b Stop codon.
The analysis of the base composition at each codon position of the 13 PCGs of R. menciana mitogenome shows that the third codon position (87.4%) is considerably higher in A + T content than the first (72.8%) and the second (70.3%) ones (Table 6). Further analysis of the base composition at third codon position among Bombycidae insects shows that the highest and lowest A + T content appeared in the R. menciana (Kim et al. 2014) and this article, respectively, whereas the C + T content of each codon position was similar.
Table 6.
Summary of base composition at each codon position of PCGs in the Bombycidae mitogenome
| Insect species | First codon position |
Second codon position |
Third codon position |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T-1 | C-1 | A-1 | G-1 | T-2 | C-2 | A-2 | G-2 | T-3 | C-3 | A-3 | G-3 | |
| R. mencianaa | 36.4 | 10.8 | 36.4 | 16.4 | 48.2 | 16.3 | 22.1 | 13.4 | 46.2 | 7.7 | 41.1 | 5.0 |
| R. menciana | 37.5 | 9.2 | 37.9 | 15.4 | 49.1 | 15.4 | 22.4 | 13.1 | 50.0 | 3.1 | 45.2 | 1.6 |
| B. mori C108 | 37.3 | 9.7 | 37.0 | 16.0 | 48.4 | 16.2 | 22.0 | 13.4 | 49.0 | 4.4 | 43.9 | 2.7 |
| B. mori Xiafang | 37.3 | 9.6 | 37.1 | 15.9 | 48.4 | 16.2 | 22.0 | 13.3 | 48.9 | 4.6 | 43.8 | 2.7 |
| B. mandarina Qingzhou | 37.3 | 9.6 | 37.2 | 15.9 | 48.4 | 16.2 | 22.0 | 13.4 | 49.0 | 4.5 | 43.7 | 2.8 |
| B. mandarina Japanese | 37.4 | 9.6 | 37.2 | 15.8 | 48.4 | 16.2 | 22.2 | 13.3 | 49.2 | 4.3 | 43.8 | 2.8 |
a This study.
Overlapping and Intergenic Spacer Regions
Eleven overlaps, a total of 34 bp, were observed in the R. menciana mitogenome (Table 3). The largest overlap, 8 bp, occurred between the trnW and trnC genes, as reported in other Bombycidae species, such as B. mandarina (Li et al. 2010a) and B. mori Dazao (Liu et al. 2013). As in other lepidopteran species, a 7 bp overlap occurred between the atp8 and atp6 genes, as well. The R. menciana mitogenome harbors 17 short non-coding regions of 1–52 bp, for a total of 149 bp, as in the other insects from Bombycidae, the longest spacer is located between the trnQ and nad2 genes (Table 3).
Transfer and rRNA Genes
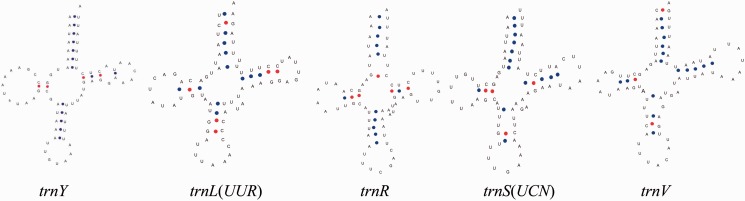
As in other lepidopteran insects, all 22 tRNA genes with characteristic cloverleaf secondary structure in the R. menciana mitogenome were predicted using the tRNAscan-SE Search Server. The length of tRNA genes ranged from 64 bp (trnI) to 71 bp (trnK) and the A + T content ranged from 73.53% (trnL(UUR)) to 92.54% (trnE) (Table 3). A total of six mismatches were found in five tRNA genes, two in amino acid acceptor stems, three in anticodon stems, and one in pseudouridine (TΨC) stems (Fig. 2). The mismatched bases show significant nucleotide bias, four U-U, one A-G, and one U-G.
Fig. 2.
Putative secondary cloverleaf structures for the tRNA genes of the R. menciana mitogenome with mismatch bases. The blue point and red point indicate Watson–Crick base pairing A-U and G-C, respectively, and the blank indicate the mismatch bases. Six mismatches (four U-U, one A-G, and one U-G) lies in five tRNA genes (two in amino acid acceptor stems, three in anticodon stems and one in pseudouridine) (TΨC).
Two rRNA genes were identified on the N-strand in the R. menciana mitogenome: the rrnL gene, located between trnL(CUN) and trnV genes, and the rrnS gene, between the trnV gene and the A + T-rich region (Table 3). The length of rrnL and rrnS genes was 1,365 bp and 782 bp, and their A + T content was 83.37% and 84.4%, respectively.
A+T-Rich Region
The A + T-rich region of R. menciana mitogenome was exactly same as that of Kim et al. (2014). The A + T-rich region was 360-bp long and located between the rrnS and trnM genes. The A + T content of the region was 91.11%, lower than the other Bombycidae insects (94.42–95.55%) (Yukuhiro et al. 2002; Hu et al. 2010; Li et al. 2010b; Liu et al. 2013). Several structures conserved in other Bombycidae mitogenomes were also observed in the R. menciana A + T-rich region (Fig. 3). The conserved “ATAGA + poly T” motif with 17 consecutive Ts was located 24 bp downstream of the rrnS gene. A microsatellite (ATAT)n element and a 12-bp poly-A region, commonly observed in other lepidopteran mitogenomes, were also found immediately upstream of the trnM gene. We also identified 2.6 tandem repeats elements of 37 bp in the R. menciana A + T-rich region (Fig. 3).
Fig. 3.
Alignments of the A + T-rich region in Bombycidae. The thread underlined and thick overlined indicate the tandom repeat in R. menciana, and B. mori and B. mandarina, respectively.
Phylogenetic Analysis
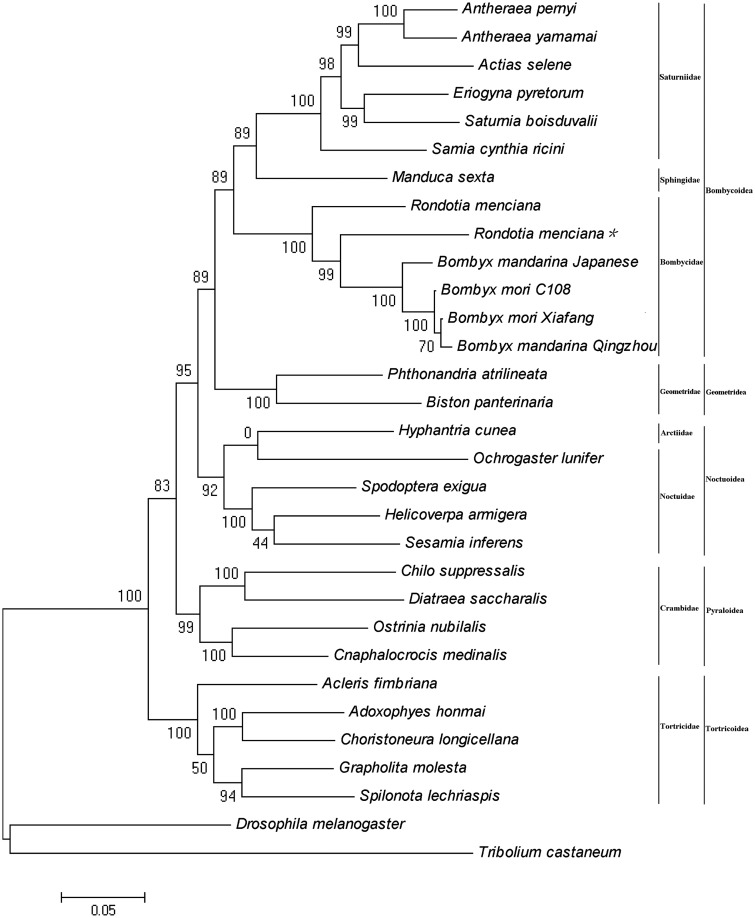
In this study, the concatenated nucleotides sequences of 13 PCGs of 29 mitogenomes were used to reconstruct the phylogenetic relationships by ML method (Fig. 4). These 29 mitogenomes represent five superfamilies within the lepidopteran suborder: Bombycoidea, Geometroidea, Noctuoidea, Pyraloidea, and Tortricidea. The results show that the phylogenetic relationships among these five superfamilies are Tortricidea + (Pyraloidea + (Noctuoidea + (Geome troidea + Bombycoidea))), the relationship between Geometridae and Bombycoidea was close. The phylogenetic relationships inside Bombycoidea and Bombycidae were Bombycidae + (Sphingidae + Saturniidae)) and R. menciana + (B. mori + B. mandarina), respectively.
Fig. 4.
Phylogenetic analysis inferred from the concatenated nucleotides sequences of 13 PCGs of mitogenome using Mega 5.05 software and ML method. D. melanogaster and T. castaneum were used as outgroups. The numbers above the branches specify bootstrap percentages (1,000 replicates). *This study.
Discussions
The complete mitogenome of R. menciana with a circular molecule of 15,301 bp was determined using the PCR method, which is the shortest in known complete mitogenomes of Bombycidae. The gene organization and order of R. menciana mitogenome are identical to the studied lepidopteran mitogenomes (Cameron and Whiting 2008, Liu et al. 2013, Yang et al. 2013). The order of the trnM tRNA gene in the lepidopteran mitogenomes is A + T-rich region, trnM, trnI, trnQ, and nad2, whereas the deduced ancestral gene order is A + T-rich region, trnI, trnQ, trnM, nad2 (Boore et al. 1998). This observation suggests that lepidopteran insects may have acquired the typical gene orientation and order independently after diverging from the ancestral insect.
The A + T content (78.87%) of the R. menciana mitogenome is lower than the other Bombycidea insects (Yukuhiro et al. 2002; Cameron and Whiting 2008; Hong et al. 2008; Jiang et al. 2009; Kim et al. 2009; Hu et al. 2010; Li et al. 2010a; Liu et al. 2012b, 2013; Kim et al. 2014) (Table 4). The AT skewness of the R. menciana mitogenome was 0.050 (Table 4), higher than the R. menciana mitogenome (0.021) of Kim et al. (2014), and lower than that (0.054∼0.059) of the B. mandarina and B. mori, indicating a higher occurrence of A compared with T nucleotides in Bombycidae. Different from the Bombycidae, there were higher occurrence of T compared with A nucleotides in the two families of Sphingidae and Saturniidae (Table 4). The GC skewness of the R. menciana mitogenome was −0.260, indicating a heavy bias toward C versus G nucleotides and much more negative than observed for other Bombycoidea mitogenome (<0.236).
There was an incomplete stop codon of a single T in R. menciana coxII gene. The incomplete stop codon had been found in several invertebrate mitochondrial genes, which seems a common phenomenon of mitochondrion genes (Jiang et al. 2009, Liu et al. 2013, Yang et al. 2013). The relative synonymous codon usage exhibits extensive similarity with other lepidopteran mitogenomes in previous study (Salvato et al. 2008). The most frequent codons in R. menciana are composed of T or a combination of A and T, especially the third codon position. The observed differences in nucleotide composition may caused by the constraints on A + T content in the third codon position, which is more relaxed than those in the first and second codon positions due to degenerated genetic code (Taanman 1999). The C + T content in each of the codon positions were similar, which is agree with the point that the high A + T content in insect mitogenome were caused by the mutation of C to T (The Honeybee Genome Sequencing Consortium 2006).
The length of the intergenic spacer regions (149 bp in 17 regions) is longer than that of Manduca sexta (115 bp in 13 regions) (Cameron and Whiting 2008) and Ac. selene (137 bp in 13 regions) (Liu et al. 2012a) and is shorter than that of C. boisduvalii (194 bp in 16 regions) (Hong et al. 2008) and Bombycidae insects (about 338 bp) (Yukuhiro et al. 2002; Hu et al. 2010; Li et al. 2010a; Liu et al. 2013), which has been suggested to be constitutively synapomorphic and restricted to Ditrysia mitogenomes (Cameron and Whiting 2008, Hao et al. 2012). Similar to the mitogenome of the other insects, the tRNA genes have typical clover-leaf structure. However, the anticodon stem of trnS(UCN) could not form a stable stem-loop structure for the two U-U mismatches. The phenomenon of two U-G mismatches occurred also in the anticodon stem of trnL(CUN) of E. pyretorum (Jiang et al. 2009), B. mori Dazao (Liu et al. 2013), and Ac. selene (Liu et al. 2012a).
The exactly same A + T-rich region occurred between the R. menciana mitogenome of this article and Kim et al. (2014). The length of 360 bp was shorter than that of Bombycidae insects (494 bp∼747 bp) (Yukuhiro et al. 2002; Hu et al. 2010; Li et al. 2010a; Liu et al. 2013) and longer slightly than that of A. yamamai (334 bp) (Kim et al. 2009) and C. boisduvalii (330 bp) (Hong et al. 2008). The A + T content of the region was 91.11%, higher than that of Ac. selene (87.91%) (Liu et al. 2012a), A. pernyi (90.4%) (Li et al. 2011), A. yamamai (90.40%) (Kim et al. 2009), and lower than those from Bombycidae insects (94.42–95.55%). There are some common structures in the A + T-rich region of R. menciana mitogenome, such as ATAGA + poly-T, and tandem repeats elements. The conserved motif of “ATAGA + poly T” stretch at the 24 bp downstream of the rrnS gene is thought to be the origin of the DNA replication (Taanman 1999, Jiang et al. 2009). There are 2.6 tandem repeats elements of 37 bp in the R. menciana A + T-rich region, whereas three 32-bp and three 37-bp tandem repeats elements in B. mori C108 and B. mandarina Qingzhou. In B. mori Dazao, the A + T-rich region harbors two 31-bp repeat elements and three 36-bp repeat elements (Liu et al. 2013). The sequence and location of tandem repeats elements among Bombycidae mitogenomes are nonconserved. The existence of tandem repeats elements maybe caused mainly by the replication slippage.
Mitogenomes are effective markers for deep-level phylogenetic studies in the Lepidoptera. In our analysis, Bombycidae (B. mori, B. mandarina, and R. menciana), Sphingidae (M. sexta), and Saturniidae (Ac. selene, A. pernyi, A. yamamai, S. cynthia ricini, E. pyretorum, and C. boisduvalii) were clustered in one branch of the phylogenetic tree, and the relationship of the three family was Bombycidae + (Sphingidae + Saturniidae), which is consistent with the morphological data and some previous studies (Zwick 2008, Zwick et al. 2011, Liu et al. 2013). The relationship of Geometridae is closer with Bombycoidea in our analyses, which is similar to the study by Yang et al. (2013).
Acknowledgments
This study was supported by the Youth Science and Technology New Star Program of Shaanxi Province, China (2013KJXX-96) and the Science and Technology Research and Development Program of Shaanxi Province, China (2011K01-40).
References Cited
- Avise J. C. 1987. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 18: 489–522. [Google Scholar]
- Ballard J.W.O. 2000. Comparative genomics of mitochondrial DNA in Drosophila simulans. J. Mol. Evol. 51: 64–75. [DOI] [PubMed] [Google Scholar]
- Ballard J.W.O., Rand D. M. 2005. The population biology of mitochondrial DNA and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 36: 621–642. [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore J. L. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore J. L., Lavrov D. V., Brown W. M. 1998. Gene translocation links insects and crustaceans. Nature 392: 667–668. [DOI] [PubMed] [Google Scholar]
- Cameron S. L., Whiting M. F. 2008. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene 408: 112–123. [DOI] [PubMed] [Google Scholar]
- Chai H. N., Du Y. Z. 2012. The complete mitochondrial genome of the pink stem borer, Sesamia inferens, in comparison with four other noctuid moths. Int. J. Mol. Sci. 13: 10236–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Goddard J. M., Martin S. C., Fauron C. M., Wolstenholme D. R. 1982. Drosophila mitochondrial DNA: a novel gene order. Nucleic Acids Res. 10: 6619–6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates B. S., Sumerford D. V., Hellmich R. L., Lewis L. C. 2005. Partial mitochondrial genome sequences of Ostrinia nubilalis and Ostrinia furnicalis. Int. J. Biol. Sci. 1: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y. M., Xiang Z. H. 1993. Cytogenetical studies on Rondotia menciana M. J. Southwest Agric. Univ. 6: 565–568. [Google Scholar]
- Friedrich M., Muqim N. 2003. Sequence and phylogenetic analysis of the complete mitochondrial genome of the flour beetle Tribolium castanaeum. Mol. Phylogenet. Evol. 26: 502–512. [DOI] [PubMed] [Google Scholar]
- Gong Y. J., Shi B. C., Kang Z. J., Zhang F., Wei S. J. 2012. The complete mitochondrial genome of the oriental fruit moth Grapholita molesta (Busck) (Lepidoptera: Tortricidae). Mol. Biol. Rep. 39: 2893–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J., Sun Q., Zhao H., Sun X., Gai Y., Yang Q. 2012. The complete mitochondrial genome of Ctenoptilum vasava (Lepidoptera: Hesperiidae: Pyrginae) and its phylogenetic implication. Comp. Funct. Genomics 2012: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M. Y., Lee E. M., Jo Y. H., Park H. C., Kim S. R., Hwang J. S., Jin B. R., Kang P. D., Kim K. G., Han Y. S., et al. 2008. Complete nucleotide sequence and organization of the mitogenome of the silk moth Caligula boisduvalii (Lepidoptera: Saturniidae) and comparison with other lepidopteran insects. Gene 413: 49–57. [DOI] [PubMed] [Google Scholar]
- Hu X. L., Cao G. L., Xue R. Y., Zheng X. J., Zhang X., Duan H. R., Gong C. L. 2010. The complete mitogenome and phylogenetic analysis of Bombyx mandarina strain Qingzhou. Mol. Biol. Rep. 37: 2599–2608. [DOI] [PubMed] [Google Scholar]
- Jiang S. T., Hong G. Y., Yu M., Li N., Yang Y., Liu Y. Q., Wei Z. J. 2009. Characterization of the complete mitochondrial genome of the giant silkworm moth, Eriogyna pyretorum (Lepidoptera: Saturniidae). Int. J. Biol. Sci. 5:351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Park J. S., Kim M. J., Kang D. P., Kim S. G., Jin R. B., Soo Y., Kim I. 2012. Complete nucleotide sequence and organization of the mitochondrial genome of eri-silkworm, Samia cynthia ricini (Lepidoptera: Saturniidae). J. Asia. Pac. Entomol. 15: 162–173. [Google Scholar]
- Kim M. J., Jun J., Kim I. 2014. Complete mitochondrial genome of the mulberry white caterpillar Rondotia menciana (Lepidoptera: Bombycidae). Mitochondrial DNA 25: 1–3 [DOI] [PubMed] [Google Scholar]
- Kim S. R., Kim M. I., Hong M. Y., Kim K. Y., Kang P. D., Hwang J. S., Han Y. S., Jin B. R., Kim I. 2009. The complete mitogenome sequence of the Japanese oak silkmoth, Antheraea yamamai (Lepidoptera: Saturniidae). Mol. Biol. Rep. 36: 1871–1880. [DOI] [PubMed] [Google Scholar]
- Li D., Guo Y., Shao H., Tellier L. C., Wang J., Xiang Z., Xia Q. 2010a. Genetic diversity, molecular phylogeny and selection evidence of the silkworm mitochondria implicated by complete resequencing of 41 genomes. BMC Evol. Biol. 10:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang X., Fan Z., Yue B., Huang F., King E., Ran J. 2011. Structural characteristics and phylogenetic analysis of the mitochondrial genome of the sugarcane borer, Diatraea saccharalis (Lepidoptera: Crambidae). DNA Cell Biol. 30: 3–8. [DOI] [PubMed] [Google Scholar]
- Li Y. P., Song W., Shi S. L., Liu Y. Q., Pan M. H., Dai F. Y., Lu C., Xiang Z. H. 2010b. Mitochondrial genome nucleotide substitution pattern between domesticated silkmoth, Bombyx mori, and its wild ancestors, Chinese Bombyx mandarina and Japanese Bombyx mandarina. Genet. Mol. Biol. 33: 186–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao F., Wang L., Wu S., Li Y. P., Zhao L., Huang G. M., Niu C. J., Liu Y. Q., Li M. G. 2010. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int. J. Biol. Sci. 6: 172–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. N., Zhu B. J., Dai L. S., Wei G. Q., Liu C. L. 2012a. The complete mitochondrial genome of the wild silkworm moth, Actias selene. Gene 505: 291–299. [DOI] [PubMed] [Google Scholar]
- Liu Q. N., Zhu B. J., Dai L. S., Liu C. L. 2013. The complete mitogenome of Bombyx mori strain Dazao (Lepidoptera: Bombycidae) and comparison with other lepidopteran insects. Genomics 101: 64–73. [DOI] [PubMed] [Google Scholar]
- Liu Y. Q., Li Y. P., Wang H., Xia R. X., Chai C.-L., Pan M. H., Lu C., Xiang Z. H. 2012b. The complete mitochondrial genome of the wild type of Antheraea pernyi (Lepidoptera: Saturniidae). Ann. Entomol. Soc. Am. 105: 498–505. [Google Scholar]
- Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. S., Shin K. S., Kim M. S., Park H., Cho S., Kim C. B. 2006. The mitochondrial genome of the smaller tea tortrix Adoxophyes honmai (Lepidoptera: Tortricidae). Gene 373: 52–57. [DOI] [PubMed] [Google Scholar]
- Mahendran B., Ghosh S. K., Kundu S. C. 2006. Molecular phylogeny of silk-producing insects based on 16S ribosomal RNA and cytochrome oxidase subunit I genes. J. Genet. 85: 31–38. [DOI] [PubMed] [Google Scholar]
- Perna N. T., Kocher T. D. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41: 353–358 [DOI] [PubMed] [Google Scholar]
- Salvato P., Simonato M., Battisti A., Negrisolo E. 2008. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genomics 9:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taanman J. W. 1999. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta 1410:103–123. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Honeybee Genome Sequencing Consortium. 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443:931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme D. R. 1992. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 141: 173–216. [DOI] [PubMed] [Google Scholar]
- Wu Q. L., Gong Y. J., Gu Y., Wei S. J. 2013. The complete mitochondrial genome of the beet armyworm Spodoptera exigua (Hübner) (Lepodiptera: Noctuidae). Mitochondrial DNA 24: 31–33. [DOI] [PubMed] [Google Scholar]
- Xu M., Zhang Y., Zhu X. F., Yan S. J., Chen J., Wang P. S. 1994. Studies on biological characters and control of mulberry white caterpillar, Rondotia menciana Moore. Acta Sericologica Sinica 20: 136–140. [Google Scholar]
- Yang L., Wei Z. J., Hong G. Y., Jiang S. T., Wen L. P. 2009. The complete nucleotide sequence of the mitochondrial genome of Phthonandria atrilineata (Lepidoptera: Geometridae). Mol. Biol. Rep. 36: 1441–1449. [DOI] [PubMed] [Google Scholar]
- Yang X., Xue D., Han H. 2013. The complete mitochondrial genome of Biston panterinaria (Lepidoptera: Geometridae), with phylogenetic utility of mitochondrial genome in the Lepidoptera. Gene 515: 349–358. [DOI] [PubMed] [Google Scholar]
- Yin J., Hong G. Y., Wang A. M., Cao Y. Z., Wei Z. J. 2010. Mitochondrial genome of the cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae) and comparison with other Lepidopterans. Mitochondrial DNA 21: 160–169. [DOI] [PubMed] [Google Scholar]
- Yin J., Wang A. M., Hong G. Y., Cao Y. Z., Wei Z. J. 2011. Complete mitochondrial genome of Chilo suppressalis (Walker) (Lepidoptera: Crambidae). Mitochondrial DNA 22: 41–43. [DOI] [PubMed] [Google Scholar]
- Yin Y., Qu F., Yang Z., Zhang X., Yue B. 2014. Structural characteristics and phylogenetic analysis of the mitochondrial genome of the rice leafroller, Cnaphalocrocis medinalis (Lepidoptera: Crambidae). Mol. Biol. Rep. 41: 1109–1116. [DOI] [PubMed] [Google Scholar]
- Yukuhiro K. S., Itoh H., Shimizu K., Banno Y. 2002. Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori. Mol. Biol. Evol. 19: 1385–1389. [DOI] [PubMed] [Google Scholar]
- Zhang D. X., Hewitt G. M. 1997. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 25: 99–120. [Google Scholar]
- Zhang D. X., Szymura J. M., Hewitt G. M. 1995. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 40: 382–391. [DOI] [PubMed] [Google Scholar]
- Zhao J. L., Zhang Y. Y., Luo A. R., Jiang G. F., Cameron S. L., Zhu C. D. 2011. The complete mitochondrial genome of Spilonota lechriaspis Meyrick (Lepidoptera: Tortricidae). Mol. Biol. Rep. 38:3757–3764. [DOI] [PubMed] [Google Scholar]
- Zwick A. 2008. Molecular phylogeny of Anthelidae and other bombycoid taxa (Lepidoptera: Bombycoidea). Syst. Entomol. 33: 190–209. [Google Scholar]
- Zwick A., Regier J. C., Mitter C., Cummings M. P. 2011. Increased gene sampling yields robust support for higher-level clades within Bombycoidea (Lepidoptera). Syst. Entomol. 36: 31–43. [Google Scholar]