Abstract
Ferritin is a 24-subunit molecule, made up of heavy chain (HC) and light chain (LC) subunits, which stores and controls the release of dietary iron in mammals, plants, and insects. In mosquitoes, dietary iron taken in a bloodmeal is stored inside ferritin. Our previous work has demonstrated the transport of dietary iron to the ovaries via ferritin during oogenesis. We evaluated the localization of ferritin subunits inside CCL-125 [Aedes aegypti Linnaeus (Diptera: Culicidae), yellow fever mosquito] and 4a3b [Anopheles gambiae Giles (Diptera: Culicidae), African malaria mosquito] cells under various iron treatment conditions to further elucidate the regulation of iron metabolism in these important disease vectors and to observe the dynamics of the intracellular ferritin subunits following iron administration. Deconvolution microscopy captured 3D fluorescent images of iron-treated mosquito cells to visualize the ferritin HC and LC homologue subunits (HCH and LCH, respectively) in multiple focal planes. Fluorescent probes were used to illuminate cell organelles (i.e., Golgi apparatus, lysosomes, and nuclei) while secondary probes for specific ferritin subunits demonstrated abundance and co-localization within organelles. These images will help to develop a model for the biochemical regulation of ferritin under conditions of iron exposure, and to advance novel hypotheses for the crucial role of iron in mosquito vectors.
Keywords: 4a3b, Aedes aegypti, CCL-125, deconvolution microscopy
Mosquitoes transmit pathogens that cause numerous infectious diseases including dengue, yellow fever, and malaria. These diseases exact a high cost on millions of lives and are the cause of abundant morbidity in endemic regions (WHO 2013a,b,2014). Infection occurs when a female blood feeds on a host to acquire sufficient nutrients for egg production (Telang et al. 2013; Jason Pitts et al. 2014). Although transmission rates are low, numbers of infected individuals are very high reflecting the numbers of disease vectors. The bloodmeal is iron rich (Zhou et al. 2007). The majority of iron absorbed from the diet is transported to the ovaries for egg production (Zhou et al. 2007), and egg numbers and progeny survivorship may fall when the insects receive an iron limited meal (Kogan 1990; Jason Pitts et al. 2014). We are interested in the potential for interference with iron metabolism or iron limitation as an insect control strategy.
We previously reported that following dietary iron intake, mosquitoes produce ferritin that is secreted into hemolymph from the midgut loaded with iron, and that ferritin, rather than transferrin, is the primary iron transport protein in these animals (Zhou et al. 2007). Earlier work in the yellow fever mosquito, Aedes aegypti Linnaeus (Diptera: Culicidae), demonstrated the presence of two ferritin subunits, heavy and light chain homologues (HCH and LCH, respectively) that are similar to the heavy and light chains subunits of mammals and other animals (Pham et al. 2000; Geiser et al. 2003). Synthesis of either homologue is subject to positive transcriptional control by iron (Dunkov et al. 2002; Pham et al. 2003; Pham and Chavez 2005), while translation of the HCH is subject to positive control by iron via an iron responsive element (IRE) found in the message (Zhang et al. 2002). In contrast to mammals, no IRE is found in the message for the LCH (Geiser et al. 2003).
In mammals, ferritin is found in cell cytoplasm, increases in response to iron exposure and serves as the primary iron storage protein (Arosio and Levi 2010; Linder 2013). Ferritin also is the primary iron storage protein in mosquitoes and expression increases in response to iron (Zhou et al. 2007; Geiser et al. 2009). However, unlike mammalian ferritin that is localized to the cytoplasm, Ae. aegypti ferritin is found primarily in the membranes of animal tissues and larval epithelial CCL-125 cells (Geiser et al. 2007). Cells show a linear uptake of iron in direct proportion to iron level of the culture medium (Geiser et al. 2006). The majority of the iron is stored in the membranes and membrane iron exceeds cytoplasmic iron by an order of magnitude. Iron exposure will increase both cytoplasmic and membrane ferritin. However, membrane ferritin plateaus because CCL-125 cells secrete iron-loaded ferritin into the culture medium, and thereby, limit cellular iron levels.
Iron can participate in the Fenton and Haber–Weiss reactions producing toxic free radicals. Ferritin is considered cytoprotective against oxidative stress because it converts ferrous to ferric and stores ferric in a complex that prevents free radical formation (Arosio and Levi 2010). Because female mosquitoes receive a high iron load in the bloodmeal, while males of the species survive on nectars with little iron, we are studying how mosquitoes cope with a high iron load. In addition to the studies in larval CCL-125 cells, we evaluated the effects of iron exposure in a second mosquito cell line, Anopheles gambiae Giles [(Diptera: Culicidae), African malaria mosquito] 4a3b cells. This cell line originates from larvae and is thought to be derived from hemocytes, immune-like phagocytic cells found in the open circulatory system of insects (Muller et al. 1999). Unlike our previous studies in CCL-125 cells, 4a3b cells accumulate high levels of iron and do not secrete iron-loaded ferritin to limit cellular iron accumulation (Geiser et al. 2009).
Materials and Methods
Cell Culture and Experimental Protocols
An. gambiae larval hemocyte-like cells (4a3b), a generous gift from Dr. Michael R. Strand (University of Georgia, Athens, GA), were maintained in Anopheles medium: Schneider’s media (Catalogue # 21720024, Invitrogen Corporation, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (Catalogue # 100-500, Gemini Bio-Products, Calabasas, CA) and 1% antibiotics/antimycotics (Catalogue # 15240062, Invitrogen), as stock cultures in vented 75 cm2 tissue culture flasks (Corning Inc., Corning, NY) incubated in a water-jacketed incubator with 10% humidity and a 95% air-5% CO2 atmosphere at 28°C. Confluent cells were split 1:2 and cells were 80% confluent in 3 d. All experiments were performed on cells at >80% confluence under sterile conditions and BSL2 containment protocols. At the start of each experiment, the complete medium was removed and the cells were washed twice with Hank’s Balanced Salt Solution (HBSS; Catalogue # 12009805, Invitrogen). Schneider’s media was added and the cells were incubated for 1 h. Following this incubation the medium was replaced with fresh Schneider’s media and supplemented with HBSS (0, control), 50–500 μM ferric ammonium citrate (F, FAC; Catalogue # F5879, Sigma, St Louis, MO, 18.3% iron, ∼1 μg Fe/μg FAC) in HBSS, 200 μM FAC and 200 μM deferoxamine mesylate salt (DFO; Catalogue # D9533, Sigma) or 500 μM FAC plus 500 μM DFO (F/D, FAC/DFO) in HBSS, or 200 μM DFO or 500 μM DFO (D) in HBSS, and incubated for 18 h. Since not all cells at the time of harvest adhere, the medium was removed from the flask of cells, transferred to a 15 ml conical tube, and centrifuged at 900 g for 10 min, 4°C. The supernatant was removed, flash frozen in liquid nitrogen and stored at −80°C for media protein analysis. The remaining cells in the flasks were scraped into 3 ml HBSS, added to the cell pellet from the medium and suspended. The cell suspension was centrifuged at 900 g for 10 min, 4°C. The supernatant was removed and the cells were suspended in 5 ml fresh HBSS. Samples of the cell suspensions from each treatment group were immediately taken to perform cell viability, total cell number, and calcein quench assays. One ml (∼1.5 × 106 cells) of the cell suspension was taken from each sample and centrifuged at 9000 g for 2 min, 4°C. The supernatant was removed and the cell pellet was frozen in liquid nitrogen and stored at −80°C for RNA isolation. The remaining cell suspensions were centrifuged at 900 g for 10 min, 4°C. The supernatant was removed and the cell pellets were suspended in hypotonic buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, freshly added 1× Protease Inhibitor Cocktail Set I (Catalogue # 539131, EMD Millipore, Billerica, MA) and 0.5 mM DTT), transferred to a fresh 1.5 ml microcentrifuge tube and frozen in liquid nitrogen and stored at −80°C for cell protein extraction.
We conducted a high iron experiment to verify the results from the moderate iron experiments. For this experiment, 4a3b cells were treated with 0 μM FAC, or 500 μM FAC, FAC/DFO or DFO. Cell treatment and analyses were done as indicated herein. All experiments were conducted in triplicate.
RNA Isolation
Total RNA was isolated from ∼1.5 × 106 4a3b cells using the RNeasy Mini Kit (Catalogue # 74124, Qiagen Inc., Valencia, CA). Purified total RNA was treated with DNase I (Catalogue # AM2222, Invitrogen) according to the manufacturer’s instructions for 15 min at 25°C and for 10 min at 65°C. The DNase-treated total RNA was used for real-time RT-PCR.
Media and Cell Extracts
Media samples were concentrated 5-fold by centrifuging in Centricon Ultracel YM-30 centrifugal filter devices (EMD Millipore) at 7,500 g for ∼20 min, 4°C. The concentrated media samples were frozen in small aliquots and held at −80°C until use. Media protein concentrations were determined by the method of Bradford (1976).
Cytoplasmic extracts were prepared as previously described with some modification (Ausubel et al. 1998). Briefly, cell pellets suspended in hypotonic buffer and stored at −80°C were frozen and thawed twice on ice to rupture the cell membranes. The cell suspensions were centrifuged at 100,000 g for 30 min, 4°C; the supernatants were frozen in small aliquots and held at –80°C until use and the supernatant protein was determined by Bradford assay that measures protein concentration in the range of 1 to 1500 µg/ml.
The remaining cell pellets were suspended in hypotonic buffer, frozen in small aliquots, and held at −80°C until use. These pellets contain plasma membranes, Golgi and other cell membrane components and are referred to as membrane extracts. In the moderate iron exposure experiments, we also attempted to extract the membrane proteins using a detergent as previously described by others (Patton et al. 2005). Briefly, membrane extracts were thawed on ice and Triton x-100 (Catalogue # X-100, Sigma) was diluted in each extract to a final concentration of 0.476%. Samples were incubated for 1 h, 4°C and then centrifuged at 20,000 g for 5 min, 4°C. Triton x-100-treated supernatants were frozen in small aliquots and held at −80°C until use. Triton x-100-treated, membrane extract protein concentrations were determined by the SDS-Lowry method (Markwell et al. 1978). However, when cytoplasmic extracts were found to yielded too little protein for assay, the cytoplasmic extracts were recombined with the Triton x-100-treated membrane extracts, and such samples are referred to as cell extracts. The final concentrations of these extracts were determined by the SDS-Lowry method.
Iron Measurements
Calcein fluorescent quench by iron was used as an indirect measure of iron uptake into cells. Fluorescent quench was measured in triplicate for all flasks using the LIVE/DEAD Viability/Cytotoxicity Assay per manufacturer’s instructions (Catalogue # L3224, Invitrogen) by adding calcein to a final concentration of 1 µM and incubating for 45 min, 28°C. The calcein fluorescent quench measurements were corrected to a percentage of control (i.e., HBSS-treated cells).
Iron Inductively Coupled Plasma-Mass Spectrometry (ICP-MS) was conducted as previously described (Geiser et al. 2006) and used as a direct measurement of iron entry into cells. Briefly, cell extracts were prepared by diluting 20 μl of each sample in 5 ml of 1% HNO3. MilliQ H20 and 1% HNO3 were used as negative controls for iron contamination of diluted samples and equipment. The Elan Dynamic Reaction Cell (DRC) II (Perkin Elmer SCIEX, Shelton, CT) was used by the Arizona Laboratory for Emerging Contaminants (ALEC) at the University of Arizona (Tucson, AZ) to quantitate iron in our samples. The iron ICP-MS data was transformed to μg Fe/μg cell protein.
Real-Time RT-PCR
DNase-treated total RNA was used for real-time RT-PCR. Reverse transcription was done according to the manufacturer’s instructions using M-MuLV Reverse Transcriptase from the First Strand cDNA Synthesis Kit (Catalogue # K1612, Fermentas, Inc.). The primers for the PCR reactions were designed to obtain specific PCR products of similar size for the ORF of each message: Ferritin Heavy Chain Homologue (HCH, 118 bp): 5′-TTCGGA ATCGTTGCCCTGATGTTC-3′ and 3′-CTGCTTACCTTGA TGTA C TTGGCAT-5′; Ferritin Light Chain Homologue (LCH, 139 bp): 5′-GATGGCAAAGATG AATGT TGTG-3′ and 3′-GTCGAAAGACAGCGCAAGG-5′; and Ribosomal Protein S7 (RPS7, 160 bp): 5′-GGTGTTCGGTTCCAA GGTG-3′ and 3′-CACTCCAGCTCAAGTTGTTG-5′. Real-time RT-PCR reactions were conducted using iQ SYBR Green Supermix (Catalogue # 12002500, BIORAD, Hercules, CA) with the buffers provided at: 94°C, 3 min, 1 cycle; 94°C, 10 s; 60°C, 30 s and 72°C, 30 s, 40 cycles; with a melt curve over a temperature range starting at 55°C and ending at 95°C in a MyiQ Cycler (BIORAD). PCR product quality was monitored using post-PCR melt curve analysis. A standard curve for each product showed the experimental samples fell within the linear range. Primer pair efficiencies for each message are indicated as follows: HCH, 100%; LCH 100%; RPS7, 105%. Data was analyzed by MyiQ Optical System Software (Version 1, BIORAD) and fold change was quantified using the Pfaffl method to calculate for relative quantification (Pfaffl 2001) utilizing RPS7 transcript as the housekeeping gene. All PCR products were cloned and sequenced to determine that the product sequence represents that of the desired message.
Immunoblots
Cellular proteins from the moderate iron exposure experiments were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using Criterion TGX Any kD Precast Gels (Catalogue # 567-1123, BIORAD) under denaturing conditions for ∼163 Vh at RT. Protein samples were boiled for 20 min in loading buffer containing β-mercaptoethanol (βME) prior to applying samples onto gels. Concentrated media proteins from these experiments were measurable by the Bradford method; however, levels were so low that resolved proteins were undetected by immunoblot. Cell extracts from these experiments showed insufficient total protein concentration such that the gels were loaded by the maximum volume, 32.8 μl. Actin, as an internal control for loading, was undetectable in the cell extracts, so purified FLAG-containing bacterial alkaline phosphatase protein (Catalogue # P7457, Sigma, 25 ng) was added as an exogenous loading control.
Proteins from the high iron experiment were resolved by SDS-PAGE using 17.5% homogeneous slab gels under denaturing conditions for ∼1370 Vh, 4°C. Protein samples were boiled for 20 min in loading buffer containing βME prior to applying samples onto gels. Cell extracts were loaded onto gels at 10 μg total protein and the concentrated media samples at 3 μg total protein. Actin was used as an internal control for loading of cell extracts, and since no secreted control protein is available for use with medium, FLAG (Sigma, 25 ng) was added as an exogenous loading control. Protein extract from 48 h post-blood fed (PBF) Anopheles stephensi Liston [(Diptera: Culicidae), Indo-Pakistan malaria mosquito] females (10 μg) served as a positive control (+) for ferritin and actin (see mosquito rearing protocol in Supplementary Methods).
Proteins were transferred to nitrocellulose membranes in the Electrophoretic Blotting System (C.B.S. Scientific Company, Inc., Del Mar, CA). Efficient transfer of proteins was confirmed by SYPRO Ruby protein blot stain (Catalogue # 179-3127, BIORAD) and Spectra Multicolor Broad Range Protein Ladder (Catalogue # 26634,Thermo, Waltham, MA) or Kaleidoscope molecular weight markers (Catalogue # 161-0375, BIORAD), as designated. The nitrocellulose membranes were cut for analysis of the individual proteins of interest. The HCH and LCH nitrocellulose membranes were blocked overnight in Tris-buffered saline (140 mM NaCl, 2.7 mM KCl, 24.8 mM Tris base) and 0.1% Tween-20 (TTBS; Catalogue # P1379, Sigma), pH 7.4 with 1% non-fat dry milk (NFDM) and 3% bovine serum albumin (BSA), 4°C. The FLAG and actin ntirocellulose membranes were blocked overnight in phosphate-buffered saline (140 mM NaCl, 2.7 mM KCl, 5.4 mM Na2HPO4·7H2O, 1.8 mM KH2PO4) and 0.05% Triton x-100 (PTX), pH 7.4 with 7% NFDM and 3% BSA, 4°C. After blocking, the nitrocellulose membranes were incubated with anti-An. gambiae HCH-specific rabbit serum (diluted in 50% glycerol; 1:250 v/v; antigen epitope: ASEEREHGMKLIEY), anti-An. gambiae LCH-specific rabbit serum (diluted in 50% glycerol; 1:125 v/v; antigen epitope: EKLYRKISDKAWAD), anti-FLAG-specific rabbit serum (1:4000 v/v; Catalogue # F7425, Sigma) or anti-β-actin-specific mouse serum (1:2000 v/v; Catalogue # ab8224, Abcam, Cambridge, MA) diluted in the appropriate incubation buffer system for 2 h, RT. Specificity of An. gambiae antiserums is demonstrated in Supplementary Fig. 3SA and SB. The HCH, LCH, and FLAG nitrocellulose membranes were detected with anti-rabbit alkaline phosphatase-conjugated antibody (1:1000 v/v; Catalogue # 711-055-152, Jackson Immuno, West Grove, PA) according to the manufacturer’s protocol. The actin nitrocellulose membranes were detected with anti-mouse phosphatase-conjugated antibody (1:1000 v/v; Catalogue # 715-055-150, Jackson Immuno). Digital images were accessed using the VersaDoc 3000 Imaging System (BIORAD).
Immunocytochemistry
Both 4a3b and Ae. aegypti larval epithelial-like cells (CCL-125) obtained from the American Type Culture Collection (Manassas, VA) were cultured to perform immunocytochemistry (ICC). 4a3b cells were maintained as described previously while CCL-125 cells were maintained in Aedes medium: 75% DMEM high glucose (Catalogue # 11965092, Invitrogen) and 25% Sf-900 II SFM (Catalogue # 10902096, Invitrogen) supplemented with 15% heat-inactivated fetal bovine serum (Gemini Bio-Products) and 0.15% antibiotics/antimycotics (Invitrogen), as stock cultures in vented 75 cm2 tissue culture flasks (Corning Inc.) incubated in a water-jacketed incubator with 10% humidity and a 95% air-5% CO2 atmosphere, 28°C. Confluent cells were split 1:2 and cells were 80% confluent when split.
For ICC experiments, 4a3b and CCL-125 cells were grown until 80% confluent on #1.5, 10 mm, round, glass coverslips (Ted Pella, Inc., Redding, CA) under sterile conditions and BSL2 containment protocols. At the start of each experiment the complete medium was removed and the cells were washed twice with HBSS. The appropriate serum-free, antibiotics/antimycotics-free mosquito cell medium was placed on the cells and the cells were incubated for 1 h. Following this incubation the medium was replaced with fresh serum-free, antibiotics/antimycotics-free mosquito cell medium and supplemented with 100 μM FAC (Sigma) in HBSS (FAC) or 100 μM FAC with 100 μM DFO (Sigma) in HBSS (F/D) and incubated for 18 h, 28°C.
After treatment, 4a3b and CCL-125 cells were rinsed in HBSS and a portion of the live cells were stained with 5 μM BODIPY TR C5-ceramide (Catalogue # B34400, Invitrogen) for 30 min, 28°C. In mammals, this ceramide analog produces selective staining of the Golgi complex for visualization by fluorescent microscopy (Celis 1998; Cooper et al. 1999; Pagano et al. 1991). All cells were then fixed in 2% paraformaldehyde/HBSS solution for 20 at RT before permeabilization with 0.2% Triton x-100 (Sigma) for 10 min, RT. All fixed cells were stained for 5 min with 300 nM DAPI (Catalogue # D3571, Invitrogen), 28°C. The portion of fixed cells that were not stained with BODIPY TR C5-ceramide, were stained with 50 nM LysoTracker (Catalogue # L7528, Invitrogen) diluted in HBSS for 45 min, 28°C.
To reduce non-specific binding of antibodies, stained 4a3b and CCL-125 cells were incubated in 2% BSA/1% donkey serum/HBSS for 30 min, RT. 4a3b cells were incubated with anti-An. gambiae HCH-specific rabbit serum (1:150 v/v) or anti-An. gambiae LCH-specific rabbit serum (1:150 v/v) in HBSS for 1 h, RT and washed twice in HBSS for 15 min each. CCL-125 cells were incubated with anti-Ae. aegypti HCH-specific rabbit serum (1:150 v/v; antigen epitope: ATLKKMKKSAPKL) or anti-Ae. aegypti LCH-specific rabbit serum (1:150 v/v; antigen epitope: YNLDSIIKEDKTKD) in HBSS for 1 h, RT and washed twice in HBSS for 15 min each. Specificity of Ae. aegypti antiserums are demonstrated in Supplementary Fig. 3SC. Following the primary antibody incubation, 4a3b and CCL-125 cells were incubated with Cy2-conjugated goat anti-rabbit IgG (1:200 v/v; Catalogue # unavailable, Thermo) in HBSS for 45 min, RT. Coverslips with 4a3b and CCL-125 cells were preserved on glass slides with ProLong Gold antifade reagent (Catalogue # P36930, Invitrogen) according to the manufacturer’s protocol. Images of 4a3b and CCL-125 cells were captured using a DeltaVision RT Restoration Microscope System (Applied Precision, Inc., Issaquah, WA) with a research-grade inverted Olympus IX 70 microscope (Center Valley, PA; including a cooled CCD camera, fiber optic coupled mercury arc lamp, high-speed filter changers, electronic shutters, and a high precision x-y-x positioning system) in the Cellular Imaging Facility Core at the University of Arizona (Tucson, AZ). Images were analyzed using ImageJ 1.47 bundled with 64-bit Java for Windows (National Institutes of Health, Bethesda, MD). Overlaid images were merged and processed using ImageJ.
Statistical Analyses
Treatment differences were determined by one-way analysis of variance using the Tukey’s multiple comparisons test or one-tailed unpaired t-test for comparison of selected data sets (Graph Pad Software, Inc., San Diego, CA). Experiments were conducted simultaneously in triplicate, labeled as A, B and C, and the data for a given variable were analyzed at the same time.
Results
An. gambiae ferritin subunits are most similar to those of Ae. aegypti. Like Aedes, An. gambiae HCH and LCH genes lay in a head-to-head configuration and share a small promoter region (Supplementary Fig. 1S) (Geiser et al. 2003). The An. gambiae HCH transcript has a predicted IRE in the 5′UTR, whereas the LCH transcript does not (Supplementary Fig. 1S). The IRE of the HCH transcript is predicted to be alternatively spliced. The deduced amino acid sequence for the HCH subunit has a putative peptide signal sequence and a consensus ferroxidase site (Supplementary Fig. 2SA), while the deduced amino acid sequence for the LCH subunit shows a putative signal sequence, a consensus porphyrin binding pocket, and some predicted sites for formation of salt bridges (Supplementary Fig. 2SB).
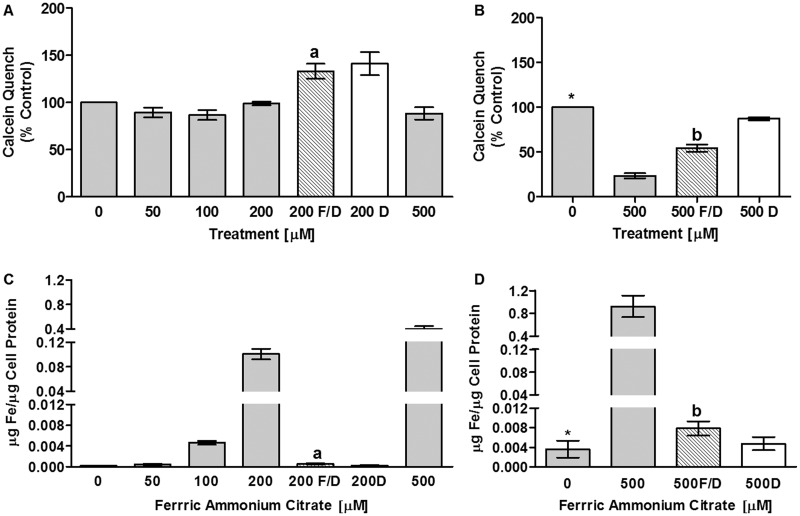
Calcein fluorescent quench by iron is used as an indirect measurement of cytoplasmic iron acquisition. In An. gambiae 4a3b cells, cytoplasmic iron did not increase significantly with increasing iron dose (Fig. 1A and B). Fluorescence declined with 200 and 500 μM FAC treatment relative to fluorescence for cells rescued from iron exposure with DFO (FAC/DFO) indicating iron was responsible for the quench observed in cells treated with FAC. 4a3b cells have little cytoplasm (Supplementary Fig. 5S) relative to Ae. aegypti CCL-125 cells, and extract was insufficient for analyses. Therefore, we measured 4a3b cell-associated iron by iron ICP-MS, and report that, despite little change in the cytoplasmic pools as shown by calcein fluorescence quench, cell-associated iron dramatically increases with iron dose particularly at higher concentrations (Fig. 1C and D). The increase in iron is mitigated by addition of DFO with FAC (Fig. 1C and D). Although 4a3b cells accumulate iron, iron treatment does not adversely influence total cell number or cell viability (Supplementary Fig. 4S). These findings concur with those for CCL-125 cells where iron increases in both cytoplasmic and membrane extracts up to 200 μM FAC with the greatest increase in membrane iron (Geiser et al. 2009). However, they differ from CCL-125 cells in that iron levels of CCL-125 cells plateau in cytoplasm and decrease in membrane extracts at 500 μM FAC (Geiser et al. 2009). These studies document that 4a3b cells take up and retain iron at high levels following iron exposure.
Fig. 1.
Iron enters and is retained by 4a3b cells. Iron uptake into 4a3b cells was measured by calcein fluorescent quench (A & B) and iron ICP-MS (C & D). Data is pooled for two separate experiments each completed in triplicate. A. Calcein quench was not changed by treatment with 0–500 μM (FAC); C. Iron levels as measured by ICP-MS were increased by 0–500 μM FAC (P < 0.0001); aSignificantly different (SD) from 200 μM FAC (P < 0.007); bSD from 500 μM FAC (P < 0.005); *SD from 500 μM FAC (P < 0.02).Key: 0–500 = 0–500 μM FAC; 200 or 500 F/D = 200 or 500 μM FAC/DFO; 200 or 500 D = 200 or 500 μM DFO; FAC = ferric ammonium citrate; DFO = deferoxamine mesylate salt.
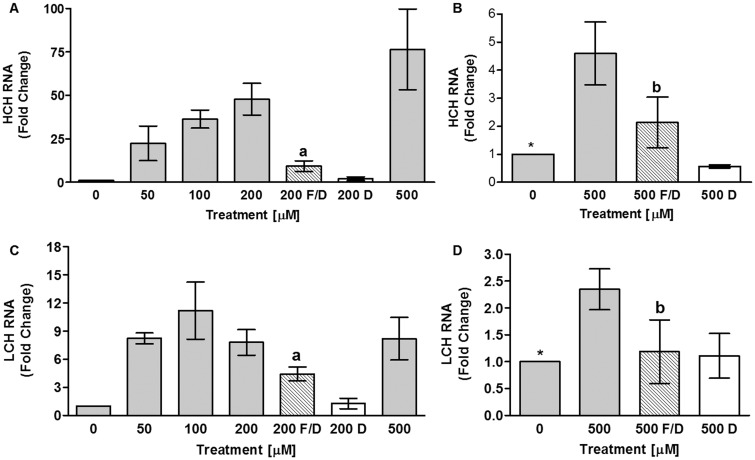
Further documentation of iron entry into 4a3b cells is demonstrated by increases in HCH mRNA expression (Fig. 2A and B), and the absence of this effect when cells are rescued by DFO treatment. HCH mRNA expression appears to follow a linear trend of iron entry into cells (Fig. 2A). In CCL-125 cells, HCH mRNA initially increases with iron at lower doses and then plateaus (Geiser et al. 2006; Geiser et al. 2009). Iron entry into 4a3b cells also increases LCH mRNA expression (Fig. 2C and D). LCH mRNA expression does not follow the linear pattern of cell-associated iron, but increases and is maximal at 50 μM FAC (Fig. 2C). This pattern differs from that for CCL-125 cells, where LCH mRNA increases linearly with FAC dose up to 200 μM FAC, and then plateaus (Geiser et al. 2009).
Fig. 2.
Iron entry into 4a3b cells increases expression of HCH and LCH mRNA. The effect of iron uptake into 4a3b cells on ferritin subunit transcript expression was measured by real-time RT-PCR. Data is pooled for two separate experiments each done in triplicate. HCH mRNA is changed by 0–500 μM FAC (P < 0.02) as is LCH mRNA (P < 0.05), but the patterns differ. aSD from 200 μM FAC (P < 0.05); bSD from 500 μM FAC (P ≤ 0.0663); *SD from 500 μM FAC (P < 0.05). Key: 0–500 = 0–500 μM FAC; 200 or 500 F/D = 200 or 500 μM FAC/DFO; 200 or 500 D = 200 or 500 μM DFO; FAC = ferric ammonium citrate; DFO = deferoxamine mesylate salt.
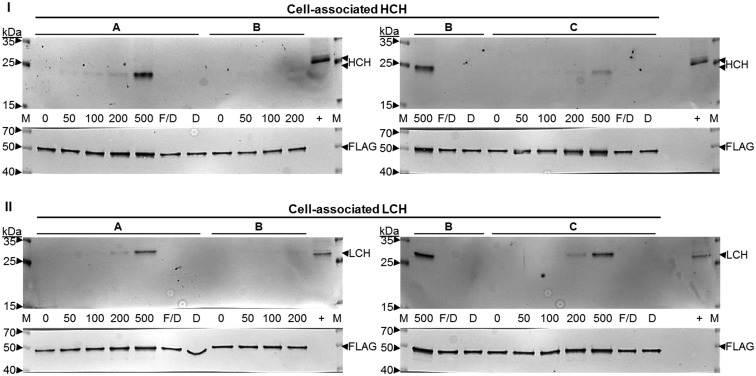
Antiserum specific for the HCH and LCH were obtained to detect the individual ferritin subunits in tissues and cells (Supplementary Fig. 3SA and SB). Iron entry into 4a3b cells increases cell-associated HCH and LCH protein (Figs. 3 and 4). Both subunits increase with FAC dose to 500 μM (Figs. 3 and 4). This differs from findings for CCL-125 cells, where ferritin increases and is maximal at 25 μM FAC (Geiser et al. 2006; Geiser et al. 2009). 4a3b cell-associated HCH expression follows the pattern of cell-associated iron levels and HCH mRNA expression (Figs. 1, 2A and B). LCH expression follows the pattern of iron entry into cells, and not mRNA expression that is maximal at 50 μM FAC (Figs. 1, 2C and D).
Fig. 3.
Iron entry into 4a3b cells increases cell-associated HCH and LCH protein. The effect of iron uptake into 4a3b cells on cell-associated ferritin subunit expression was measured by separation of cell extract protein (32.8 μl) using SDS-PAGE and detection of the HCH (I) and LCH (II) subunits by immunoblot assay. Data are triplicates from one experiment and are representative. Key: M = Spectra Protein Ladder (Thermo); 0–500 = 0–500 μM FAC; 200 or 500 F/D = 200 or 500 μM FAC/DFO; 200 or 500 D = 200 or 500 μM DFO;+ = Control Anopheles stephensi 48 h post-blood fed (PBF) female (10 μg protein); FAC = ferric ammonium citrate; DFO = deferoxamine mesylate salt
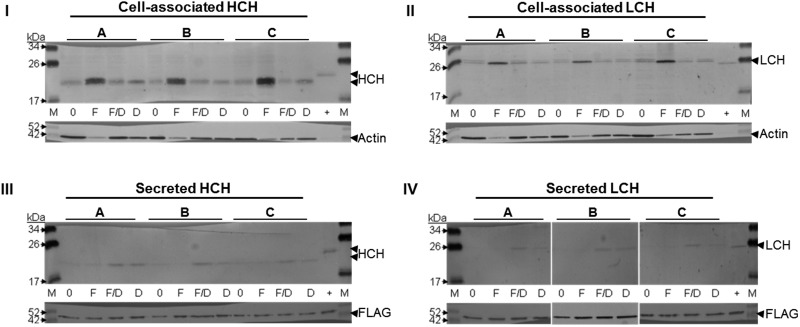
Fig. 4.
High concentrations of iron stimulate 4a3b cell-associated HCH and LCH protein without increasing ferritin secretion. The effect of iron uptake into 4a3b cells on cell-associated (I & II; 10 μg) and secreted (III & IV; 3 μg) ferritin subunit expression was measured by separation of cell extract and concentrated media protein using SDS-PAGE and detection with HCH (I & III) and LCH (II & IV) subunit-specific antiserum by immunoblot assay. Data are triplicates from one experiment and are representative. Key: M = Spectra Protein Ladder (Thermo); 0 = 0 μM FAC; F = 500 μM FAC; F/D = 500 μM FAC/DFO; D = 500 μM DFO; + = Control An. stephensi 48 h PBF female (10 μg); FAC = ferric ammonium citrate; DFO = deferoxamine mesylate salt
CCL-125 cells secrete iron-loaded ferritin in direct proportion to iron dose (Geiser et al. 2006; Geiser et al. 2009). We attempted to evaluate ferritin in culture medium of 4a3b cells. We found protein levels of 4a3b cell culture medium extremely low and maximal concentration of the medium for cells treated with moderate iron dose yielded insufficient protein for subunit analysis by immunoblotting. However, we were able to obtain results for the medium for cells from the 500 μM FAC experiment and results indicate that even at this high iron dose, 4a3b cells secrete little ferritin into the culture medium (Fig. 4III and IV). Surprisingly, detection of the subunits was greater for the rescue (FAC/DFO) and chelator (DFO) treated cells suggesting that in the absence of iron some constitutive secretion of HCH and LCH subunits from 4a3b cells occurs (Fig. 4III and IV). These results were so strikingly different from those of CCL-125 cells, that we visualized the location of the respective ferritin subunits in either cell type.
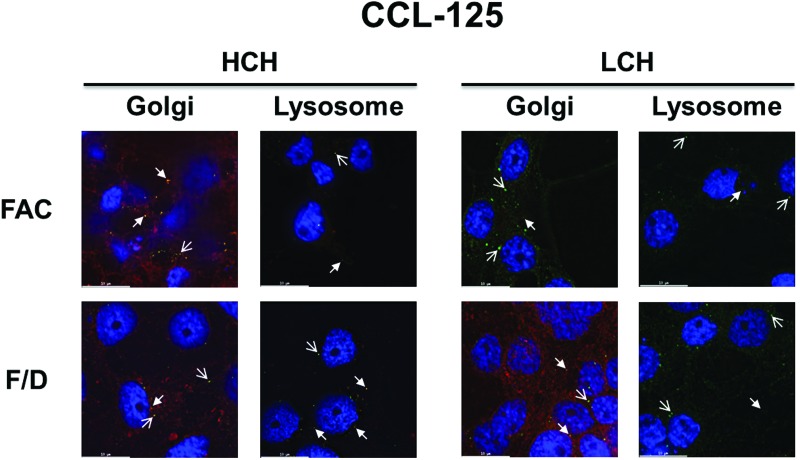
In CCL-125 cells, both ferritin subunits are constitutively expressed regardless of iron status (green) (Geiser et al. 2006; Geiser et al. 2007; Geiser et al. 2009) and seem to coalesce with fluorescent-labeled organelles (closed arrows). The subunits also appear to be found in individual, non-organelle labeled, punctate vesicles (open arrows; Supplementary Figs. 5S, 6S and 7S). HCH and LCH subunit co-localization with the Golgi appears to be similar with 100 μM FAC treatment when compared with the cells treated with 100 μM FAC/DFO (closed arrows; Fig. 5) potentially indicating the consistent presence of the subunit in this organelle. HCH co-localization with lysosomal vesicles appears reduced (closed arrows) in cells treated with FAC relative to those treated with 100 μM FAC/DFO, while LCH lysosomal association appears similar for both treatment groups (closed arrows; Fig. 5). Although in our previous work we found that both HCH and LCH increase by the administration of FAC in these cells (Geiser et al. 2006; Geiser et al. 2007; Geiser et al. 2009), the lysosomal coalescence data suggests there may be differences in expression of the ferritin subunits. From these data, we conclude that ferritin appears to exist in three major locations within CCL-125 cells, co-localized with Golgi and lysosomes (closed arrows), and in non-labeled punctate vesicles (open arrows). The subunits may differ in each location, the lysosomal pool of HCH declines with iron treatment, most likely as ferritin is secreted from the cells, and HCH and LCH are found in non-organelle labeled punctate vesicles under both treatment conditions.
Fig. 5.
Iron entry into CCL-125 cells increases HCH and LCH coalescence with the Golgi compared with lysosomes. The effect of iron uptake into CCL-125 cells on cell-associated ferritin subunit expression and location was detected using fluorescent deconvolution microscopy (100×). A representative image of each treatment group from Supplementary Fig. 7S is displayed. Key: Blue = Nucleus (DAPI); Red = Golgi (BODIPY TR C5-ceramide) or Lysosome (LysoTracker); Green = HCH and LCH (Cy2-conjugated goat anti-rabbit IgG secondary); Orange-Yellow = Co-localization (overlaid images were merged and processed using ImageJ); Open Arrow = Detection of protein alone; Closed Arrow = Detection of protein with organelle; FAC = 100 μM FAC; F/D = 100 μM FAC/DFO; FAC = ferric ammonium citrate; DFO = deferoxamine mesylate salt
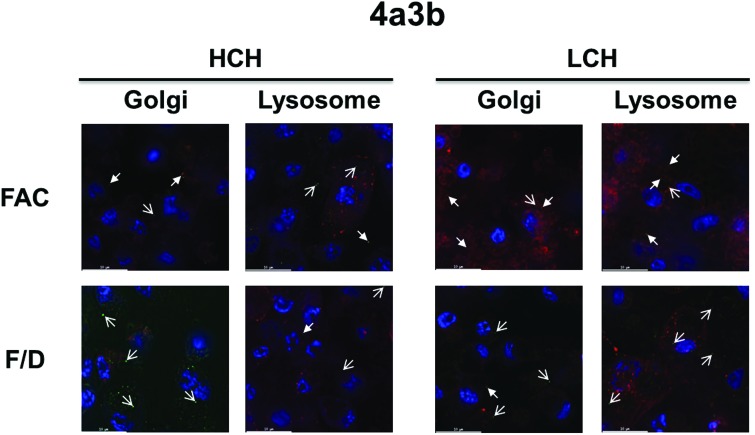
As with CCL-125 cells, in 4a3b cells both ferritin subunits appear to be constitutively expressed regardless of iron status (green; Supplementary Figs. 6S, 8S and 9S). We observed large accumulations of the ferritin subunits with 500 μM FAC treatments in 4a3b cells (Fig. 4); however, this was not clearly observed in the images of coalescence with 100 μM FAC (Fig. 6). Similar to CCL-125 cells, both ferritin subunits in 4a3b cells appear to coalesce with fluorescent labeled organelles (closed arrows) as well as in individual punctate vesicles (open arrows; Fig. 6). Ferritin subunit co-localization with the Golgi appears to be increased with 100 μM FAC treatment compared with treatment with the rescue (100 μM FAC/DFO; closed arrows; Fig. 6). However, coalescence of the LCH subunit with lysosomal vesicles (closed arrows) also appears increased with iron treatment compared with the rescue group; whereas the association of the HCH subunit with lysosomes appears to remain relatively unchanged for both treatment groups.
Fig. 6.
Iron entry into 4a3b cells increases the accumulation of HCH and LCH ferritin subunits in the Golgi and lysosomes. The effect of iron uptake into 4a3b cells on cell-associated ferritin subunit expression and location was detected using fluorescent deconvolution microscopy (100×). A representative image of each treatment group from Supplementary Fig. 9S is displayed. Key: Blue = Nucleus (DAPI); Red = Golgi (BODIPY TR C5-ceramide) or Lysosome (LysoTracker); Green = HCH and LCH (Cy2-conjugated goat anti-rabbit IgG secondary); Orange-Yellow = Co-localization (overlaid images were merged and processed using ImageJ); Open Arrow = Detection of protein alone; Closed Arrow = Detection of protein with organelle; FAC = 100 μM FAC; F/D = 100 μM FAC/DFO; FAC = ferric ammonium citrate; DFO = deferoxamine mesylate salt
Discussion
Cells of both mosquito lines show relatively low increases in cytoplasmic iron concentration with high levels of iron uptake, and both exhibit significant up-regulation of expression of the ferritin subunits in response to iron exposure. Our prior work demonstrated that iron taken into CCL-125 cells is subsequently secreted in ferritin, thus, protecting the cells from iron overload (Geiser et al. 2009). In contrast, high levels or iron exposure in 4a3b cells results in continuous iron uptake that leads to accumulation of cell-associated ferritin and iron retention. The increase in ferritin is due in part to an increase in transcription. The differences in iron handling and ferritin regulation between CCL-125 and 4a3b cells may be due to differences in cell origin and/or cell function. Although these differences could reflect species differences, we think this less likely. The CCL-125 cells are larval epithelial-like cells and secrete ferritin, whereas the 4a3b cells are larval hemocyte-like and do not secrete ferritin. Because ferritin is not secreted in 4a3b cells, we expanded our work to visualize ferritin in 4a3b (non-secretory) cells and CCL-125 (secretory) cells using deconvolution microscopy.
Ferritin subunits associated with the Golgi may represent a site for modification of the ferritin subunits, molecule assembly, and/or iron loading, as well as preparation for secretion. Aedes subunits have glycosylation sites and some mammalian ferritin subunits are glycosylated (Zaman and Verwilghen 1981; van Gelder et al. 1996; Geiser et al. 2003; Kannengiesser et al. 2009). Although it remains unknown where assembly and iron loading of the ferritin molecule occurs within the cell (Theil 2013); this might occur in the Golgi in insect cells (Nichol and Locke 1999). However, ferritin subunits also appear to be associated with lysosomal vesicles. This could represent a pool of ferritin held at a lower pH to maintain iron solubility for loading into the molecule. Alternatively, this pool could represent ferritin held for degradation and turnover of the molecule for iron release as was observed in human fibroblasts (Laskar et al. 2012). Another explanation is that ferritin secretion occurs from the lysosomal compartment as demonstrated in murine bone marrow-derived macrophages (Cohen et al. 2010). However, where iron loading occurs is unknown. Of interest, although there is some coalescence of both subunits with Golgi and lysosomes in both cell types in both treatment groups, ferritin subunits also consistently appear in punctate vesicles that do not coalesce with either Golgi or lysosomal markers. These conclusions are based on the entire z-stacks of the 3D images as shown in Supplementary Figs. 7S and 9S. What these pools of ferritin represent, we do not know and neither do we know their origin. We speculate that this is a form for iron storage.
Little is known about the role of hemocytes in mosquitoes. There are several types of hemocytes found in hemolymph, numbers are limited and the cell types are detected at different ratios among mosquito species (Castillo et al. 2006; Araujo et al. 2008). These cells function in the innate immune response to pathogen invasion through cellular mechanisms like phagocytosis and the secretion of humoral factors including antimicrobial peptides, complement-like proteins and components that regulates melanization (Rodrigues et al. 2010; Coggins et al. 2012). It is interesting to note that hemolymph ferritin decreased in An. gambiae in response to bacterial challenge (Paskewitz and Shi 2005), this may be a protective mechanism to limit the availability of iron to pathogens in the hemolymph. However, the relocation of this ferritin remains unclear. In regards to iron metabolism, the levels and retention of iron we observed could suggest that hemocytes might function as a reservoir for iron and sequestering of iron in an immune response. Apoferritin has been identified in the vacuolar system of insect hemocytes (Locke 1991). Alternatively, hemocytes could serve as a putative mechanism for detoxifying and protecting tissues as insects have an open circulatory system and a transient increase in hemocyte abundance and activation was observed in female mosquitoes following a bloodmeal (Castillo et al. 2006; Bryant et al. 2014). These factors may explain why 4a3b cells accumulate iron and do not secrete ferritin with increased exposure to this mineral.
Finally, it is interesting to consider the regulatory mechanism(s) that allow ferritin to be secreted from one cell type when exposed to iron, but inhibits secretion in another cell type. What is the signaling mechanism for positional regulation and secretion from these cells? CI-976, a lysophospholipid acyltransferase antagonist, blocks ferritin secretion in CCL-125 cells (Geiser et al. 2009) by inhibiting COP II vesicle formation, inducing Golgi membrane tubulation, and blocking the endocytic recycling pathway (Brown et al. 2008; Geiser et al. 2009). However, in animal studies we have been unable to prevent ferritin secretion and iron transport from the mosquito gut with either CI-976 (D. G. and J. W., Unpublished data) or RNAi of ferritin subunits (J. Bhakta and J. W., Unpublished data). Because the subunits are transcribed and translated similarly in different cell types from different species under the same environmental iron conditions, clearly, positional regulation is a key and probably a rate limiting feature of ferritin metabolism in mosquitoes. This aspect of the regulation of mosquito iron metabolism warrants further investigation.
SUPPLEMENTARY DATA
Supplementary data are available at Journal of Insect Science online.
Acknowledgments
This work was supported by funds from the Agricultural Experiment Station, the College of Agriculture and Life Sciences, the Undergraduate Biology Research Program, and the Honors College at the University of Arizona. In addition, we would like to thank Douglas W. Cromey and Dr. David Elliott from the University of Arizona Cellular Imaging Facility Core for their help and assistance with technical microscopy.
References Cited
- Araujo H. C., Cavalcanti M. G., Santos S. S., Alves L. C., Brayner F. A. 2008. Hemocytes ultrastructure of Aedes aegypti (Diptera: Culicidae). Micron. 39: 184–189. [DOI] [PubMed] [Google Scholar]
- Arosio P., Levi S. 2010. Cytosolic and mitochondrial ferritins in the regulation of cellular iron homeostasis and oxidative damage. Biochim. Biophys. Acta. 1800: 783–792. [DOI] [PubMed] [Google Scholar]
- Ausubel F., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Stuhl K. 1998. Current protocols in molecular biology. John Wiley and Sons, Inc, New York. [Google Scholar]
- Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brown W. J., Plutner H., Drecktrah D., Judson B. L., Balch W. E. 2008. The lysophospholipid acyltransferase antagonist CI-976 inhibits a late step in COPII vesicle budding. Traffic. 9: 786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant W. B., Michel K., Lombardo F., Ghani Y., Kafatos F. C., Christophides G. K. 2014. Blood feeding induces hemocyte proliferation and activation in the African malaria mosquito, Anopheles gambiae Giles. J. Exp. Biol. 217: 1238–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo J. C., Robertson A. E., Strand M. R. 2006. Characterization of hemocytes from the mosquitoes Anopheles gambiae and Aedes aegypti. Insect Biochem. Mol. Biol. 36: 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis J. E. 1998. Cell biology: a laboratory handbook, 2nd ed. Academic Press, San Diego. [Google Scholar]
- Coggins S. A., Estevez-Lao T. Y., Hillyer J. F. 2012. Increased survivorship following bacterial infection by the mosquito Aedes aegypti as compared to Anopheles gambiae correlates with increased transcriptional induction of antimicrobial peptides. Dev. Comp. Immunol. 37: 390–401. [DOI] [PubMed] [Google Scholar]
- Cohen L. A., Gutierrez L., Weiss A., Leichtmann-Bardoogo Y., Zhang D. L., Crooks D. R., Sougrat R., Morgenstern A., Galy B., Hentze M. W., et al. 2010. Serum ferritin is derived primarily from macrophages through a nonclassical secretory pathway. Blood. 116: 1574–1584. [DOI] [PubMed] [Google Scholar]
- Cooper M. S., D’Amico L. A., Henry C. A. 1999. Confocal microscopic analysis of morphogenetic movements. Methods Cell. Biol. 59: 179–204. [DOI] [PubMed] [Google Scholar]
- Dunkov B. C., Georgieva T., Yoshiga T., Hall M., Law J. H. 2002. Aedes aegypti ferritin heavy chain homologue: feeding of iron or blood influences message levels, lengths and subunit abundance. J. Insect Sci. 2: 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser D. L., Chavez C. A., Flores-Munguia R. F., Winzerling J. J., Pham D. Q. 2003. Aedes aegypti ferritin: a cytotoxic protector against iron and oxidative challenge? Eur. J. Biochem. 270: 1–8. [DOI] [PubMed] [Google Scholar]
- Geiser D. L., Zhang D., Winzerling J. J. 2006. Secreted ferritin: mosquito defense against iron overload? Insect Biochem. Mol. Biol. 36: 177–187. [DOI] [PubMed] [Google Scholar]
- Geiser D. L., Mayo J. J., Winzerling J. J. 2007. The unique regulation of Aedes aegypti larval cell ferritin by iron. Insect Biochem. Mol. Biol. 37: 418–429. [DOI] [PubMed] [Google Scholar]
- Geiser D. L., Shen M. C., Mayo J. J., Winzerling J. J. 2009. Iron loaded ferritin secretion and inhibition by CI-976 in Aedes aegypti larval cells. Comp. Biochem. Physiol. 152: 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jason Pitts R., Baldini F., Gabrieli P., South A., Valim C., Mancini F., Catteruccia F. 2014. A blood-free protein meal supporting oogenesis in the Asian tiger mosquito, Aedes albopictus (Skuse). J. Insect Physiol. 11: e1001695. [DOI] [PubMed] [Google Scholar]
- Kannengiesser C., Jouanolle A. M., Hetet G., Mosser A., Muzeau F., Henry D., Bardou-Jacquet E., Mornet M., Brissot P., Deugnier Y., et al. 2009. A new missense mutation in the L ferritin coding sequence associated with elevated levels of glycosylated ferritin in serum and absence of iron overload. Haematologica. 94: 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan P. H. 1990. Substitute blood meal for investigating and maintaining Aedes aegypti (Diptera: Culicidae). J. Med. Entomol. 27: 709–712. [DOI] [PubMed] [Google Scholar]
- Laskar A., Ghosh M., Khattak S. I., Li W., Yuan X. M., Zhang Y., Mikhael M., Xu D., Li Y., Soe-Lin S., et al. 2012. Degradation of superparamagnetic iron oxide nanoparticle-induced ferritin by lysosomal cathepsins and related immune response. Nanomedicine. 7: 705–717. [DOI] [PubMed] [Google Scholar]
- Linder M. C. 2013. Mobilization of stored iron in mammals: a review. Nutrients. 5: 4022–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke M. 1991. Apoferritin in the vacuolar system of insect hemocytes. Tiss. Cell. 23: 367–375. [DOI] [PubMed] [Google Scholar]
- Markwell M. A., Haas S. M., Bieber L. L., Tolbert N. E. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87: 206–210. [DOI] [PubMed] [Google Scholar]
- Muller H. M., Dimopoulos G., Blass C., Kafatos F. C. 1999. A hemocyte-like cell line established from the malaria vector Anopheles gambiae expresses six prophenoloxidase genes. J. Biol. Chem. 274: 11727–11735. [DOI] [PubMed] [Google Scholar]
- Nichol H., Locke M. 1999. Secreted ferritin subunits are of two kinds in insects molecular cloning of cDNAs encoding two major subunits of secreted ferritin from Calpodes ethlius. Insect Biochem. Mol. Biol. 29: 999–1013. [DOI] [PubMed] [Google Scholar]
- Pagano R. E., Martin O. C., Kang H. C., Haugland R. P. 1991. A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J. Cell. Biol. 113: 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paskewitz S. M., Shi L. 2005. The hemolymph proteome of Anopheles gambiae. Insect Biochem. Mol. Biol. 35: 815–824. [DOI] [PubMed] [Google Scholar]
- Patton S. M., Pinero D. J., Surguladze N., Beard J., Connor J. R. 2005. Subcellular localization of iron regulatory proteins to Golgi and ER membranes. J. Cell. Sci. 118: 4365–4373. [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham D.Q.-D., Brown S. E., Knudson D. L., Winzerling J. J., Dodson M. S., Shaffer J. J. 2000. Structure and location of the ferritin gene of the yellow fever mosquito Aedes aegypti. Eur. J. Biochem. 267: 3885–3890. [DOI] [PubMed] [Google Scholar]
- Pham D.Q.-D., Shaffer J. J., Chavez C. A., Douglass P. L. 2003. Identification and mapping of the promoter for the gene encoding the ferritin heavy-chain homologue of the yellow fever mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 33: 51–62. [DOI] [PubMed] [Google Scholar]
- Pham D. Q., Chavez C. A. 2005. The ferritin light-chain homologue promoter in Aedes aegypti. Insect Mol. Biol. 14: 263–270. [DOI] [PubMed] [Google Scholar]
- Rodrigues J., Brayner F. A., Alves L. C., Dixit R., Barillas-Mury C. 2010. Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science. 329: 1353–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telang A., Rechel J. A., Brandt J. R., Donnell D. M. 2013. Analysis of ovary-specific genes in relation to egg maturation and female nutritional condition in the mosquitoes Georgecraigius atropalpus and Aedes aegypti (Diptera: Culicidae). J. Insect Physiol. 59: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil E. C. 2013. Ferritin: the protein nanocage and iron biomineral in health and in disease. Inorg. Chem. 52: 12223–12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelder W., Huijskes-Heins M. I., Klepper D., van Noort W. L., Cleton-Soeteman M. I., van Eijk H. G. 1996. Isolation and partial characterization of two porcine spleen ferritin fractions with different electrophoretic mobility. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 115: 191–199. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO). 2013a. Malaria Fact Sheet. N°94. WHO Media Centre Geneva, Switzerland. [Google Scholar]
- WHO. 2013b. Yellow Fever Fact Sheet. N°100. WHO Media Centre Geneva, Switzerland. [Google Scholar]
- WHO. 2014. Dengue and Severe Dengue Fact Sheet. N°117. WHO Media Centre Geneva, Switzerland. [Google Scholar]
- Zaman Z., Verwilghen R. L. 1981. Non-enzymic glycosylation of horse spleen and rat liver ferritins. Biochim. Biophys. Acta. 669: 120–124. [DOI] [PubMed] [Google Scholar]
- Zhang D., Dimopoulos G., Wolf A., Minana B., Kafatos F. C., Winzerling J. J. 2002. Cloning and molecular characterization of two mosquito iron regulatory proteins. Insect Biochem. Mol. Biol. 32: 579–589. [DOI] [PubMed] [Google Scholar]
- Zhou G., Kohlhepp P., Geiser D., Frasquillo Mdel C., Vazquez-Moreno L., Winzerling J. J. 2007. Fate of blood meal iron in mosquitoes. J. Insect Physiol. 53: 1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]