Abstract
Autophagy is an evolutionarily conserved mechanism in both animals and plants, which has been shown to be involved in various essential developmental processes in plants. Nicotiana tabacum is considered to be an ideal model plant and has been widely used for the study of the roles of autophagy in the processes of plant development and in the response to various stresses. However, only a few autophagy-related genes (ATGs) have been identified in tobacco up to now. Here, we identified 30 ATGs belonging to 16 different groups in tobacco through a genome-wide survey. Comprehensive expression profile analysis reveals an abroad expression pattern of these ATGs, which could be detected in all tissues tested under normal growth conditions. Our series tests further reveal that majority of ATGs are sensitive and responsive to different stresses including nutrient starvation, plant hormones, heavy metal and other abiotic stresses, suggesting a central role of autophagy, likely as an effector, in plant response to various environmental cues. This work offers a detailed survey of all ATGs in tobacco and also suggests manifold functions of autophagy in both normal plant growth and plant response to environmental stresses.
Keywords: autophagy, tobacco, gene expression, signalling, environmental stresses
1. Introduction
Autophagy is an evolutionarily conserved mechanism for recycling of the cellular cytoplasmic contents or breaking down damaged materials in a cell, which plays essential roles in the remobilization of cytoplasmic components during nutrient starvation conditions.1 Identification of genes regulating autophagy in 1990s opened up the possibility to understand the molecular mechanism underlying autophagy and to explore the potential roles of autophagy in different physiological processes.2 In the past two decades, a series of autophagy-related genes (ATGs) required for autophagy have been characterized in yeast and mammals, and several important roles of autophagy in various developmental events, such as adaptation to starvation, regulation of metabolism, differentiation of cell types, clearance of damaged organelles, suppression of tumour, have been discovered.2,3 In addition, analyses of the networks of autophagy-related proteins revealed a canonical molecular pathway regulating the process of autophagy in yeast and mammals.4
Although remarkable progresses have been made in our understanding of the molecular mechanisms underlying autophagic pathways in yeast and mammals, molecular mechanisms and potential roles of autophagy in plants are still largely unknown. Recently, a great effort has been put on relevant studies and made it an energetic field. On the basis of sequence similarity to ATGs required for autophagy in yeast and mammals, a set of ATGs have been identified in Arabidopsis thaliana5,6 and Oryza sativa,7 respectively, and several roles of autophagy in plant development have been elucidated, e.g. leaf starch degeneration,8 tracheary element differentiation,9 hypertensive cell death,10 senescence,11,12 stress responses,13 life span extension,14 maintenance of peroxisomal quality,15–17 and anther development.18 In addition, core molecular machinery of autophagy was approved to be conserved among plants, yeast and mammals.4,6 However, the overall and specific roles of autophagy under normal and stress conditions, and their regulatory pathway underlying autophagy in plants, are still largely unknown.
Nicotiana tabacum, a traditional model plant, is assumed to originate from a hybridization event between ancestors of Nicotiana sylvestris and Nicotiana tomentosiformis ∼200,000 yrs ago.19 It is considered to be an ideal model plant for the study of autophagy in the processes of plant development and in the response to various severe environmental factors.20–22 However, only a few ATGs have been identified in tobacco. To facilitate our understanding of the roles of autophagy in plant development and molecular mechanism underlying it, it is essential to identify ATGs in tobacco. A systematic survey of ATGs in draft genomes of N. tabacum and expression sequence tags deposited in NCBI was performed, and comprehensive analyses of the expression patterns of ATGs under both normal and stress conditions were also carried out to gain insights into their putative roles in plant development under normal and in plant response to ill-suited environments.
2. Methods and materials
2.1. Plant materials
Nicotiana tabacum L. cv. Petite Havana SR1 plants were grown under 16 h/8 h light/dark cycles, at 25°C in greenhouse.
2.2. Identification ATGs in tobacco
To identify ATGs based on the draft genomes of N. tabacum,23 the program tBlastn using autophagy-related protein sequences in A. thaliana and O. sativa was performed in National Center for Biotechnology Information (NCBI). The DNA fragments and EST sequences related to ATGs were collected, respectively. Sequence assembling and open reading frame (ORF) analysis of each contig were performed using ContigExpress and OMEGA, respectively. After ORF analysis, BLASTP analysis with intact or partial deduced protein sequences of each contig was performed. The contigs corresponding to autophagy-related protein sequences based on the returning information were selected as candidates for further study.
2.3. Isolation of full-length cDNA of each ATG in tobacco
After ORF and BLASTP analyses of each contig, full-length cDNA of ATG candidates was obtained through an electronic cloning method or a rapid amplification of cDNA ends (RACE) approach. Total RNA isolated from anthers and leaves was used as a template to synthesize first-strand cDNA with the SMART RACE cDNA Amplification Kit (Clontech), and all reactions were performed according to the manufacturer's instructions. And then, full-length cDNA of each ATG was further confirmed through RT-PCR with specific primers at the 5′ and 3′ end, respectively.
2.4. Protein sequence and phylogenetic analysis
To analyse the relationships of autophagy-related genes identified in N. tabacum with that in A. thaliana and O. sativa, a multiple sequence alignment of ATG protein sequences was conducted with Clustal X ver. 1.81 program using the default multiple alignment parameters. The tree was constructed with MEGA 5.1 software using a maximum parsimony method.
2.5. RNA isolation and RT-PCR
Total RNA was extracted from leaf, root, stem, anther and pollen using TRI Reagent Solution (Ambion), and total RNA of seeds at different stages were extracted with RNAqueous™ (Ambion). All total RNA were treated with RNase-free DNase I (Promega) and cDNA were synthesized using PrimeScript Reverse Transcriptase (TaKaRa) under the condition recommended by the manufacturer procedure. Semi-quantitative RT-PCR was carried out in a 50-µl PCR mixture containing 5 µl of 10× Ex Taq buffer, 2.5 mM MgCl2, 200 µM dNTPs, 0.2 µM of primers, 1.2 U of Ex Taq DNA polymerase (Takara) and cDNA prepared from different tissues. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control for normalization of cDNA prepared from different tissues. Detailed PCR conditions are described as follows: initial step for denaturation at 94°C for 2 min; then 35 cycles of denaturation at 94°C for 30 s, annealing at Tm −5°C for 30 s; extension at 72°C for 1 min and a final extension at 72°C for 5 min. Quantitative real-time reverse transcription PCR (RT-qPCR) was conducted for expression pattern analysis according to the previous procedure.24
2.6. Different stress treatments for tobacco seedlings
For different stress treatments, tobacco seeds were germinated in modified MS medium at 28°C for 2 weeks. Two-week-old seedlings were then transferred to different conditions for stress treatment. For carbon starvation, 2-week-old seedlings were transferred into modified MS medium without sucrose for 4, 16, 24 and 48 h, respectively. For nitrogen starvation, 2-week-old seedlings were transferred into nitrogen-free MS medium, in which NH4NO3 and KNO3 were replaced by KCl. For salt treatment, 2-week-old seedlings were transferred into modified MS medium containing 250 mM NaCl for 4 h. For cold treatment, 2-week-old seedlings were cultured in modified MS medium in 4°C for 4 h. For drought treatment, seedlings were kept in filter paper for 4 h at 28°C. For dark treatment, 2-week-old seedlings were kept in dark for 48 h at 28°C. For hormone treatment, 2-week-old seedlings were transferred into modified MS medium containing 1 µM naphthalene acetic acid (NAA), 5 µM 2,4-dichlorophenoxyacetic acid (2,4-D), 25 µM abscisic acid (ABA) or 5 µM gibberellic acid (GA3), 500 µM salicylic acid (SA) and 500 µM jasmonic acid (JA), for 24 h, respectively. For heavy metal treatment, 2-week-old seedlings were transferred into modified MS medium containing 40 µM CdCl2, 40 µM NiSO4, 40 µM ZnCl2, 20 µM CuSO4 or 100 µM MnCl4 for 24 h, respectively.
3. Results
3.1. Collection and identification of ATGs in tobacco
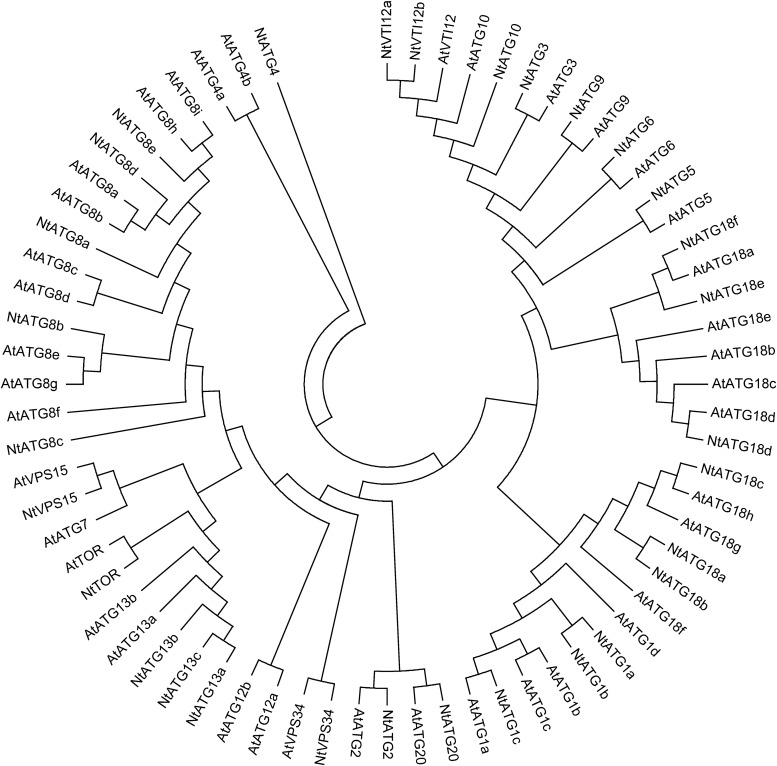
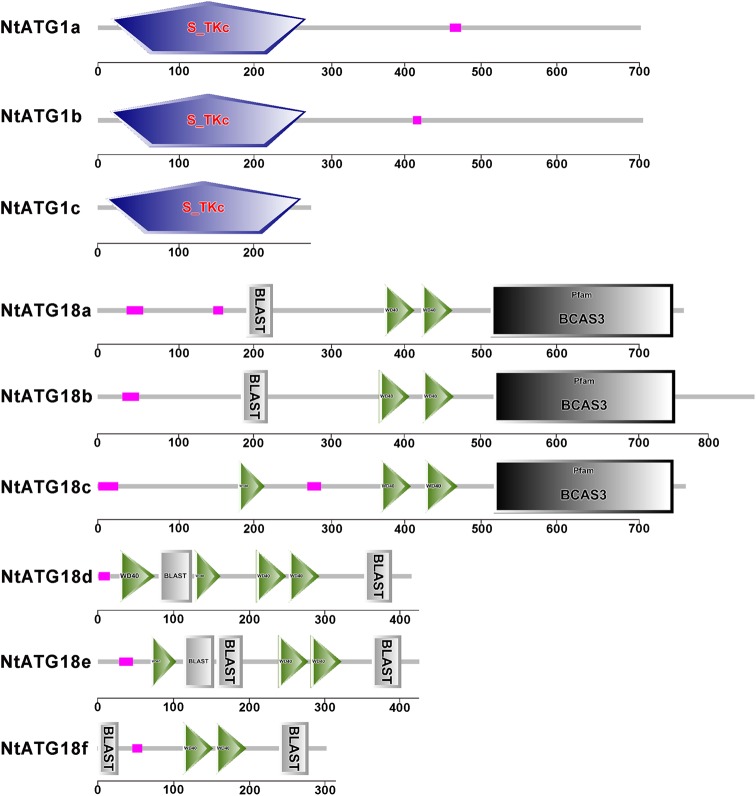
To identify ATGs in tobacco, the tBlastn program using different autophagy-related protein sequences from A. thaliana and O. sativa was performed. Returned sequences related to ATGs were collected and assembled using ContigExpress, and redundant sequences were omitted manually. Then, a total of 30 individual contigs related to ATGs in tobacco were obtained. Full-length cDNA of them were obtained through an electronic cloning method or a RACE technique, and were further confirmed through RT-PCR with gene-specific primers. The detailed information of each gene was described in Table 1. To confirm that the 30 putative ATG homologues in tobacco are indeed ATGs, the deduced ATG protein sequences were analysed in the Pfam database and their sequence similarities to known ATG proteins in A. thaliana were analysed. The returned information of each ATG in the Pfam database were listed in Table 1, suggesting that all 30 ATGs in tobacco could be considered as true ATGs. Phylogenetically, each ATG protein sequence exhibits high similarities to their homologue in A. thaliana (Fig. 1 and Supplementary Fig. S1). However, several ATG groups (ATG1, ATG8, ATG10 and ATG18) in A. thaliana and N. tabacum were separated by ATGs in O. sativa in the phylogenetic tree (Supplementary Fig. S1), indicating the diversification of ATG evolution in different ATG groups. Generally, predicated 30 ATGs in tobacco could be divided into 12 different ATG groups and 4 relatives, and among them, the following groups comprise multiple isoforms: ATG1, ATG8, ATG13, ATG18 and VTI12. The composition of domains in each subgroup is similar with two exceptions (ATG1 and ATG18). ATG1c shows a similar serine/threonine-protein kinase domain at N-terminal to that of ATG1a and b, but lacks the C-terminal structure as shown in ATG1a and b (Fig. 2). A similar phenomenon was also observed in the ATG18 group. Two different subgroups could be divided according to the C-terminal structure. They show a similar N-terminal structure, but lacking of a C-terminal BCAS3 domain in ATG18d, ATG18e and ATG18f (Fig. 2). The composition of conserved domain in other groups is similar to that in Arabidopsis, indicating that the core machinery of autophagy is conserved in different angiosperms.
Table 1.
The ATGs in tobacco
| Group | Gene name | Accession no. | ORF (bp) | Predicated protein information |
||||
|---|---|---|---|---|---|---|---|---|
| No. of amino acids | Mw (kDa) | PI | Signal peptide | Predicted function | ||||
| ATG1 | NtATG1a | KR336556 | 2,091 | 696 | 77.3 | 7.67 | – | Serine/threonine-protein kinase |
| NtATG1b | KR336557 | 2,073 | 690 | 76.7 | 6.13 | – | Serine/threonine-protein kinase | |
| NtATG1c | KR336558 | 849 | 283 | 31.9 | 6.50 | – | Serine/threonine-protein kinase | |
| ATG2 | NtATG2 | KR336559 | 5,943 | 1,980 | 217.2 | 5.27 | – | Autophagy-related protein 2 |
| ATG3 | NtATG3 | KR336560 | 945 | 314 | 35.6 | 4.7 | – | Autophagocytosis-associated protein 3 |
| ATG4 | NtATG4 | KR336561 | 1,188 | 396 | 43.7 | 4.75 | – | Peptidase family C54 |
| ATG5 | NtATG5 | KR336562 | 1,116 | 371 | 41.3 | 5.06 | – | Autophagy protein Apg5 |
| ATG6 | NtATG6 | KR336563 | 1,674 | 557 | 5.57 | 62.8 | – | Autophagy protein Apg6 |
| ATG8 | NtATG8a | KR336564 | 360 | 119 | 13.7 | 9.1 | – | Autophagy protein Atg8 |
| NtATG8b | KR336565 | 369 | 122 | 14 | 6.61 | – | Autophagy protein Atg8 | |
| NtATG8c | KR336566 | 369 | 122 | 14 | 7.85 | – | Autophagy protein Atg8 | |
| NtATG8d | KR336567 | 372 | 123 | 14.1 | 6.61 | – | Autophagy protein Atg8 | |
| NtATG8e | KR336568 | 372 | 123 | 14.1 | 6.61 | – | Autophagy protein Atg8 | |
| ATG9 | NtATG9 | KR336569 | 2,574 | 857 | 99 | 6.32 | – | Autophagy protein Apg9 |
| ATG10 | NtATG10 | KR336570 | 756 | 251 | 28.3 | 4.85 | – | Autophagocytosis-associated protein |
| ATG13 | NtATG13a | KR336571 | 1,827 | 608 | 67.4 | 8.62 | – | Autophagy-related protein 13 |
| NtATG13b | KR336572 | 1,815 | 604 | 66.7 | 8.68 | – | Autophagy-related protein 13 | |
| NtATG13c | KR336573 | 1,827 | 608 | 67.5 | 8.62 | – | Autophagy-related protein 13 | |
| ATG18 | NtATG18a | KR336574 | 2,316 | 771 | 83.1 | 8.63 | – | WD-40 repeat containing protein |
| NtATG18b | KR336575 | 2,586 | 861 | 93.1 | 8.33 | – | WD-40 repeat containing protein | |
| NtATG18c | KR336576 | 2,310 | 769 | 83.8 | 7.30 | – | WD-40 repeat containing protein | |
| NtATG18d | KR336577 | 1,251 | 416 | 45.8 | 7.15 | – | WD-40 repeat containing protein | |
| NtATG18e | KR336578 | 1,281 | 426 | 47.8 | 8.27 | – | WD-40 repeat containing protein | |
| NtATG18f | KR336579 | 912 | 303 | 33.5 | 8.88 | – | WD-40 repeat containing protein | |
| ATG20 | NtATG20 | KR336580 | 1,206 | 401 | 46.1 | 7.68 | – | Sorting nexin 1-like protein |
| VTI12 | NtVTI12a | KR336581 | 666 | 221 | 25 | 9.18 | – | Vesicle transport v-SNARE protein |
| NtVTI12b | KR336582 | 666 | 221 | 24.8 | 8.47 | – | Vesicle transport v-SNARE protein | |
| VPS15 | NtVPS15 | KR336583 | 4,659 | 1,552 | 173.1 | 6.32 | – | Serine/threonine-protein kinase |
| VPS34 | NtVPS34 | KR336584 | 2,445 | 814 | 93.2 | 6.33 | – | Phosphoinositide 3-kinase |
| TOR | NtTOR | KR336585 | 7,782 | 2,593 | 291.8 | 6.90 | – | Serine/threonine-protein kinase |
Figure 1.
Phylogenetic relationship of ATGs from N. tabacum and A. thaliana. The tree was calculated with MEGA 5.1 software using the maximum parsimony method.
Figure 2.
The structure divergence of ATG1s and ATG18s. This figure is available in black and white in print and in colour at DNA Research online.
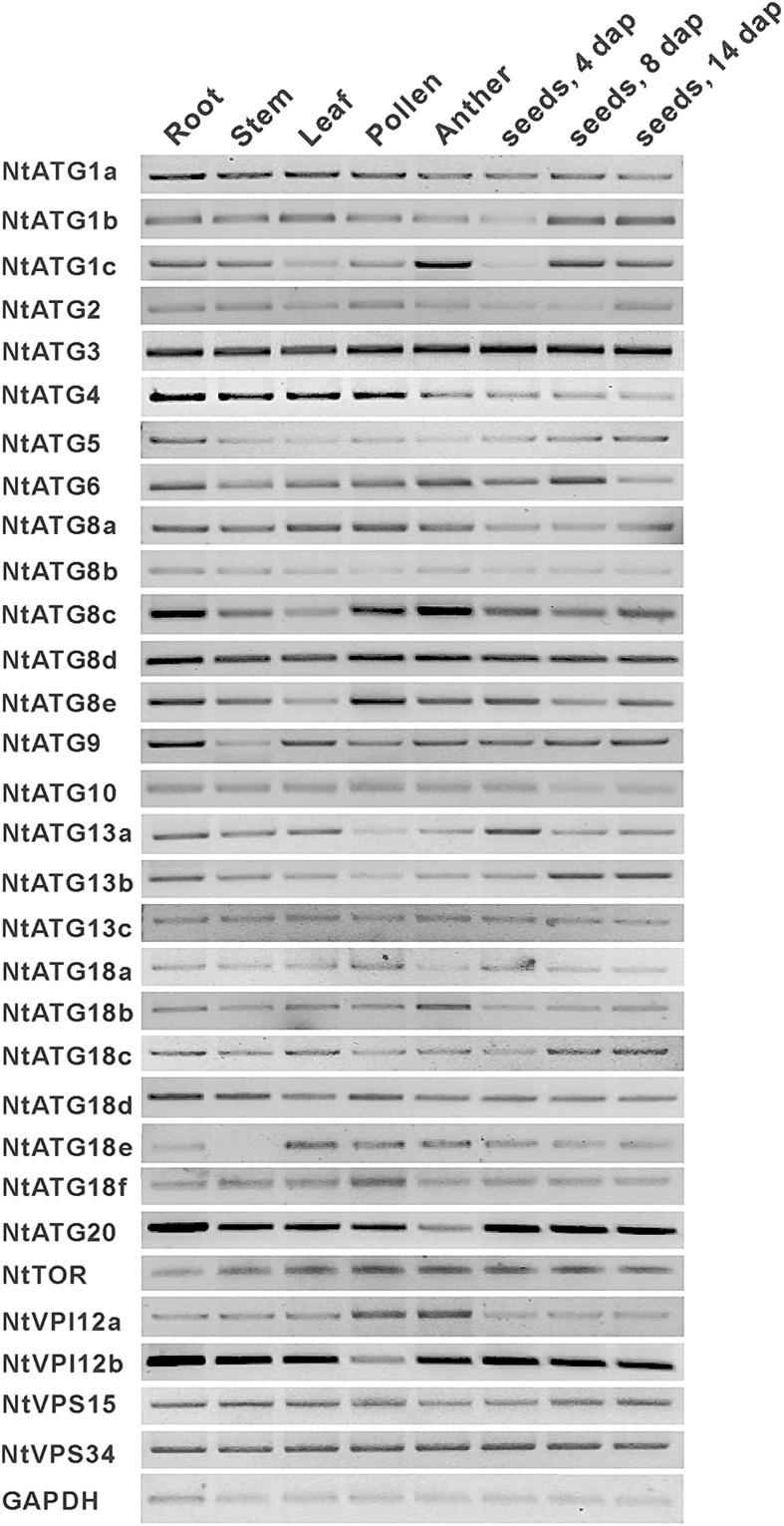
To further confirm the existence of predicated ATGs in tobacco, cDNA prepared from root, stem, leaf, pollen, anther and seed at different stages were selected as templates for RT-PCR. House-keeping gene GAPDH was used as the control for PCR. The transcripts of 30 predicated ATGs could be detected in different tissues of tobacco as shown in Fig. 3. Interestingly, all of them show a universal expression pattern in the eight tissues tested. These data suggest that all predicted ATGs are exactly existed in tobacco and autophagy may play a house-keeping role in the process of plant development.
Figure 3.
RT-PCR examination of the transcripts of ATGs in N. tabacum. The cDNA prepared from root, stem, leaf, pollen, anther, seeds (4, 8 and 14 days after pollination, respectively) were selected as templates for PCR. GAPDH was used as the control.
3.2. Expression profile of NtATGs under normal growth environments
There are growing evidences, suggesting that autophagy plays critical roles in the processes of plant development under normal conditions such as leaf starch degradation,8 root cell growth,25 rubisco degradation during leaf senescence,26 programmed cell death (PCD) of suspensor in somatic embryos of Picea abies,27 and maintenance of peroxisomal quality.15–17 To explore their potential roles in the processes of plant development, it is essential to study their expression pattern under normal growth conditions. RT-qPCR experiments were thus carried out, based on the cDNA from different tissues including leaf, stem, root, petal, sepal, anther, pollen, pistil, ovule and seeds at different developmental stages. The relative expression level of each ATG in different tissues was detected and compared with each other. Heat map analysis based on the relative expression level of each ATG was also performed, and an overview of the expression profile of ATGs in tobacco is presented in Fig. 4. Each ATG in tobacco exhibits a relatively broad expression pattern, which differ from the expression pattern of ATGs in O. sativa.7 Some tissue specifically expressed ATGs, such as OsATG1b and OsATG8d, have been identified.7 In contrast to that in O. sativa, the transcripts of each ATG in tobacco could be detected in all tissues tested, indicating a universal role of autophagy in the process of plant development. However, majority of ATGs show a relatively higher expression level in pollen/anther compared with that in other tissues tested. And, four homologues of ATG8 (NtATG8a, NtATG8b, NtATG8d and NtATG8e) show a relatively high expression level among all ATGs tested, which was thought to be required for autophagosome formation and reliable markers for the induction and progression of autophagy. Whereas four ATG18 group members (NtATG18a, NtATG18b, NtATG18c and NtATG18e) in ATG9 recycling complex show a relatively low expression level among all ATGs tested.
Figure 4.
Expression profile of NtATGs in different tissues under normal conditions of plant growth. Expression profile of NtATGs in tobacco, which is constructed based on the relative expression level of each ATGs in different tissues. The expression level was normalized to GAPDH (AJ133422), Polyubiquitin (GQ281244), Actin (GQ281246) and Elongation factor 1a (AF120093). Blue box indicates the lower transcriptional level of ATGs, whereas red box indicates the higher expression level of ATGs. Scale bar represents fold change (log2 value).
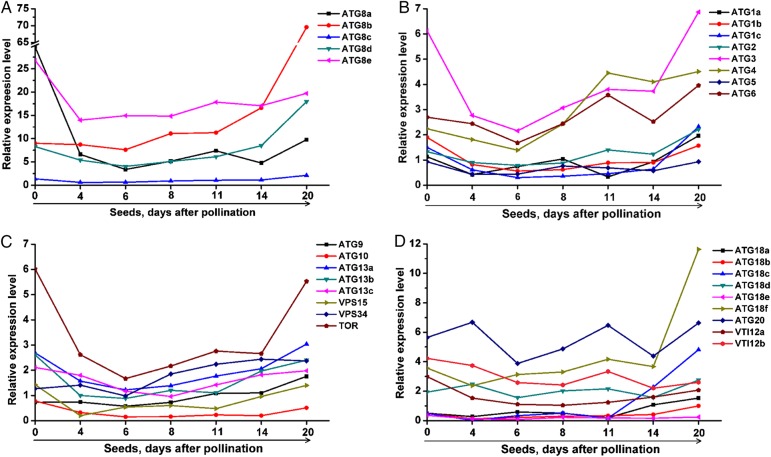
Another striking characteristic of the expression profile is the temporal variations of ATGs during the process of seed formation. Three expression characteristics of ATGs could be observed in the whole process of seed development. The first is that the expression level of majority ATGs decreased upon fertilization with a few exceptions (ATG8b, ATG9, ATG18d, ATG20 and NtVPS34). The second is that the expression levels of most ATGs are relatively stable during embryogenesis (seeds at 4–14 days after pollination). The third is that the expression peak of ATGs was found at the stage of seed maturation (Fig. 5). All these data suggest that autophagy may function at different developmental stages during the process of seed formation.
Figure 5.
Dynamic changes in NtATGs during the process of seed development. Expression profile of NtATGs in tobacco, which is constructed based on the relative expression level of each ATG in seeds at different developmental stages. The expression level was normalized to GAPDH (AJ133422), Polyubiquitin (GQ281244), Actin (GQ281246) and Elongation factor 1a (AF120093). Data represent relative expression level (log2 value). This figure is available in black and white in print and in colour at DNA Research online.
3.3. Expression pattern analysis of NtATGs under nitrogen and carbon starvation
Autophagy is thought to function in protein breakdown and recycling of amino acids for survival in response to nutrient starvation, which is evolutionarily conserved from yeast to mammals. Some ATGs in plants have been approved to participate in regulating nutrient recycling under starvation conditions.28 To explore the potential roles of NtATGs during starvation conditions, their relative expression levels were investigated in seedlings that were treated in the condition of carbon and nitrogen starvation, respectively. To analyse the relative expression levels of NtATGs under carbon and nitrogen starvation, seedlings after treated for 4, 16, 24 and 48 h were collected, respectively. The expression levels of NtATGs in the plants were quantified and compared with the control to uncover the key ATGs involved in response to the starvation (Fig. 6 and Table 2). Our analysis showed that 17 NtATGs (NtATG1a, NtATG2, NtATG5, NtATG6, NtATG9, NtATG13a, NtATG13b, NtATG13c, NtATG18a, NtATG18b, NtATG18c, NtATG18d, NtATG18e, NtATG18f, NtVPS15, NtVPS34 and NtTOR) were up-regulated (>2-fold expression change) and 1 NtATG (NtATG8d) was down-regulated (>2-fold expression change) in seedlings after carbon starvation 24 h. In addition, nine NtATGs (NtATG1a, NtATG2, NtATG9, NtATG13c, NtATG18a, NtATG18c, NtATG18e, NtVPS15 and NtVPS34) show dramatically changed expression (>8-fold expression change).
Figure 6.
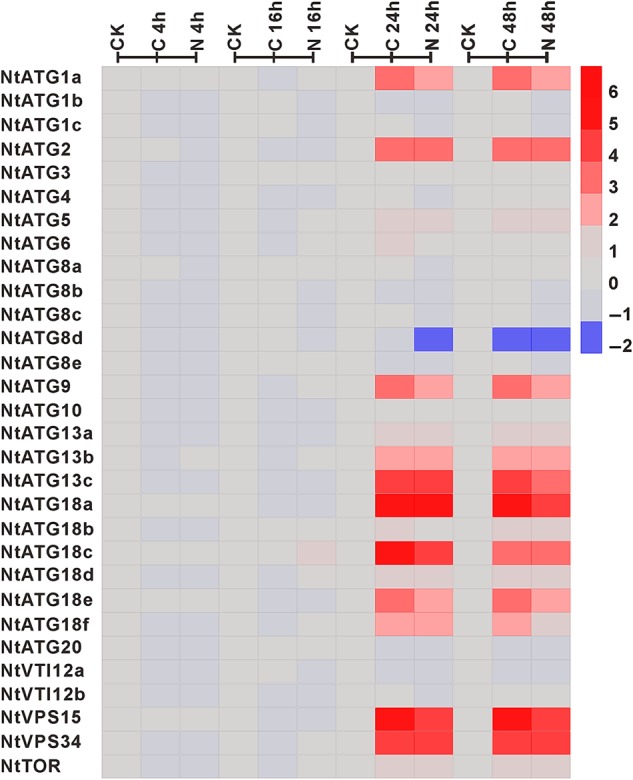
Expression profile of NtATGs in seedlings under sucrose and nitrogen starvation. Relative expression levels of each ATG in seedlings under sucrose and nitrogen starvation were normalized to GAPDH (AJ133422), Polyubiquitin (GQ281244), Actin (GQ281246) and Elongation factor 1a (AF120093). The expression level of each ATG was calculated and compared with that in seedlings under normal growth conditions. The expression level of each ATG in seedlings under normal growth conditions was indicated with grey box (0). Blue box indicates the lower transcriptional level of ATGs, whereas red box indicates the higher expression level of ATGs. Scale bar represents fold change (log2 value).
Table 2.
Overview of ATGs in tobacco response to different stresses
| Name | Starvation |
Hormone |
Heavy metal |
Other stress |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| –C | –N | NAA | 2,4-D | ABA | GA3 | SA | JA | Cu2+ | Ni2+ | Zn2+ | Cd2+ | Mn2+ | Dark | Cold | Drought | Salt | |
| NtATG1a | +++ | +++ | No | − | − | −− | + | ++ | + | +++ | +++ | +++ | ++ | +++ | ++ | + | +++ |
| NtATG1b | No | No | No | No | No | No | No | No | No | No | No | No | No | No | −−− | − | − |
| NtATG1c | No | No | No | No | No | No | No | No | No | +++ | No | No | No | + | − | No | No |
| NtATG2 | +++ | +++ | ++ | ++ | ++ | No | No | + | +++ | +++ | +++ | ++ | +++ | +++ | + | + | +++ |
| NtATG3 | No | No | No | No | No | No | No | No | No | No | No | No | No | No | − | No | No |
| NtATG4 | No | No | No | No | No | No | No | No | No | No | No | No | No | No | − | No | No |
| NtATG5 | + | + | + | + | No | − | No | No | No | + | + | No | + | − | No | No | + |
| NtATG6 | + | No | No | No | No | No | No | No | No | No | No | No | No | No | − | No | No |
| NtATG8a | No | No | No | No | No | No | No | No | No | No | No | − | No | ++ | −− | No | No |
| NtATG8b | No | No | No | No | No | No | No | No | No | No | − | − | No | + | − | No | No |
| NtATG8c | No | No | No | No | No | No | No | No | No | No | No | No | No | No | + | − | No |
| NtATG8d | − | − | No | No | No | No | No | No | No | No | No | No | No | + | − | No | No |
| NtATG8e | No | No | No | No | No | No | No | No | No | No | No | No | No | No | − | No | No |
| NtATG9 | +++ | +++ | ++ | ++ | ++ | No | No | + | +++ | +++ | +++ | +++ | +++ | − | ++ | ++ | +++ |
| NtATG10 | No | No | No | No | No | No | No | No | No | No | No | No | No | No | − | − | − |
| NtATG13a | + | + | No | No | No | No | No | No | + | + | + | No | + | + | No | No | + |
| NtATG13b | ++ | ++ | No | No | No | − | No | + | + | + | ++ | + | + | ++ | No | No | + |
| NtATG13c | +++ | +++ | +++ | ++ | ++ | No | No | + | +++ | +++ | +++ | +++ | +++ | +++ | + | ++ | +++ |
| NtATG18a | +++ | +++ | No | No | No | −− | + | +++ | + | +++ | +++ | + | ++ | +++ | + | + | +++ |
| NtATG18b | + | + | No | No | No | No | No | ++ | No | No | No | No | No | No | − | No | No |
| NtATG18c | +++ | +++ | No | No | No | − | ++ | +++ | + | + | +++ | + | + | +++ | ++ | + | ++ |
| NtATG18d | + | + | No | No | No | − | No | No | + | + | + | No | + | −−− | − | No | No |
| NtATG18e | +++ | +++ | No | No | No | −−− | + | + | No | No | ++ | No | No | No | No | No | + |
| NtATG18f | ++ | ++ | + | No | No | No | No | No | + | + | ++ | + | + | + | No | No | ++ |
| NtATG20 | No | No | No | No | No | No | No | No | No | No | No | No | No | + | − | No | No |
| NtVTI12a | No | No | No | No | No | No | − | No | No | No | No | No | No | ++ | − | − | −−− |
| NtVTI12b | No | No | No | No | No | No | No | No | No | No | No | No | No | No | No | No | No |
| NtVPS15 | +++ | +++ | No | No | No | −−− | No | + | ++ | +++ | +++ | +++ | +++ | +++ | No | No | +++ |
| NtVPS34 | +++ | +++ | ++ | ++ | ++ | −− | No | + | +++ | +++ | +++ | +++ | +++ | +++ | ++ | + | +++ |
| NtTOR | + | + | No | No | No | No | No | No | + | + | + | +++ | + | No | No | No | + |
‘+’ and ‘−’ mean that the expression of ATGs were up-regulated and down-regulated by different stresses, respectively. The number of ‘+’ or ‘−’ means different fold change of relative expression levels. ‘+’ or ‘−’ means >2-fold change. ‘++’ or ‘−−’ means >4-fold change. ‘+++’ or ‘−−−’ means >8-fold change.
The duration of starvation treatment for the expression level changes is another focus in our study. The relative expression levels of all NtATGs in seedlings show no visible change after 4- and 16-h carbon starvation treatment. However, the relative expression levels of most NtATGs in seedlings increased dramatically after 24-h carbon starvation treatment and with a little further decrease in seedlings after 48-h treatment, indicating that the response of autophagy to starvation signals usually occurs after 16- to 24-h starvation treatment. More interestingly, all NtATGs show a similar response pattern under nitrogen starvation with that in carbon starvation treatment, indicating a common response mechanism to carbon and nitrogen starvation stresses.
3.4. Differential expression of NtATGs in response to stress treatments
To gain insights into the potential roles of NtATGs in response to various stress treatments, their expression dynamics were investigated through RT-qPCR in tobacco seedlings subjected to dark, cold, drought and salt treatments. Generally, all of the NtATGs, except NtVTI12b in tobacco, are responsive to different stress treatments, and the relative expression levels of them changed significantly upon different stresses (Fig. 7 and Table 2). However, the response of NtATGs to different stresses varied significantly. Only six ATGs (NtATG1a, NtATG2, NtATG13c, NtATG18a, NtATG18c and NtVPS34) were commonly up-regulated by all the four different stress treatments, indicating a common role these NtATGs in dark, cold, drought and salt stresses. Whereas the expression of some NtATGs (NtATG3, NtATG4, NtATG6, NtATG8e, NtATG18b, NtATG18e and NtTOR) were responsive to some specific stress.
Figure 7.
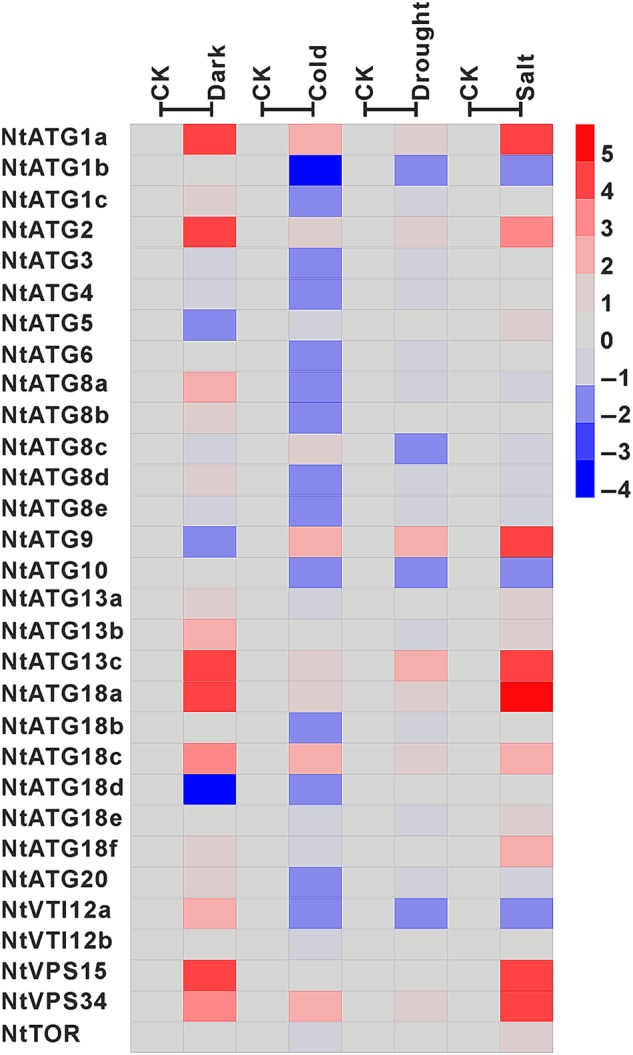
Expression profile of NtATGs in seedlings under dark, cold, drought and salt stresses. Relative expression levels of each ATG in seedlings under dark, cold, drought and salt stresses were normalized to GAPDH (AJ133422), Polyubiquitin (GQ281244), Actin (GQ281246) and Elongation factor 1a (AF120093). The expression level of each ATG was calculated and compared with that in seedlings under normal growth conditions. The expression level of each ATG in seedlings under normal growth conditions was indicated with grey box (0). Blue box indicates the lower transcriptional level of ATGs, whereas red box indicates the higher expression level of ATGs. Scale bar represents fold change (log2 value).
In response to dark treatment, the expression level of 19 NtATGs shows significantly changes. Most of them (NtATG1a, NtATG1c, NtATG2, NtATG8a, NtATG8b, NtATG8d, NtATG13a, NtATG13b, NtATG13c, NtATG18a, NtATG18c, NtATG18f, NtATG20, NtVTI12a, NtVPS15 and NtVPS34) are up-regulated in seedlings under dark conditions (>2-fold expression change) and three ATGs (NtATG5, NtATG9 and NtATG18d) were down-regulated (>2-fold expression change). Among them, only six NtATGs (NtATG1c, NtATG8a, NtATG8b, NtATG8d, NtATG20 and NtVTI12a) and two NtATGs (NtATG5 and NtATG9) were specifically up-regulated and down-regulated by dark treatment, respectively. Unlike dark treatment, a larger number of NtATGs (NtATG1b, NtATG1c, NtATG3, NtATG4, NtATG6, NtATG8a, NtATG8b, NtATG8d, NtATG8e, NtATG10, NtATG18b, NtATG18d, NtATG20 and NtVTI12a) were down-regulated by cold stress (>2-fold expression change). For drought treatment, a relatively small number of ATGs (11 of 30 ATGs) show their response to drought stress (seven ATGs up-regulation and four ATGs down-regulation). For salt treatment, 14 NtATGs (NtATG1a, NtATG2, NtATG5, NtATG9, NtATG13a, NtATG13b, NtATG13c, NtATG18a, NtATG18c, NtATG18e, NtATG18f, NtVPS15, NtVPS34 and NtTOR) and 3 NtATGs (NtATG1b, NtATG10 and NtVTI12a) were up-regulated and down-regulated, respectively.
3.5. Differential expression of NtATGs in response to heavy metal stress
Heavy metals are major environmental pollutants, which are highly reactive and toxic to living cells. Correspondingly, organisms have adapted to multiple heavy metal stresses by developing various response mechanisms. In human haematopoietic stem cells, autophagy has been approved to be implicated in the response to Cd2+ stress.29 However, the role of autophagy in response to heavy metal stress in plants still remains unknown. To survey the putative roles of autophagy in plant response to heavy metal stress, it is a prerequisite to extensively analyse their expression dynamics in response to different heavy metal stresses. Thus, the relative expression levels of 30 ATGs were analysed in seedlings treated with five different heavy metals including Cu2+, Ni2+, Zn2+, Cd2+ and Mn2+ with a comparison to that in untreated seedlings (Fig. 8 and Table 2). Expression profile analysis data showed that the expression of 18 ATGs in seedlings are regulated by at least one heavy metal (>2-fold expression change), and the expression levels of other 12 ATGs (NtATG1b, NtATG3, NtATG4, NtATG6, NtATG8c, NtATG8d, NtATG8e, NtATG10, NtATG18b, NtATG20, NtVTI12a and NtVTI12b) show no visible changes. Among the 18 heavy metal-responsive ATGs, the expression levels of 11 ATGs are commonly up-regulated with 7 exceptions (NtATG1c, NtATG5, NtATG8a, NtATG8b, NtATG13a, NtATG18d and NtATG8e) in seedlings after treated with five different heavy metals, indicating a common mechanism of ATGs in the response to different heavy metal treatments. Interestingly, the expression of ATGs is more sensitive to Zn2+ treatment than to other heavy metals. However, how these heavy metals regulate autophagy remains to be elucidated in the further studies.
Figure 8.
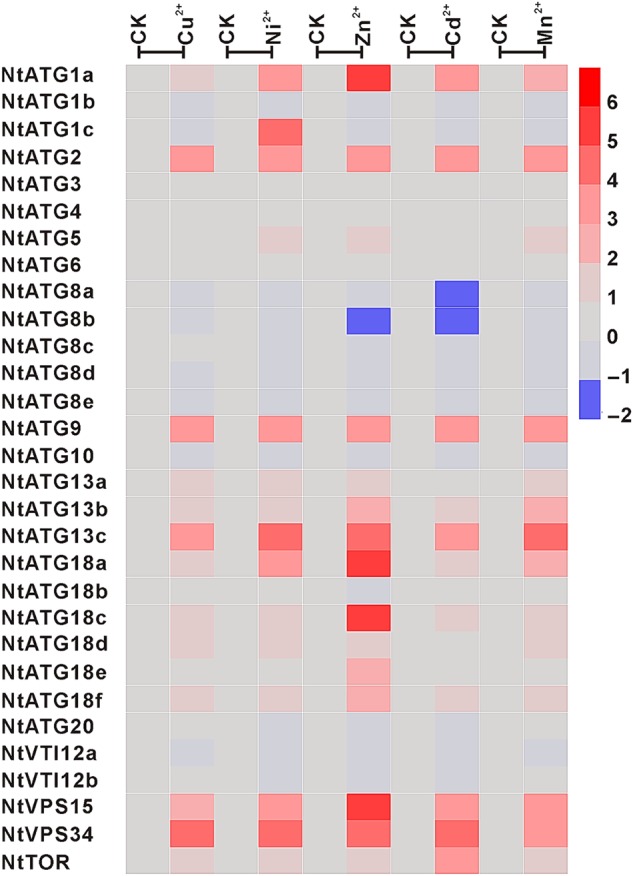
Expression profile of NtATGs in seedlings under heavy metal stresses. Relative expression levels of each ATG in seedlings under heavy metal stresses were normalized to GAPDH (AJ133422), Polyubiquitin (GQ281244), Actin (GQ281246) and Elongation factor 1a (AF120093). The expression level of each ATG was calculated and compared with that in seedlings under normal growth conditions. The expression level of each ATG in seedlings under normal growth conditions was indicated with grey box (0). Blue box indicates the lower transcriptional level of ATGs, whereas red box indicates the higher expression level of ATGs. Scale bar represents fold change (log2 value).
3.6. Differential expression of NtATGs in response to plant hormone treatment
Plant hormones are a group of small signal molecules that exert powerful effects on a variety of developmental processes such as embryogenesis,30 root development,31 PCD32 and many other aspects of plant development by ‘regulating gene’ expression. In a recent report, SA signalling pathway was shown to be required for early senescence in atg mutant.12 However, the relationship between plant hormones and autophagy is still largely unknown. To test whether plant hormones could regulate the expression pattern of ATGs, the responses of NtATGs to different plant hormones were analysed in the present study. The relative expression levels of each ATG under six different hormone treatments including NAA, 2,4-D, ABA, GA3, SA and JA were investigated by RT-qPCR, and heat map analysis was conducted based on the relative expression level of each ATG. The data revealed that the expression levels of several ATGs in seedlings changed significantly when treated with different plant hormones. Notably, different ‘hormones’ have ‘diverse effects on the regulation of’ ATG ‘expression’ (Fig. 9 and Table 2). After GA3 treatment, the ‘expression levels of nine’ ATGs (NtATG1a, NtATG5, NtATG13b, NtATG18a, NtATG18c, NtATG18d, NtATG18e, NtVPS15 and NtVPS34) in seedlings were down-regulated significantly (>2-fold expression change). However, the expression levels of ATGs were usually up-regulated with a few exceptions in seedlings by other five hormones tested. After NAA treatment, the expression levels of six ATGs (NtATG2, NtATG5, NtATG9, NtATG13c, NtATG18f and NtVPS34) showed a significant increase (>2-fold expression change). The expression levels of five ATGs (NtATG2, NtATG5, NtATG9, NtATG13c and NtVPS34) and four ATGs (NtATG2, NtATG9, NtATG13c and NtVPS34) were increased significantly (>2-fold expression change) after 2, 4-D and ABA treatments, respectively. In addition, 4 ATGs (NtATG1a, NtATG18a, NtATG18c and NtATG18e) and 11 ATGs (NtATG1a, NtATG2, NtATG9, NtATG13b, NtATG13c, NtATG18a, NtATG18b, NtATG18c, NtATG18e, NtVPS15 and NtVPS34) were up-regulated (>2-fold expression change) by SA and JA, respectively. That is to say, these five plant hormones (NAA, 2,4-D, ABA, SA and JA) usually exert opposite effects on the regulation of ATG expression compared with GA3, implying that they may have common mechanism on regulating autophagy occurrences in the process of plant development, but differ from that of GA3.
Figure 9.
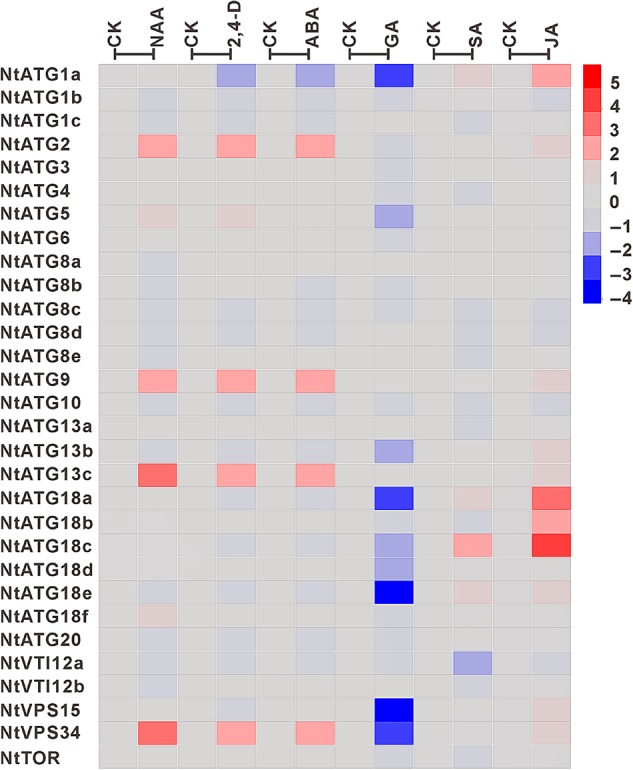
Expression profile of NtATGs in seedlings under different hormone treatments. Relative expression levels of each ATG in seedlings under different hormone treatments were normalized to GAPDH (AJ133422), Polyubiquitin (GQ281244), Actin (GQ281246) and Elongation factor 1a (AF120093). The expression level of each ATG was calculated and compared with that in seedlings under normal growth conditions. The expression level of each ATG in seedlings under normal growth conditions was indicated with grey box (0). Blue box indicates the lower transcriptional level of ATGs, whereas red box indicates the higher expression level of ATGs. Scale bar represents fold change (log2 value).
4. Discussion
Autophagy is an evolutionarily conserved cellular degradation process among yeast, mammals and plants.4,6 A set of genes involved in this process are called ATGs and has been extensively studied in yeast and mammals. More than 30 ATGs have been identified until now, and analyses of them unveiled multiple roles of autophagy and diverse aspects of molecular mechanism underlying it.3 On the basis of sequence similarity to autophagy-related proteins in yeast and mammals, 39 ATGs belonging to 20 different groups in A. thaliana5,6 and 33 ATGs belonging to 13 different groups in O. sativa7 were identified, respectively, suggesting the core components of autophagy are conserved among different species. In addition, these ATGs could be divided into four major functional groups including ATG1 kinase complex group, ATG9 recycling complex, phosphoinositide-3-kinase (PI3K) complex and ubiquitin-like conjugation system. However, the roles of them in plant development and in response to dynamic environment, as well as conservation of autophagy between monocotyledon and dicotyledon, are still largely unknown.
Nicotiana tabacum has been used as an ideal model plant for the study of autophagy in plants for many years.20–22 However, only a few ATGs have been identified up to now in tobacco. To facilitate our understanding of the molecular mechanism regulating autophagy, it is necessary to identify all the ATGs in tobacco genome. Thus, in the present study, 30 ATGs belonging to 16 different classes according to the sequence similarity to ATGs of A. thaliana have been identified. No homologues of ATG7 and ATG12 have been identified in the present study based on the draft genomic sequences of N. tabacum deposited in NCBI, and no EST sequences covering ATG7 and ATG12 were found according to available data. Most of the ATGs (ATG2, ATG3, ATG4, ATG5, ATG6, ATG9, ATG10, ATG20, VPS15 and VPS34) comprise single member. Only a few of them (ATG1, ATG8, ATG13, ATG18 and VTI12) contain multiple members, ranging from two to six in different groups (Table 1). However, majority of ATGs in O. sativa have multiple isoforms with four exceptions (ATG5, ATG7, ATG12 and ATG16), indicating that plants may have species-specific mechanism regulating autophagy. In N. tabacum, the compositions of domain within each class are similar with two exceptions (ATG1 and ATG18), suggesting possible functional redundancy of different members in the same group. ATG1c shows a similar serine/threonine-protein kinase domain at N-terminal with 1a and 1b, but lacks a similar C-terminal structure that in ATG1a and 1b. Meanwhile, all members in the ATG18 group show a similar N-terminal structure, but lacking of a C-terminal BCAS3 domain in ATG18d, ATG18e and ATG18f, suggesting the functional divergence of different members in the same class. In a recent work, two different members in the Cysteine Proteases ATG4 group (AtAtg4a and AtAtg4b) of A. thaliana were approved to selectively cleavage different AtAtg8 substrates. AtAtg4a is more active when processing substrates AtAtg8a, AtAtg8c, AtAtg8d and AtAtg8i better than AtAtg4b in vitro, but displays similar levels with AtAtg4b when processing other AtAtg8 substrates,33 suggesting the function divergence of ATGs in the same group. In the present study, we found that the responses of ATGs in the same group to different environmental signals are distinct. Hence, the conserved and specific functions of ATGs in the same group need to be elucidated in the further study.
During past decades, autophagy has been shown to play roles in the adaption of plant to different stresses, including nutrient starvation, oxidative stress, drought, salt and other abiotic stresses in plants.6 Among them, nitrogen deficiency, starvation and darkness stress in relation to autophagy were most extensively explored, and have been used as model systems for the study of plant autophagy.34 It was reported that autophagy could be induced by nutrient starvation and is critical for nutrient remobilization.35 The relative expression levels of some ATGs were increased significantly under the condition of nutrient starvation. In A. thaliana, the transcriptional levels of ATGs in the ATG8 conjugation pathway were transiently unregulated in a co-ordinated manner by starvation.36 In O. sativa, the expression levels of ATGs were also reported to be induced by starvation stresses.7 However, not all ATGs in the ATG8 conjugation pathway were synergistically unregulated in O. sativa. In the present study, the responses of 30 ATGs in tobacco to carbon and nitrogen starvation were also investigated. Surprisingly, none of ATGs in the ATG8 conjugation pathway was induced by carbon and nitrogen starvation. However, we found that ATGs in ATG1 kinase, ATG9 recycling and PI3K complex were coordinately unregulated by both carbon and nitrogen starvation, e.g. ATG13s in ATG1 complex and ATG18s in ATG9 complex and three members of PI3K complex, which play critical roles in the initial stages of autophagy. Early study in A. thaliana showed that eight ATG18 genes were differentially expressed in response to different growth conditions, and only ATG18a was induced by both sucrose and nitrogen starvation, which is likely required for autophagosome formation.11 However, six members of ATG18 gene are commonly unregulated by both carbon and nitrogen starvation in N. tabacum, introducing a possibility that different members of ATG18 may cooperate in starvation induced autophagy. In addition, the responses of ATGs to different stresses including starvation, cold, drought, salt and heavy metals were also detected and compared with one another. We found that several ATGs (ATG1a, ATG2, ATG13c, ATG18a, ATG18c and VPS34) are commonly up-regulated by all abiotic stresses tested. Interestingly, all these ATGs also belong to ATG1 kinase, ATG9 recycling and PI3K complex, which suggested that a central role of ATG1 kinase, ATG9 recycling and PI3K complex as an effector in plant response to different stresses. In contrast, we also found that several ATGs are responsive only to specific stress in the present study. Interestingly, the conserved autophagy machinery could play its role in responses to different stresses, whereas several ATGs only response to specific stresses. This seems contradictory and remains a question with great interesting for further study. Two possible mechanisms might explain this phenomenon. The first is that the transcription of each ATG in different conditions may be controlled by a set of different transcriptional factors, which show different responses to various stress conditions. Another possibility is that external signals could regulate the autophagy via regulating the partners of ATGs, rather than regulate ATG expression itself. In a recent report, an ATG3-interacting protein cytosolic glyceraldehyde-3-phosphate dehydrogenases (GAPCs) has been identified, and the interaction between them was affected by ROS, which was reported to have the ability to induce autophagy.37 In this case, ATGs may not directly response to some stresses.
Plant hormones, as the major regulators in the different processes of plant development, have also been shown to be involved in the regulation of PCD.38 Two different classes of PCD, vacuolar cell death and necrosis, could be distinguished according to morphological characteristics.39 Necrosis usually occurs under abiotic stresses, whereas vacuolar cell death, an autophagy-like process, is common during the processes of plant development.39 Four major phytohormones, including SA, JA, GA3 and ABA, have been reported as PCD-signal regulators. SA has been approved as a well-known mediator in hypersensitive cell death,40 and JA usually functions in biotic stress-induced PCD.41–43 However, the links of plant hormones to autophagy-like vacuolar cell death remain to be elucidated. There are a few reports, indicating that GA3 and ABA were involved in the regulation of vacuolar cell death.38 In addition, the relationship between phytohormones and autophagy is also largely unknown. There are only some pioneer researches related to the relationship between phytohormones and autophagy. For example, autophagy was approved to be involved in the response to ABA.44 In a recent report, SA signalling pathway was also shown to be required for early senescence in atg mutant.12 In the present study, the responses of ATGs to ABA and SA were analysed first. The expression levels of several ATGs increased in seedlings after treatment by ABA and SA, respectively. However, no ATG gene responded to both ABA and SA synchronously, suggesting that ABA and SA regulate autophagy via different mechanisms. In addition, the responses of ATGs to other four phytohormones including NAA, 2, 4-D, GA3 and JA were comprehensively studied and compared with one another. The data implied that some ATGs are response to phytohormones and most of them are regulated by NAA, 2, 4-D and ABA, implying a common role of autophagy in the response to different phytohormones. The exact role of autophagy in plant response to phytohormones and the linkage between autophagy and phytohormone signals are worthy to be elucidated in the further works.
On the other hand, there are growing evidences indicating additional crucial roles of autophagy in plant development under normal conditions, including leaf starch degradation,8 lifespan extension,14 vacuolar cell death of embryo suspensor,27 lipid metabolism regulation, anther development18 and maintenance of peroxisomal quality.15–17 In the present study, expression profile analysis of ATGs in tobacco shows that the transcripts of all ATGs could be detected in all tissues tested, but the transcriptional levels of ATGs vary among different tissues especially in the processes of seed formation, which have not yet been analysed in both O. sativa and A. thaliana. The expression profile analysis results implied that autophagy has more extensive roles than we thought in plant development under normal conditions, particularly in seed development. However, clear roles of autophagy in seed development are largely unknown up to date. There are only two examples implying the role of autophagy in embryo development. In P. abies, autophagy has been reported to be necessary for vacuolar cell death of embryo suspensor, which is essential for normal embryogenesis. Silencing of ATG5 or ATG6 abrogates normal suspensor formation and consequent embryo development.27 Autophagy has also been reported to have a role in microspore embryogenesis.45 Electron microscopy observation and dansylcadaverine staining revealed that autophagy exclusively occurs in embryogenic microspores, but not in other microspore-derived structures.45 Hence, the exact roles of autophagy in seed formation including embryogenesis, endosperm development and seed coat development are worthy to be elucidated in the further study.
In addition, our expression profile analysis of ATGs in tobacco revealed that a relatively higher expression level of most ATGs in pollen/anther compared with that in other tissues tested, suggesting a potential role of autophagy in male gametophyte development. It was reported that AtATG6, one of the components of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex, has been approved to be essential for pollen germination.46–48 Two other members of the PtdIns3K complex, VPS1549 and VPS34,50 were also shown to be important for pollen germination in A. thaliana. However, ATG6, VPS15 and VPS34 commonly exist in two separate complexes that regulate either autophagy or the vacuolar protein sorting pathway. In addition, the knockout mutants of other ATG genes in A. thaliana are fertile and display normal life cycle, indicating that the defects in pollen germination of ATG6, VPS15 and VPS34 are likely not attributing to the deficiency of autophagic activity in pollen. However, the contribution of autophagy to plant developmental processes may vary among different species. In P. abies, autophagy has been reported to be necessary for vacuolar cell death of embryo suspensor.27 In addition, ATG7-knockout mutant with defects in autophagy and sporophytic male sterility has also been identified in rice.18 Hence, the comparison of the roles of autophagy in different systems might be a charming work for understanding molecular mechanism underlying autophagy.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
The work was financially supported by the National Natural Science Foundation of China (31400171 and 31270362). Funding to pay the Open Access publication charges for this article was provided by National Natural Science Foundation of China.
Supplementary Material
References
- 1.Bassham D.C. 2007, Plant autophagy—more than a starvation response, Curr. Opin. Plant Biol., 10, 587–93. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N., Levine B.. 2010, Autophagy in mammalian development and differentiation, Nat. Cell Biol., 12, 823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boya P., Reggiori F., Codogno P.. 2013, Emerging regulation and functions of autophagy, Nat. Cell Biol., 15, 713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y.. 2009, Dynamics and diversity in autophagy mechanisms: lessons from yeast, Nat. Rev. Mol. Cell Biol., 10, 458–67. [DOI] [PubMed] [Google Scholar]
- 5.Il Kwon S., Park O.K.. 2008, Autophagy in plants, J Plant Biol., 51, 313–20. [Google Scholar]
- 6.Han S., Yu B., Wang Y., Liu Y.. 2011, Role of plant autophagy in stress response, Protein Cell, 2, 784–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia K., Liu T., Ouyang J., Wang R., Fan T., Zhang M.. 2011, Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.), DNA Res., 18, 363–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Yu B., Zhao J. et al. 2013, Autophagy contributes to leaf starch degradation, Plant Cell, 25, 1383–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon S.I., Cho H.J., Park O.K.. 2010, Role of Arabidopsis RabG3b and autophagy in tracheary element differentiation, Autophagy, 6, 1187–9. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y., Schiff M., Czymmek K., Talloczy Z., Levine B., Dinesh-Kumar S.P.. 2005, Autophagy regulates programmed cell death during the plant innate immune response, Cell, 121, 567–77. [DOI] [PubMed] [Google Scholar]
- 11.Xiong Y., Contento A.L., Bassham D.C.. 2005, AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana, Plant J., 42, 535–46. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimoto K., Jikumaru Y., Kamiya Y. et al. 2009, Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis, Plant Cell, 21, 2914–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong Y., Contento A.L., Nguyen P.Q., Bassham D.C.. 2007, Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis, Plant Physiol., 143, 291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minina E.A., Sanchez-Vera V., Moschou P.N. et al. 2013, Autophagy mediates caloric restriction-induced lifespan extension in Arabidopsis, Aging Cell, 12, 327–9. [DOI] [PubMed] [Google Scholar]
- 15.Farmer L.M., Rinaldi M.A., Young P.G., Danan C.H., Burkhart S.E., Bartel B.. 2013, Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation, Plant Cell, 25, 4085–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibata M., Oikawa K., Yoshimoto K. et al. 2013, Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis, Plant Cell, 25, 4967–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshimoto K., Shibata M., Kondo M. et al. 2014, Organ-specific quality control of plant peroxisomes is mediated by autophagy, J. Cell Sci., 127, 1161–8. [DOI] [PubMed] [Google Scholar]
- 18.Kurusu T., Koyano T., Hanamata S. et al. 2014, OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development, Autophagy, 10, 878–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sierro N., van Oeveren J., van Eijk M.J. et al. 2013, Whole genome profiling physical map and ancestral annotation of tobacco Hicks Broadleaf, Plant J., 75, 880–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moriyasu Y., Ohsumi Y.. 1996, Autophagy in tobacco suspension-cultured cells in response to sucrose starvation, Plant Physiol., 111, 1233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takatsuka C., Inoue Y., Matsuoka K., Moriyasu Y.. 2004, 3-methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions, Plant Cell Physiol., 45, 265–74. [DOI] [PubMed] [Google Scholar]
- 22.Yano K., Hattori M., Moriyasu Y.. 2007, A novel type of autophagy occurs together with vacuole genesis in miniprotoplasts prepared from tobacco culture cells, Autophagy, 3, 215–21. [DOI] [PubMed] [Google Scholar]
- 23.Sierro N., Battey J.N., Ouadi S. et al. 2014, The tobacco genome sequence and its comparison with those of tomato and potato, Nature Commun., 5, 3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao P., Zhou X.M., Zou J. et al. 2014, Comprehensive analysis of cystatin family genes suggests their putative functions in sexual reproduction, embryogenesis, and seed formation, J. Exp. Bot., 65, 5093–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y.. 2006, AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells, Plant Cell Physiol., 47, 1641–52. [DOI] [PubMed] [Google Scholar]
- 26.Ono Y., Wada S., Izumi M., Makino A., Ishida H.. 2013, Evidence for contribution of autophagy to rubisco degradation during leaf senescence in Arabidopsis thaliana, Plant Cell Environ., 36, 1147–59. [DOI] [PubMed] [Google Scholar]
- 27.Minina E.A., Filonova L.H., Fukada K. et al. 2013, Autophagy and metacaspase determine the mode of cell death in plants, J. Cell Biol., 203, 917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D.. 2005, Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways, Plant Physiol., 138, 2097–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Gioacchino M., Petrarca C., Perrone A. et al. 2008, Autophagy as an ultrastructural marker of heavy metal toxicity in human cord blood hematopoietic stem cells, Sci. Total Environ., 392, 50–8. [DOI] [PubMed] [Google Scholar]
- 30.Friml J., Vieten A., Sauer M. et al. 2003, Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis, Nature, 426, 147–53. [DOI] [PubMed] [Google Scholar]
- 31.Guo J., Wei J., Xu J., Sun M.X.. 2014, Inducible knock-down of GNOM during root formation reveals tissue-specific response to auxin transport and its modulation of local auxin biosynthesis, J. Exp. Bot., 65, 1165–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volz R., Heydlauff J., Ripper D., von Lyncker L., Gross-Hardt R.. 2013, Ethylene signaling is required for synergid degeneration and the establishment of a pollen tube block, Dev. Cell, 25, 310–6. [DOI] [PubMed] [Google Scholar]
- 33.Woo J., Park E., Dinesh-Kumar S.P.. 2014, Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases, Proc. Natl. Acad. Sci. USA, 111, 863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bassham D.C., Laporte M., Marty F. et al. 2006, Autophagy in development and stress responses of plants, Autophagy, 2, 2–11. [DOI] [PubMed] [Google Scholar]
- 35.Guiboileau A., Yoshimoto K., Soulay F., Bataille M.P., Avice J.C., Masclaux-Daubresse C.. 2012, Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis, New Phytol., 194, 732–40. [DOI] [PubMed] [Google Scholar]
- 36.Rose T.L., Bonneau L., Der C., Marty-Mazars D., Marty F.. 2006, Starvation-induced expression of autophagy-related genes in Arabidopsis, Biol. Cell, 98, 53–67. [DOI] [PubMed] [Google Scholar]
- 37.Han S., Wang Y., Zheng X. et al. 2015, Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana, Plant Cell, 27, 1316–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bethke P.C., Lonsdale J.E., Fath A., Jones R.L.. 1999, Hormonally regulated programmed cell death in barley aleurone cells, Plant Cell, 11, 1033–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Doorn W.G., Beers E.P., Dangl J.L. et al. 2011, Morphological classification of plant cell deaths, Cell Death Differ., 18, 1241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love A.J., Milner J.J., Sadanandom A.. 2008, Timing is everything: regulatory overlap in plant cell death, Trends Plant Sci., 13, 589–95. [DOI] [PubMed] [Google Scholar]
- 41.Overmyer K., Tuominen H., Kettunen R. et al. 2000, Ozone-sensitive arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death, Plant Cell, 12, 1849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao M.V., Lee H., Creelman R.A., Mullet J.E., Davis K.R.. 2000, Jasmonic acid signaling modulates ozone-induced hypersensitive cell death, Plant Cell, 12, 1633–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tuominen H., Overmyer K., Keinanen M., Kollist H., Kangasjarvi J.. 2004, Mutual antagonism of ethylene and jasmonic acid regulates ozone-induced spreading cell death in Arabidopsis, Plant J., 39, 59–69. [DOI] [PubMed] [Google Scholar]
- 44.Vanhee C., Batoko H.. 2011, Autophagy involvement in responses to abscisic acid by plant cells, Autophagy, 7, 655–6. [DOI] [PubMed] [Google Scholar]
- 45.Corral-Martinez P., Parra-Vega V., Segui-Simarro J.M.. 2013, Novel features of Brassica napus embryogenic microspores revealed by high pressure freezing and freeze substitution: evidence for massive autophagy and excretion-based cytoplasmic cleaning, J. Exp. Bot., 64, 3061–75. [DOI] [PubMed] [Google Scholar]
- 46.Fujiki Y., Yoshimoto K., Ohsumi Y.. 2007, An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination, Plant Physiol., 143, 1132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin G., Ma Z., Zhang L. et al. 2007, Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development, Cell Res., 17, 249–63. [DOI] [PubMed] [Google Scholar]
- 48.Harrison-Lowe N.J., Olsen L.J.. 2008, Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana, Autophagy, 4, 339–48. [DOI] [PubMed] [Google Scholar]
- 49.Xu N., Gao X.Q., Zhao X.Y., Zhu D.Z., Zhou L.Z., Zhang X.S.. 2011, Arabidopsis AtVPS15 is essential for pollen development and germination through modulating phosphatidylinositol 3-phosphate formation, Plant Mol. Biol., 77, 251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee Y., Kim E.S., Choi Y. et al. 2008, The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development, Plant Physiol., 147, 1886–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.