Abstract
Jakrem hot water spring is located in the West Khasi Hill District of the state of Meghalaya, and is one of the most popular hot springs of the state. There is a populist belief among the inhabitants and people that the hot spring water has got curative properties against various skin ailments. This is the first report on V3 hyper-variable region of 16S rDNA metagenome sequence employing Illumina platform to profile the microbial community of this less known hot spring from Meghalaya, India. Metagenome comprised of 10, 74,120 raw sequences with a sequence length of 151 bp and 56.35% G + C content. Metagenome sequence information is now available at NCBI, SRA database accession no. SRP056897. A total of 8, 77, 364 pre-processed reads were clustered into 694 OTUs (operational taxonomical units) comprising of 14 bacterial phyla including unknown phylum demonstrating 49 families. Hot spring bacterial community is dominated by Firmicutes (61.60%), Chloroflexi (21.37%), Cyanobacteria (12.96%) and unclassified bacteria (1.2%) respectively.
Keywords: Alkaline hot spring, Metagenome, Illumina sequencing, Microbial diversity, Firmicutes
| Specifications | |
|---|---|
| Organism/cell line/tissue | Metagenome of Jakrem hot spring |
| Sex | Not applicable |
| Sequencer or array type | Illumina |
| Data format | Analyzed |
| Experimental factors | Environmental sample |
| Experimental features | V3 hypervariable region of 16S rDNA was sequenced using paired end Illumina Mi-Seq technology, sequence analyzed by QIIME data analysis package. |
| Consent | Not applicable |
| Sample source location | Microbial/algal mat, Jakrem hot spring, Shillong, Meghalaya, India (25°24.450′ N and 91°32.414′ E, Elevation 4756 ft.) |
Direct link to deposited data
http://www.ncbi.nlm.nih.gov/Traces/sra_sub/sub.cgi?subid=436405&from=list&action=show:submission.
Experimental design, materials and methods
The thermal springs harbor population of microorganisms with great commercial importance and interest to the researchers and industry working on enzymes, sugars, compatible solutes and antibiotics [1]. Diversity analysis of such extreme environments has got ample attention due to their diverse and unique ecology, chemistry and opportunity they provide to identify rare compounds and genes [2]. Despite the possibility of the presence of novel microbes with high economic and industrial use very few reports are available on the microbial diversity of hot springs from the Indian subcontinent. Cultivation-dependent studies are valuable for isolating novel organisms and exploring their properties but the cultivation independent methods offer a more comprehensive assessment of microbial diversity [3]. The study was designed to investigate community composition by using metagenomic approach and a clone library based study of hot spring microbial mat samples. The findings revealed the presence of a large number of sequence reads from bacterial taxa Arthronema which may represent novel species within this genus which was reported for the first time in association with hot spring.
India is one of the tectonically active areas in the world inhabiting many hot springs. The Jakrem hot spring (25°24.450′ N and 91°32.414′ E) is one of the less explored hot water springs in Meghalaya, India. The temperature and pH of the hot spring are reported to be 46 °C and 9 respectively. The only report available on this less explored hot water spring revealed the presence of species of Mastigocladus and Microcoleus of Cyanophyceae [4]. In the present investigation, the total community DNA was isolated from the microbial mat samples of the spring using a FastDNA spin kit (MP Biomedicals, LLC, USA). The freshly extracted DNA was purified using 0.5% low gelling temperature agarose. Final DNA concentration was quantified using a microplate reader (BMG Labtech, Jena, Germany).
The V3 region of the 16S rRNA gene was amplified using 341F/518R primer combination (5′CCTACGGGAGGCAGCAG 3′; 5′ATTACCGCGGCTGCTGG 3′). Amplicon was excised and purified by QIA quick Gel Extraction kit (Qiagen, Valencia, CA, USA) according to the manufacturer's manual. Purified amplicon was paired-end sequenced on an Illumina Mi-Seq platform. QIIME data analysis package was used for 16S rRNA data analysis [5]. Quality check on raw sequences was performed as per base quality score distributions, average base content per read and GC distribution in the reads. Singletons, the unique OTU that did not cluster with other sequences, were removed as it might be a consequence of sequencing errors and can result in spurious OTUs. Chimeras were also removed using UCHIME, and pre-processed consensus V3 sequences were grouped into operational taxonomic units (OTUs) using the clustering program UCLUST at a similarity threshold of 0.97 [6], [7]. All the pre-processed reads were used to identify the OTUs using QIIME program for constructing a representative sequence for each OTU. The representative sequence was finally aligned against Greengenes core set of sequences using PyNAST program [8]. Representative sequence for each OTU was classified using RDP classifier and Greengenes OTU database and the sequences of those not classified were categorized as unknown.
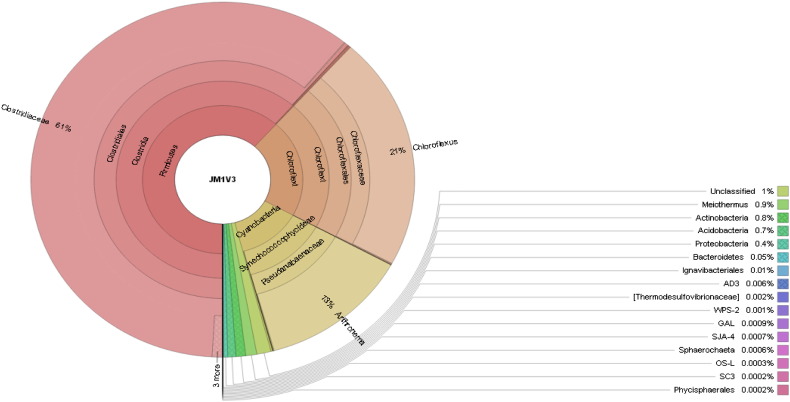
In the sample studied, the classified OTUs represent 13 distinct phyla dominated by Firmicutes (61.60%), Chloroflexi (21.37%) and Cyanobacteria (12.96%) (Fig. 1). Firmicutes grow chemo-organo-trophically on a number of organic substrates particularly in alkaline springs [9]. Firmicutes constituted 138 OTUs i.e. 19.88% of total OTUs and 5, 40,441 reads which are 61.59% of the total reads. Chloroflexi constitutes 58 OTUs i.e. 8.35% of total OTUs and 1, 87,546 reads which is 21.37% of total reads whereas 237 OTUs i.e. 34.14% of total OTUs belong to the unknown phylum substantiating the earlier findings that non-acidic hot springs harbor more diverse and variable bacterial phyla than the acidic springs [10]. The presence of many novel species is indicated by more than 200 unknown OTUs.
Fig. 1.
Community composition in Jakrem hot spring metagenome.
Prominent families in the metagenome were Clostridiaceae (60.92%), Chloroflexaceae (21.26%) and Pseudanabaenaceae (12. 77%). A total of 44 genera including unknown bacterial genus (63.50%), Chloroflexus (21.22%) and Arthronema (12.69%) were obtained. The existence of significant percentage of Arthronema sequences in the metagenome suggests that the microbial mat pigment richness in the microbial/algal mat of the hot spring may have massive commercial value as natural colorants in nutraceuticals, cosmetics and pharmaceutical industries. At the species level analysis, 12 bacterial species were recognized with the prominent presence of unknown species (99.38%) and Rhodococcus fascians (0.44%). More than 99% of the unknown species strongly suggest that the Jakrem hot spring harbors the wealth of uncultivable bacteria.
Nucleotide sequence accession number
Metagenome sequence data are available at NCBI Accession no. SRP056897.
Competing interests
The authors declare that there are no competing interests.
Acknowledgments
This research was funded by a grant from SERB, Govt. of India, New Delhi vide Project No. SB/FT/LS-335/2012.
References
- 1.Satyanarayana T., Raghukumar C., Shivaji S. Extremophilic microbes: diversity and perspectives. Curr. Sci. 2005;89:78–90. [Google Scholar]
- 2.Kuddus M., Ramtekke P.W. Recent developments in production and biotechnological applications of cold-active microbial proteases. Crit. Rev. Microbiol. 2012;38:380–388. doi: 10.3109/1040841X.2012.678477. [DOI] [PubMed] [Google Scholar]
- 3.Tringe S.G., Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr. Opin. Microbiol. 2008;11:442–446. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Siangbood H., Ramanujam P. A report on thermophilic cyanophyta (cyanobacteria) from Jakrem hotspring. Meghalaya. International Journal on Algae. 2011;13(2):178–185. [Google Scholar]
- 5.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 8.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou W., Wang S., Dong H., Jiang H., Briggs B.R., Peacock J.P., Huang Q., Huang L., Wu G., Zhi X., Li W., Dodsworth J.A., Hedlund B.P., Zhang C., Hartnett H.E., Dijkstra P., Hungate B.A. A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLoS ONE. 2013;8(1):e53350. doi: 10.1371/journal.pone.0053350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Z.Q., Wang F.P., Zhi X.Y., Chen J.Q., Zhou E.M., Liang F., Xiao X., Tang S.K., Jiang H.C., Zhang C.L., Dong H., Li W.J. Bacterial and archaeal diversities in Yunnan and Tibetan hot springs. China. Environ Microbiol. 2013;15(4):1160–1175. doi: 10.1111/1462-2920.12025. [DOI] [PubMed] [Google Scholar]