Abstract
Mucosal sites such as the oropharynx contain a wide range of microorganisms, collectively designated as the microbiome. The microbiome can affect behavior through a number of neurobiological and immunological mechanisms. Most previous studies have focused on the bacterial components of the microbiome. However, the microbiome also includes viruses such as bacteriophages, which are viruses that infect bacteria and alter their metabolism and replication. We employed metagenomic analysis to characterize bacteriophage genomes in the oral pharynx of 41 individuals with schizophrenia and 33 control individuals without a psychiatric disorder. This analysis was performed by the generation of more than 100000000 sequence reads from each sample and the mapping of these reads to databases. We identified 79 distinct bacteriophage sequences in the oropharyngeal samples. Of these, one bacteriophage genome, Lactobacillus phage phiadh, was found to be significantly different in individuals with schizophrenia (P < .00037, q < 0.03 adjusted for multiple comparisons). The differential levels of Lactobacillus phage phiadh remained significant when controlling for age, gender, race, socioeconomic status, or cigarette smoking (P < .006). Within the group of individuals with schizophrenia, the level of Lactobacillus phage phiadh correlated with the prevalence of immunological disorders as well as with the administration of valproate, which has been shown in animal models to alter the microbiome. The bacteriophage composition of the oropharynx in individuals with schizophrenia differs from that of controls. The biological consequences of this difference and the potential effects of altering bacteriophage levels through therapeutic interventions are worthy of further investigation.
Key words: virus, metagenome, inflammation, bacteriophage
Introduction
Human mucosal sites are not sterile but are colonized by a range of microbial organisms. These organisms, collectively termed the microbiome, have recently been the target of extensive investigations, and have been found to affect a number of biological processes, particularly ones related to the immune system.1,2 Many investigations have focused on the bacterial components of the microbiome since conserved ribosomal sequences can be utilized to amplify and characterize virtually all bacterial sequences in an efficient manner.3 However, recent studies have indicated that the microbiome also consists of viruses.4 This “virome” can be characterized by the newly developed techniques of whole genome sequencing which do not rely on the amplification of ribosomal or other shared nucleotide sequences and hence can detect a wide range of viral species.5 The application of whole genome sequencing techniques has revealed that the virome of most human mucosal surfaces consists largely of a wide range of bacteriophages, which are viruses that infect bacteria.6 Infection of bacteria by bacteriophages can result in the death of the bacteria or the integration of the phage into the bacterial genome, with subsequent effects on bacterial functioning. The bacteriophage components of the human virome have been most studied in the intestinal tract. These studies have been performed by the analysis of fecal samples or samples obtained by endoscopic techniques.7 The viromes of other human mucosal sites, such as the oropharynx, are less well characterized.8 It has been hypothesized that the collection of bacteriophages, characterized as the “phageome” plays an important, and as yet incompletely characterized, role in human health and disease.9,10
Schizophrenia is a complex, pervasive neuropsychiatric disorder with worldwide prevalence.11 Recent studies indicate that schizophrenia and related psychiatric disorders are defined by both genetic susceptibility and environmental exposures.12 Furthermore, schizophrenia and related disorders are often associated with altered immune functioning, generally manifested by increased levels of antibodies to foreign antigens and markers of systemic inflammation. Furthermore recent genetic studies of schizophrenia have indicated that genes related to the immune response are risk factors.13 However, the pathological events leading to the altered inflammatory status observed in individuals with schizophrenia have not been defined.
Numerous studies in animal models have documented the fact that the virome can alter immune functioning.14,15 Furthermore, studies have shown the microbiome can affect cognition and behavior through a number of immune-related mechanisms.16,17 We employed whole genome sequencing to characterize the virome of individuals with schizophrenia and control individuals without a psychiatric disorder and to relate the composition of the virome to disease status.
Methods
Study Population
Samples for analysis were obtained from 41 individuals with schizophrenia and 33 control individuals without a psychiatric disorder living in the Baltimore, Maryland Metropolitan area. The methods used for ascertainment of the case and control individuals as well as the inclusion and exclusion criteria have been previously described.18–20 Individuals without a history of psychiatric disorder were recruited from posted announcements at local health care facilities and universities in the same geographic area as the settings where the psychiatric participants were recruited. These control individuals were enrolled after they were screened to rule out the presence of a current or past psychiatric disorder with the Structured Clinical Interview for DSM-IV Axis I Disorders—Non-patient Edition.21 Information about demographic variables and current smoking status was obtained from participants’ self report. Participants were also asked about the presence of co-occurring medical conditions. Cases were interviewed and rated on the Positive and Negative Syndrome Scale (PANSS)22 to determine psychiatric symptom severity. The current medications of schizophrenia participants were determined from medical records. The clinical and demographic characteristics of the case and control individuals who participated in the study are presented in table 1. Patients were drawn from several different inpatient units, day hospitals, and rehabilitation programs within the Baltimore area. All of the participants provided written informed consent following explanation of the study goals and procedures. The study was approved by the Institutional Review Boards of Johns Hopkins University and Sheppard Pratt Hospital.
Table 1.
Demographic Data Related to the Study Participants
| Schizophrenia, N = 41 | Control, N = 33 | P value | |
|---|---|---|---|
| Age, years | 39.2 (9.9) | 30.9 (8.8) | <.0004 |
| Maternal education, years | 12.9 (3.3) | 14.2 (2.5) | NS |
| Body mass index | 34.1 (5.9) | 26.0 (6.4) | <.0001 |
| Gender–male | 65.9% | 57.6% | NS |
| Race-Caucasian | 48.8% | 51.5% | NS |
| Cigarette smoker | 63.4% | 3.0% | <.0001 |
Note: NS, not significant (P < .1). Data are indicated for the 41 individuals with schizophrenia and 33 controls who participated in the study except that data for body mass index was missing for 3 individuals with schizophrenia. Numbers in parentheses indicate standard deviation.
Clinical Samples
Samples were obtained by gentle swabbing of the posterior pharynx using sterile swabs (CultureSwab Liquid Stuart, Becton Dickinson). The swabs were placed in sterile saline at the collection site and shipped to the processing laboratory in a cooler. Samples were stored at −70°C immediately after they were received at the processing laboratory which was generally within several hours after the sample was obtained.
Sample Processing
DNA was extracted from throat swabs using Qiagen’s Gentra Puregene Buccal Cell Kit. The collection brush heads from the swab ends were excised and incubated at 65°C overnight in the kit cell lysis solution. Manufacturer’s instructions were followed for the remainder of the protocol except that polyacrylamide was added as a carrier during the precipitation reactions to increase DNA yield. Aliquots of 75–100ng of DNA were used to generate paired end libraries using the Nugen Ultralow DR Multiplex System following the manufacturer’s instructions (http://info.nugeninc.com/UGOvationUltralowDRMultiplexSystem1-96.html). The libraries were purified and run on the Bioanalyzer to confirm size and concentration.
Libraries were then sequenced using the Illumina Hi Seq 2000 generating a median of 202000000 individual reads (109000000, 370000000 lower, upper quartile), 100 nucleotides in length.
Sequence Analysis
Sequence reads were filtered to remove low quality sequences, resulting in a minimum length of 60 nucleotides. Human, bacterial, fungal, and parasitic sequence reads were then removed by bioinformatic filtering using the following general protocol. Sequence reads with homology to human sequences were removed in 2 stages. The first stage employed the program Bowtie (http://bowtie-bio.sourceforge.net/index.shtml). A sliding window approach was used to align a forty base pair subsequence from the reads to the human genome Build 37 (ftp://ftp.ccb.jhu.edu/pub/data/bowtie_indexes/hg18.ebwt.zip). During each iteration, reads mapping to the human genome were removed from the analysis and the subsequence used for alignment was offset by 5 bases. The reads that survived this subtraction procedure were then imported into CLC Genomics Workbench Version 6 (www.clcbio.com) and a second subtraction against the human genome build 37 and additional sets of human sequences available from the UCSC Genome Browser (http://genome.ucsc.edu/) were performed using the Reference Mapping algorithm employing global alignment with the settings of similarity = 0.4 and length = 0.4. In some cases duplicate reads were also removed by the use of CLC. The sequence reads which remained following the removal of the human sequences were filtered sequentially to remove bacterial, fungal, and, in some cases protozoal sequences by matching to NCBI Reference Sequence (RefSeq) bacterial, fungal, and protozoal databases, respectively (http://www.ncbi.nlm.nih.gov/refseq/). This filtering was performed using the Reference Mapping algorithm and default settings. The filtering resulted in the removal of a median of 96% of the original reads (90.9%–99.2%, lower, upper quartile)
The remaining sequence reads were then aligned to the Refseq Virus Database (ftp://ftp.ncbi.nlm.nih.gov/refseq/release/viral/) using the Read Mapping algorithm of the CLC Genomics Workbench Version 6 and settings of length fraction = 0.8 and similarity = 0.8 with the retention of samples which matched uniquely to a single phage reference genome. Sequence reads were also mapped against the complete host bacterial genome Lactobacillus gasseri (NC_008530.1) utilizing the same parameters. This bacterial species was selected based on the results of the phage sequence analysis as described later.
Statistical Analysis
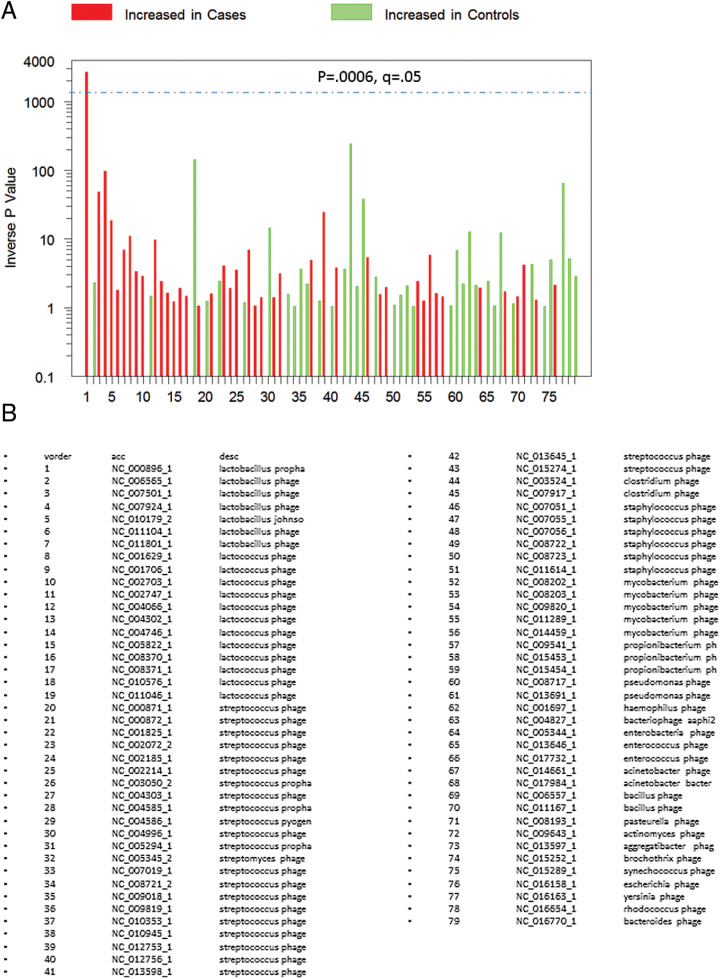
Reference sequences were employed for statistical analysis if they consisted of reference bacteriophage genomes which had at least 1 match to at least 5 of the clinical samples. There were a total of 79 complete bacteriophage genomes which fulfilled these criteria (figure 1). The number of sequence reads from the cases and control samples were initially compared for matches to each of these 79 bacteriophage genome sequences by means of 2-way analysis of variance (ANOVA) employing logarithmic transformation of the sequence reads and a minimum value of 0.1 reads to allow for the analysis of these reads following logarithmic transformation. Logarithmic transformation was employed to reduce skewness as has been employed in previous studies.23 Statistical significance was defined as P < .00063; q < 0.05 following the Bonferroni correction taking into account the N = 79 target bacteriophage genome sequences employed in the analysis.
Fig. 1.
(A) P values representing different levels of phages in 41 individuals with schizophrenia and 33 control individuals without a psychiatric disorder calculated as indicated in the text. The red and green colors indicated levels of the individual phages which are increased or decreased in cases, respectively. The dashed horizontal line indicates P < .05 corrected for multiple comparisons. The names and accession numbers of the phages corresponding to the numbers are shown in B.
This preliminary analysis identified a single bacteriophage genome which met the established criteria for significance (figure 1). The homologies of sequence reads aligned to this genome were confirmed by matching to the complete nonredundant NCBI nucleotide sequence database using the Blastn algorithm (http://blast.ncbi.nlm.nih.gov/html/blastcgihelp.html). Sequence reads which showed the greatest match to the corresponding phage genome with a probability of E ≤ 10–5 were retained for further analysis. Log transformed numbers of sequence reads meeting these criteria were further analyzed for case–control differences employing linear regression models which included the potentially confounding covariates of age, gender, race, cigarette smoking, body mass index (BMI), and maternal education, the latter being a marker of socioeconomic status. We also employed logistic regression models to compute odds ratios of having at least one sequence read which matched the target bacteriophage genome, using case status as the categorical outcome variable and the same covariates as were used in the linear regression model except that BMI was not included in the logistic regression due to the generation of asymmetric confidence intervals. Individuals with schizophrenia were also analyzed in terms of association between the presence of sequence reads homologous to the target bacteriophage genome and clinical variables related to their disease. These variables included ones related to illness duration, medications, and co-morbid clinical conditions. These analyses employed chi-square models for discrete variables and 2-way analysis of variance for continuous outcomes.
Sequence reads from the case and control individuals were further analyzed by the assembling of contigs following mapping to the target bacteriophage genome using the CLC algorithm described earlier. The contigs were also analyzed in terms of protein coding capabilities by the use of the Blastx algorithm and the nonredundant protein database (http://blast.ncbi.nlm.nih.gov/html/blastcgihelp.html)
All statistical analyses were performed using STATA Version 12, STATA Co, College Station Texas.
Results
The demographic variables of the individuals with schizophrenia and controls who participated in this study are depicted in table 1. The groups were similar in terms of gender, race, and socioeconomic status as measured by maternal education. They differed in terms of age, body mass index (BMI), and prevalence of cigarette smoking. The increased prevalence of cigarette smoking is characteristic of individuals with schizophrenia.24 The mean age of onset for the individuals with schizophrenia was 17.1 years (SD = 4.4) and duration of illness was 21.9 years (SD = 9.9).
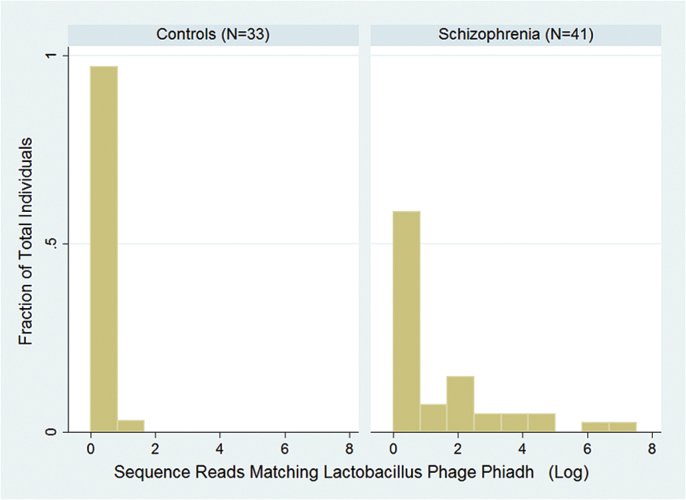
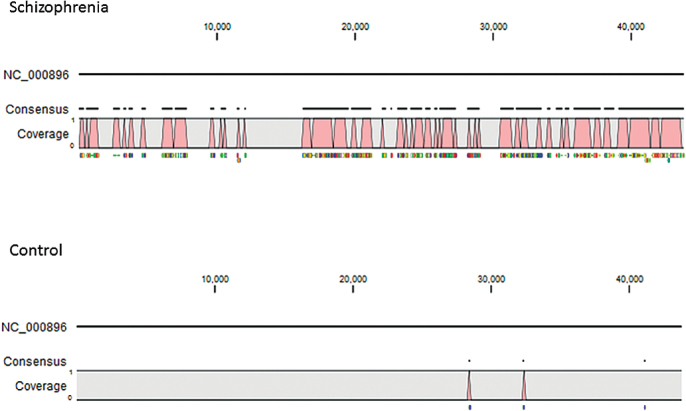
The P values related to case–control differences for the samples from the 41 individuals with schizophrenia and 33 controls mapped to 79 phage genomes are depicted in figure 1. There was a single phage, Lactobacillus phage phiadh (Refseq Accession Number NC_000896) which displayed significantly different numbers of sequence reads in cases and controls taking into account the multiple comparisons employed in the analysis (P < .00037, q < 0.029). The number of sequence reads in case and control samples mapping to the Lactobacillus phage phiadh genome and confirmed by Blastn homology analyses are depicted in figure 2. The number of sequence reads with homology to Lactobacillus phage phiadh in the individuals with schizophrenia was significantly greater than the number of matches in the controls (coefficient 1.4, 95% confidence interval [CI] 0.4–2.4, P < .009 adjusted for age, gender, race, maternal education, BMI, and cigarette smoking). The number of matches to Lactobacillus phage phiadh was not independently associated with age, gender, race, maternal education, BMI, or cigarette smoking (all P > .1). The alignments of the sequence reads from the case and control individuals to the Lactobacillus phage phiadh genome are depicted in figure 3. The case individuals had 2418 sequence reads which matched the Lactobacillus phage phiadh genome. These matches formed 107 contigs consisting of a total of 23596 nucleotides, corresponding to approximately 54% of the total genome. Translation of the sequence reads from the case individuals displayed homology to 42 different bacteriophage genes with protein coding capacities (supplementary table 1). The one control individual whose pharynx harbored sequence reads homologous to Lactobacillus phage phiadh had 3 sequence reads matching 3 separate regions of the phage genome.
Fig. 2.
Histogram depicting the number of sequence reads matching Lactobacillus phage phiadh (NC_000896) obtained from 41 individuals with schizophrenia and 33 controls. Sequence matches were originally identified by the use of the CLC homology algorithm and confirmed by Blastn homology searches against the entire nonredundant nucleotide database as described in the text.
Fig. 3.
Mapping of sequence reads to the Lactobacillus phage phiadh genome (NC_000896) from the individuals with schizophrenia (top) and controls (bottom). The nucleotide position within the phage sequence is shown at the top. The genes homologous to the sequence reads in the throat swabs are depicted in supplementary table 1.
Overall a total of 17 of 41 (41.5%) of individuals with schizophrenia had at least 1 match to Lactobacillus phage phiadh as compared to 1 of 33 (3.3%) of controls (P < .001 Fisher’s Exact Test). The odds ratio of having at least 1 pharyngeal sequence read homologous to Lactobacillus phage phiadh and being a case was 52.4, 95% CI 3.7 to >100, P < .003 adjusted for age gender, race, maternal education, and smoking status. The odds of harboring Lactobacillus phage phiadh were not independently associated with age, gender, race, or cigarette smoking (all P < .1), but were associated with maternal education (odds ratio 1.6, 95% CI 1.1–2.4, P < .013). Overall Lactobacillus phage phiadh comprised a mean of 1.9% of the total phages which were analyzed in each individual (95% CI 0–3.9%). The total number of bacteriophages analyzed did not differ between cases and controls (P > .1).
We also compared the presence of pharyngeal Lactobacillus phage phiadh with clinical variables within the group of individuals with schizophrenia (table 2). There was a marginal association between the presence of pharyngeal Lactobacillus phage phiadh and the PANSS positive score (F = 3.2, P = .082) but no association of the phage with scores on the PANSS Negative, PANSS General, or PANSS Total Scores (P > .1). There was an association between harboring Lactobacillus phage phiadh and a co-morbid immunological disease (9 of 17 vs 2 of 24, P < .012). A total of 6 individuals colonized with Lactobacillus phage phiadh had type 2 diabetes, 2 had type 1 diabetes, and 1 had Crohn’s Disease. The 2 individuals with immunological disease who were not colonized with Lactobacillus phage phiadh had type 2 diabetes. There was no association between colonization with Lactobacillus phage phiadh and gastrointestinal disease, body mass index, or other identified co-morbid condition (all P > .1) .
Table 2.
Clinical Variables in Individuals With Schizophrenia Who Did or Did Not Have Detectable Lactobacillus Phage Phiadh Sequences in Their Pharyngeal Samples
| Lactobacillus Phage Present (N = 17) | Lactobacillus Phage Not Present (N = 24) | Statistical Significance | |
|---|---|---|---|
| PANSS positive | 18.5 (3.9) | 16.3 (4.1) | P < .082 |
| PANSS negative | 18.7 (4.3) | 19.4 (5.1) | NS |
| PANSS general | 35.3 (6.1) | 33.2 (6.6) | NS |
| PANSS total | 72.6 (8.4) | 69.0 (10.6) | NS |
| RBANS total | 67.3 (14.6) | 64.8 (11.9) | NS |
| Age of onset-years | 17.6 (4.8) | 16.8 (4.2) | NS |
| Illness duration-years | 20.7 (8.8) | 22.5 (10.7) | NS |
| Body mass index | 33.6 (7.2) | 34.5 (4.8) | NS |
| Cigarette smoker | 64.7% | 62.5% | NS |
| Mouth/tooth disease | 11.7% | 16.7% | NS |
| Immunological disease | 52.9% | 8.3% | P < .012 |
| Type 2 diabetes | 35.3% | 8.3% | P < .032 |
| Valproate therapy | 0 | 25% | P < .026 |
| Atypical antipsychotic therapy | 88.2% | 91.7% | NS |
Note: NS, not significant (P > .1). The numbers in the parentheses indicate the standard deviation. Statistical analyses were performed by chi-square analysis or analysis of variance as described in the text. P > .1.
In terms of medications used for the treatment of schizophrenia, there were no significant associations with other agents used to treat schizophrenia such as the atypical antipsychotics olanzapine, clozapine, or risperidone, (all P > .1).
However, there was a significant association with the administration of valproate, an anticonvulsant mood stabilizer. None of the 6 individuals taking valproate had Lactobacillus phage phiadh in their pharynx as opposed to 17 of the 35 individuals who were not receiving valproate (χ2 = 4.98, P = .026). There were no significant associations with other medications commonly used in individuals with schizophrenia such as other anticonvulsant agents, antidepressants, or anticholinergic agents (all P < .1).
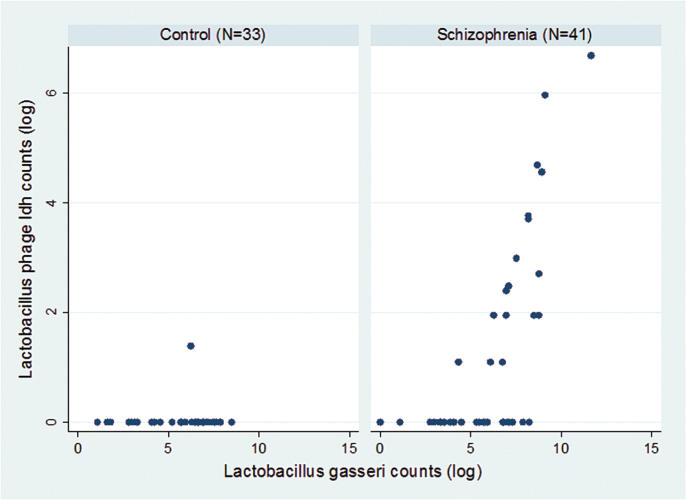
We also examined the relationship between the level of Lactobacillus phage phiadh and its host bacterium Lactobacillus gasseri (refseq NC_008530.1) in the throat swab samples. As depicted in figure 4 the levels of Lactobacillus gasseri correlated moderately with the levels of Lactobacillus phage phiadh (R 2 = .22 adjusted for age, gender, race, maternal education, diagnosis, and cigarette smoking). Overall, the levels of Lactobacillus gasseri in the case and control individuals did not differ to a statistically significant extent.
Fig. 4.
Scatterplot indicating the relationship between number of Lactobacillus phage phiadh (NC_000896) and Lactobacillus gasseri sequence reads in individuals with schizophrenia (right) and controls (left).
Discussion
We employed whole genome sequencing and bioanalytical techniques to characterize the pharyngeal virome of individuals with schizophrenia and controls. The pharynx was selected for study since it is easily accessible by noninvasive sampling techniques and biological samples from cases and controls could be collected and processed in an identical manner. In addition, several studies have documented the presence of a complex ecology of viruses within human saliva and the oral pharynx. Bacteriophages were selected as the viruses to be analyzed since they have been shown to comprise the major component of the virome measured in the oral cavity and other mucosal locations.6,25
We identified a single phage, Lactobacillus phage phiadh, which had statistically significant differences between individuals with schizophrenia and controls taking into consideration multiple comparisons as well as demographic variables such as age, gender, race, and maternal education as an indicator of socioeconomic status.. Lactobacillus phage phiadh is a temperate phage which preferentially infects the bacteria Lactobacillus gasseri. Sequence and morphological analysis of this phage has shown that it is a member of the viral family siphoviridae and has a B1 morphotype. These viruses have an icosahedral capsid of 40–76nm in diameter and a tail of 116–500nm in length. The genome of this virus consists of a single molecule of double stranded DNA which is 43785 nucleotides in length. Lactobacillus phage phiadh has been well characterized in terms of its genome organization, gene transcription, and mechanisms involving integration into its host bacteria.26,27
The primary host bacteria for Lactobacillus phage phiadh is Lactobacillus gasseri. This lactobacillus is a common component of the oral and gastrointestinal mucosae and is capable of binding to intestinal epithelium. It is a common component of foods and probiotic preparations.28,29 Lactobacillus gasseri has been shown to modulate the immune system, largely by alterations in the function of dendritic cells, enterocytes, and components of innate immunity.30,31 Administration of Lactobacillus gasseri to humans has been shown to have a number of beneficial effects on the immune system and on gastrointestinal functioning.
Within the group of individuals with schizophrenia, the presence of Lactobacillus phage phiadh was not associated with the demographic variables of age, gender, or race. There were no associations with cigarette smoking. In terms of medications used by the individuals with schizophrenia, no associations were found with antibiotic medications or antipsychotic medications used to treat this disorder. However, the level of phiadh was found to be associated with the administration of valproate, an anticonvulsant mood stabilizer commonly administered to individuals with schizophrenia. Although valproate has a number of recognized pharmacological activities, including histone deacetylase (HDAC) inhibition and NMDA receptor modulation, the mechanisms by which its administration improves symptoms in some individuals with schizophrenia is not known with certainty.32 The association between the administration of valproate and the level of Lactobacillus phage phiadh found in our study population is of interest since valproate has been shown to alter the microbiome and the intestinal levels of microbial metabolites in an animal model of autism.33
We also found that the level of Lactobacillus phage phiadh was correlated with an increased rate of co-morbid immunological disorders in the individuals with schizophrenia. This finding is of potential interest in light of the high prevalence of co-morbid immune-related disorders in individuals with schizophrenia.34
The mechanisms by which Lactobacillus phage phiadh might be associated with schizophrenia and co-morbid immunological conditions are not known with certainty. However, previous studies suggest a number of plausible mechanisms which might underlie this association. The most likely possibility is that Lactobacillus phage phiadh modulates the level of its host bacteria Lactobacillus gasseri with subsequent effects on the host immune systems. The effect of the phage in its host bacteria can occur either by the direct killing of the host or by the establishment of a long-term lysogenic state within the host bacterial genome. The fact that many individuals displayed a positive correlation between the numbers of Lactobacillus phage phiadh and its host bacteria (figure 4) suggests that in many cases the phage infection is in a lysogenic state. Phage in this state can be induced by changes in environmental conditions such as temperature, pH, and oxygen concentration, leading to reactivation of the virus and the subsequent killing of the host bacteria.
Recently, Lactobacillus phage phiadh has been found to mediate plasmid transfer among lactobacilli, indicating that this phage may exert a further impact on the ecology of bacteria within the human microbiota through the control of additional species of Lactobacilli and perhaps other bacteria as well. Colonization with Lactobacillus phage phiadh might thus have widespread effects on the ecology and evolution of the host microbiome.
It has recently been shown that bacteriophages can also affect mammalian hosts independent of their ability to modulate the level of bacteria. For example, some species of phage can directly affect the immune system through the release of their DNA from phagocytized phage-infected bacteria and interaction of this phage DNA with mammalian toll like receptors.35 Some species of bacteriophage may also be able to affect host defenses by direct interactions with mucous lining the surfaces of epithelial cells.36 It is not currently known if Lactobacillus phage phiadh has these properties or others which might directly interact with the host immune system. In light of our findings of increased levels of Lactobacillus phage phiadh in individuals with schizophrenia, further studies of the mechanisms by which this phage might interact with the host immune system are warranted. It is also not currently known what the source of Lactobacillus phage phiadh is in terms of the human virome. Possibilities include food, water, person-to-person transmission, and fomites. The source of Lactobacillus phage phiadh and other medically important phage as well as their ecology and epidemiology should be the subject of future studies.
It should also be noted that metagenomic analyses such as the ones used in this report are still under development and that alternative types of sequencing method or statistical analyses might lead to differing results and interpretations. Currently, the sample sizes of metagenomic studies are limited principally by the costs involved in the sequencing procedures. It can be anticipated that these costs will decrease in the future and that the analysis of larger and more samples will lead to a more definitive characterization of the virome in individuals with psychiatric disorders and controls.
The individuals in our study with schizophrenia were not recently diagnosed and had experienced their illness for a mean of 21.9 years. In the study of environmental factors in this population, it is difficult to know with certainty if the factors are a component of the etiopathogenesis of the psychiatric disorder or are associated with some of the effects of this disorder on the individual, eg, as a result of hospitalization, medications, oral hygiene, or the effects of lifestyle. In the case of Lactobacillus phage phiadh, the levels were not associated with length of illness, number of hospitalizations, or the administration of antipsychotic medications.
The possible association of Lactobacillus phage phiadh and disease should be addressed by studies of individuals early in the course of illness and by individuals at high risk for disease. It is of note that Lactobacillus phage phiadh was associated with an increased prevalence of immunological disorders such as diabetes in the study population. If this is confirmed interventions directed at decreasing the carriage rate of Lactobacillus phage phiadh and its host bacteria in individuals with schizophrenia might be considered, with the goal of altering the course of the disorder and preventing co-morbid immunological disorders. These might include probiotic preparations, prebiotic compounds, and the narrow spectrum antibiotics. The successful implementation of these methods might lead to new modalities for the prevention and treatment of schizophrenia and some of its co-morbid conditions.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
NIMH P50 Silvio O. Conte Center at Johns Hopkins (MH-94268) and the Stanley Medical Research Institute (907067).
Supplementary Material
Acknowledgments
The authors thank Dr Todd Klaenhammer for his critical reading of the manuscript, Dr Lori Brando for her suggestions and Ms Ann Cusic for her assistance in the preparation of the manuscript. R.Y. is a member of the Stanley Medical Research Institute Board of Directors and Scientific Advisory Board. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. The other authors have no conflicts to declare.
References
- 1. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012; 489:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011; 474:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tap J, Mondot S, Levenez F, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009; 11:2574–2584. [DOI] [PubMed] [Google Scholar]
- 4. Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. 2013; 14:654–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ogilvie LA, Bowler LD, Caplin J, et al. Genome signature-based dissection of human gut metagenomes to extract subliminal viral sequences. Nat Commun. 2013; 4:2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Paepe M, Leclerc M, Tinsley CR, Petit MA. Bacteriophages: an underestimated role in human and animal health? Front Cell Infect Microbiol. 2014; 4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A. 2013; 110:12450–12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pride DT, Salzman J, Haynes M, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012; 6:915–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dalmasso M, Hill C, Ross RP. Exploiting gut bacteriophages for human health. Trends Microbiol. 2014; 22:399–405. [DOI] [PubMed] [Google Scholar]
- 10. Abeles SR, Pride DT. Molecular bases and role of viruses in the human microbiome. J Mol Biol. 2014; 426:3892–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Whiteford HA, Degenardt L, Rehm J, et al. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet. 2013; 382:1575–1586. [DOI] [PubMed] [Google Scholar]
- 12. McOmish CE, Burrows EL, Hannan AJ. Identifying novel interventional strategies for psychiatric disorders: integrating genomics, ‘enviromics’ and gene–environment interactions in valid preclinical models. Br J Pharmacol. 2014; 171:4719–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophenia-associated genetic loci. Nature. 2014; 511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mavrommatis B, Young GR, Kassiotis G. Counterpoise between the microbiome, host immune activation and pathology. Curr Opin Immunol. 2013; 25:456–462. [DOI] [PubMed] [Google Scholar]
- 15. De Vlaminck I, Khush KK, Strehl C, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013; 155:1178–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rescigno M. Intestinal microbiota and its effects on the immune system. Cell Microbiol. 2014; 16:1004–1013. [DOI] [PubMed] [Google Scholar]
- 17. Hsiao EY, McBride SW, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013; 155:1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dickerson FB, Boronow JJ, Stallings C, Origoni AE, Ruslanova I, Yolken RH. Association of serum antibodies to herpes simplex virus 1 with cognitive deficits in individuals with schizophrenia. Arch Gen Psychiatry. 2003; 60:466–472. [DOI] [PubMed] [Google Scholar]
- 19. Dickerson F, Stallings C, Sullens A, et al. Association between cognitive functioning, exposure to Herpes Simplex Virus type 1, and the COMT Val158Met genetic polymorphism in adults without a psychiatric disorder. Brain Behav Immun. 2008; 22:1103–1107. [DOI] [PubMed] [Google Scholar]
- 20. Yolken RH, Jones-Brando L, Dunigan DD, et al. Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice. Proc Natl Acad Sci U S A. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV Axis I Disorders—Non-patient Edition (SCID-I/NP). New York: Biometrics Research, New York State Psychiatric Institute; 1998. [Google Scholar]
- 22. Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987; 13:261–276. [DOI] [PubMed] [Google Scholar]
- 23. Balamurugan R, Mary RR, Chittaranjan S, Jancy H, Shobana Devi R, Ramakrishna BS. Low levels of faecal lactobacilli in women with iron-deficiency anaemia in south India. Br J Nutr. 2010; 104:931–934. [DOI] [PubMed] [Google Scholar]
- 24. Dicvkerson F, Stallings CR, Origoni AE, et al. Cigarette smoking among persons with schizophrenia or bipolar disorder in routine clinical settings, 1999–2011. Psychiatr Serv. 2013; 64:44–50. [DOI] [PubMed] [Google Scholar]
- 25. Pride DT, Salzman J, Haynes M, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012; 6:915–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Villion M, Moineau S. Bacteriophages of lactobacillus. Front Biosci (Landmark Ed). 2009; 14:1661–1683. [DOI] [PubMed] [Google Scholar]
- 27. Altermann E, Klein JR, Henrich B. Primary structure and features of the genome of the Lactobacillus gasseri temperate bacteriophage (phi)adh. Gene. 1999; 236:333–346. [DOI] [PubMed] [Google Scholar]
- 28. Kadooka Y, Sato M, Ogawa A, et al. Effect of Lactobacillus gasseri SBT2055 in fermented milk on abdominal adiposity in adults in a randomised controlled trial. Br J Nutr. 2013; 110:1696–1703. [DOI] [PubMed] [Google Scholar]
- 29. Ogawa A, Kadooka Y, Kato K, Shirouchi B, Sato M. Lactobacillus gasseri SBT2055 reduces postprandial and fasting serum non-esterified fatty acid levels in Japanese hypertriacylglycerolemic subjects. Lipids Health Dis. 2014; 13:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luongo D, Miyamoto J, Bergamo P, et al. Differential modulation of innate immunity in vitro by probiotic strains of Lactobacillus gasseri . BMC Microbiol. 2013; 13:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Selle K, Klaenhammer TR. Genomic and phenotypic evidence for probiotic influences of Lactobacillus gasseri on human health. FEMS Microbiol Rev. 2013; 37:915–935. [DOI] [PubMed] [Google Scholar]
- 32. Park HJ, Kang WS, Paik JW, Kim JW. Effect of valproic acid through regulation of NMDA receptor-ERK signaling in sleep deprivation rats. J Mol Neurosci. 2012; 47:554–558. [DOI] [PubMed] [Google Scholar]
- 33. de Theije CG, Wopereis H, Ramadan M, et al. Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav Immun. 2014; 37:197–206. [DOI] [PubMed] [Google Scholar]
- 34. Schoepf D, Uppal H, Potluri R, Heun R. Physical comorbidity and its relevance on mortality in schizophrenia: a naturalistic 12-year follow-up in general hospital admissions. Eur Arch Psychiatry Clin Neurosci. 2014; 264:3–28. [DOI] [PubMed] [Google Scholar]
- 35. Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. 2013; 14:654–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barr JJ, Auro R, Furlan M, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A. 2013; 110:10771–10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.