Abstract
We report the 3.3-Mb draft genome of Laceyella sacchari strain GS 1-1, isolated from hot spring water sample, Chumathang, Leh, India. Draft genome of strain GS 1-1 consists of 3, 324, 316 bp with a G + C content of 48.8% and 3429 predicted protein coding genes and 75 RNAs. Geobacillus thermodenitrificans strain NG80-2, Geobacillus kaustophilus strain HTA426 and Geobacillus sp. Strain G11MC16 are the closest neighbors of the strain GS 1-1.
Keywords: Laceyella sacchari strain GS 1-1, Thermophilic, Illumina-HiSeq, CLC Bio Workbench, Rapid Annotation using Subsystems Technology (RAST)
| Specifications | |
|---|---|
| Organism/cell line/tissue | Laceyella sacchari |
| Strain(s) | GS 1-1 |
| Sequencer or array type | Sequencer; the Illumina-HiSeq 1000 |
| Data format | Processed |
| Experimental factors | Microbial strain |
| Experimental features | Draft genome of L. sacchari strain GS 1-1; assembly and annotation |
| Consent | n/a |
Direct link to the data
Direct link: http://www.ncbi.nlm.nih.gov/nuccore/ASZU00000000.
The genus Laceyella was proposed by Yoon et al. 2005 after a detailed polyphasic study and reclassification on the genus Thermoactinomyces [1]. At present the genus Laceyella has four recognized species namely; Laceyella putida [1], L. sacchari, type species of the genus Laceyella [1], Laceyella sediminis [2] and Laceyella tengchongensis [3]. Strain GS 1-1 is a Gram-positive bacteria and thermophilic bacteria. Cells are aerobic, non-acid-fast, produce endospores and chemo-organotrophic. Aerial and substrate mycelia are formed. Aerial mycelium is white. Yellow–brown or grayish-yellow soluble pigment may be produced. The cell-wall peptidoglycan contains meso-DAP. Predominant menaquinone is MK-9. Major fatty acids are iso-C15:0 and anteiso-C15:0.
L. sacchari strain GS 1-1, was isolated from hot spring of Chumathang, Leh, Ladakh, India, (N 32° 58′ E78° 15″), at the height of 4600 m above sea level. Genomic DNA was extracted from 36 h old culture using ZR Fungal/Bacterial DNA MiniPrep™ as per manufacturer's instructions. The genome of L. sacchari strain GS 1-1 was sequenced using the Illumina-HiSeq 1000 paired-end technology that produced a total of 40,874,820 paired-end reads (paired distance (insert size) ~ 330 bp) of 101 bp. CLC Bio Workbench v6.0.2 (CLC Bio, Denmark) was employed for preprocessing the data so as to trim and remove low quality sequences. A total of 40,668,128 high quality, vector filtered reads (~ 1194 times coverage) were used for assembly with CLC Bio Workbench (at word size of 45 and bubble size of 98). The final assembly contains 42 contigs of total size 3,324,316 bp with N50 contig length of 249,341 bp; the largest contig assembled measures 698,403 bp. This draft genome comprising 3,324,316 bp was annotated with the help of RAST (Rapid Annotation using Subsystem Technology) system [4] server. A total of 3429 predicted coding regions (CDSs), 6 rRNAs and 69 tRNAs were predicted.
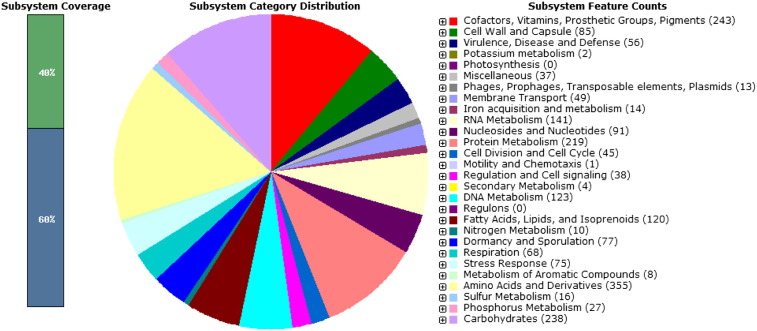
RAST indicates that strain Geobacillus thermodenitrificans strain NG80-2 (score 502), Geobacillus kaustophilus strain HTA426 (score 471) and Geobacillus sp. Strain G11MC16 (score 436) are the closest neighbors of the strain GS 1-1. The strain GS 1-1 contains the genes for glycolysis and gluconeogenesis, TCA cycle and pentose phosphate pathway. Genes of alkaline phosphatase (EC 3.1.3.1), ferroxidase (EC 1.16.3.1), manganese superoxide dismutase (EC 1.15.1.1), shikimate kinase I (EC 2.7.1.7.1), chorismate synthase (EC 4.2.3.5), alcohol dehydrogenase (1.1.1.1) and superoxide dismutase [Fe] (EC 1.15.1.1) and pathogenicity islands are also present in the genome annotation of strain GS 1-1. We have mapped all predicted 3429 CDSs to KEGG pathways [5] with the help of KASS server [6]. The genome has all the essential pathways for DNA, RNA metabolism, iron, sulfur and phosphorus acquisition and metabolism pathways (Fig. 1).
Fig. 1.
Sub-system distribution of L. sacchari strain GS 1-1 (based on RAST server).
Nucleotide sequence accession number
The L. sacchari strain GS 1-1 whole genome shot gun (WGS) project has been deposited at DDBJ/EMBL/GenBank under the project accession ASZU00000000 of the project (01) that has the accession numbers ASZU00000000 and consists of sequences ASZU01000001–ASZU01000042.
Conflict of interest
The authors declare that there is no conflict of interest on any work published in this paper.
Acknowledgments
We thank Mr. Malkit Singh for his technical assistance and Dr. Tsering Stobdan, Scientist, Defence Institute of High Altitude Research, DRDO, Leh for his help during the expedition to Chumathang, Leh. This work was funded by IMTECH—CSIR. NK is supported by a research fellowship from the University Grant Commission (UGC) Govt. of India, NK2 is supported by a research internship from the Council of Scientific and Industrial Research (CSIR), Govt. of India. This is IMTECH communication number 109/2013.
References
- 1.Yoon J.H., Kim I.G., Shin I.K., Park Y.H. Proposal of the genus Thermoactinomyces sensu stricto and three new genera, Laceyella, Thermoflavimicrobium and Seinonella, on the basis of phenotypic, phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 2005;55:395–400. doi: 10.1099/ijs.0.63203-0. [DOI] [PubMed] [Google Scholar]
- 2.Chen J.J., Lin L.B., Zhang L.L., Zhang J., Tang S.K., Wei Y.L., LI W.J. Laceyella sediminis sp. nov., a thermophilic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2012;62:38–42. doi: 10.1099/ijs.0.028282-0. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J., Tang S.K., Zhang Y.Q., YU L.Y., Klenk H.P., LI W.J. Laceyella tengchongensis sp. nov., a thermophile isolated from soil of a volcano. Int. J. Syst. Evol. Microbiol. 2010;60:2226–2230. doi: 10.1099/ijs.0.011767-0. [DOI] [PubMed] [Google Scholar]
- 4.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R.A., McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration 88 and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. KAAS: an automatic 90 genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]